Abstract
Tumor suppressor genes are classified by their somatic behavior either as caretakers (CTs) that maintain DNA integrity or as gatekeepers (GKs) that regulate cell survival, but the germ line role of these disease-related gene subgroups may differ. To test this hypothesis, we have used genomic data mining to compare the features of human CTs (n = 38), GKs (n = 36), DNA repair genes (n = 165), apoptosis genes (n = 622), and their orthologs. This analysis reveals that repair genes are numerically less common than apoptosis genes in the genomes of multicellular organisms (P < 0.01), whereas CT orthologs are commoner than GK orthologs in unicellular organisms (P < 0.05). Gene targeting data show that CTs are less essential than GKs for survival of multicellular organisms (P < 0.0005) and that CT knockouts often permit offspring viability at the cost of male sterility. Patterns of human familial oncogenic mutations confirm that isolated CT loss is commoner than is isolated GK loss (P < 0.00001). In sexually reproducing species, CTs appear subject to less efficient purifying selection (i.e., higher Ka/Ks) than GKs (P = 0.000003); the faster evolution of CTs seems likely to be mediated by gene methylation and reduced transcription-coupled repair, based on differences in dinucleotide patterns (P = 0.001). These data suggest that germ line CT/repair gene function is relatively dispensable for survival, and imply that milder (e.g., epimutational) male prezygotic repair defects could enhance sperm variation—and hence environmental adaptation and speciation—while sparing fertility. We submit that CTs and repair genes are general targets for epigenetically initiated adaptive evolution, and propose a model in which human cancers arise in part as an evolutionarily programmed side effect of age- and damage-inducible genetic instability affecting both somatic and germ line lineages.
Keywords: molecular evolution, adaptive evolution, carcinogenesis, DNA repair
Introduction
A longstanding debate in evolutionary biology concerns how species of increasing structural complexity maintain their capacity for genetic variation—and, hence, adaptation and divergence—despite a predictably increasing need for genetic fidelity (Gulick 1893; Gould JL and Gould CG 1997). Relevant to this conflict, Cope's rule postulates that increasing body size creates a short-term reproductive advantage for the individual organism (Kingsolver and Pfennig 2004) while worsening long-term extinction risk for the clade (Van Valkenburgh et al. 2004; Hone and Benton 2005). This trade-off suggests that higher evolving organisms are subject to a progressive “Red Queen”–type clash between intensifying negative selection for phenotypic stability and weakening positive selection for genotypic variability (Markov 2000)—consistent with the modest proportion (0.03%) of coding sequence estimated to have been positively selected in humans, when compared with that negatively selected (2.5–5%) in humans or positively selected in simpler species such as Drosophila (20%) (Ponting and Lunter 2006). Indeed, prevailing theory teaches that most genetic novelty results from fixation of random (nonadaptive) drift affecting neutral (Kimura 1968) or near-neutral (Ohta 1998) alleles, rejecting the Lamarckian doctrine that environmental pressures can drive (i.e., not merely fix) beneficial mutations.
In previous work, we showed that silent mutations may nonrandomly affect intragenic sites of differing functional importance (Epstein et al. 2000; Lin et al. 2003) and that such mutational patterns vary with both strand-specific transcription-related DNA repair (Tang et al. 2006) and gene expression levels (Tang and Epstein 2007). It therefore remains plausible that ambient stressors such as heat (Maresca and Schwartz 2006), starvation (Hastings et al. 2004), inflammation (Blanco et al. 2007; Lavon et al. 2007), toxins (Salnikow and Zhitkovich 2008), free radical injury (Cerda and Weitzman 1997), or other sources of DNA damage (Ponder et al. 2005) could modify gene transcription and thus alter the rate of mutations affecting fitness (Galhardo et al. 2007)—including the occasional generation of beneficial mutations (Monk 1995; Elena and de Visser 2003; Nei 2005). Clues favoring this inducible (adaptive) evolutionary paradigm over neutrality for metazoan genomes—as is already accepted for bacterial (Ponder et al. 2005; Cirz and Romesberg 2007) and plant genomes (Galloway and Etterson 2007)—include faster-than-expected rates of phenotype acquisition, close temporal correlation with environmental changes, proof of improved fitness, or convergence (Levasseur et al. 2007).
A mechanism for such non-Darwinian genomic plasticity has been suggested in recent times by the discovery of heritable epigenetic changes capable of reprogramming developmental and adult gene expression (Martin et al. 2005; Morgan et al. 2005), coupled with the predisposition of such changes to cause germ line mutations (Cooper and Krawczak 1989) or postzygotic mosaicism (Ohlsson et al. 1999) that sometimes cause disease (Andrews et al. 1996; Smith and Hurst 1998; Esteller et al. 2001). The frequency of germ line epimutations or imprinting errors—estimated to be an order of magnitude higher than that of germ line mutations (Horsthemke 2006)—can be either environmentally regulated (Dolinoy and Jirtle 2008), as illustrated by the inducibility of spermatogonial stem cell DNA hypermethylation by air pollution (Yauk et al. 2008), or parentally age dependent (Oakes et al. 2003; Perrin et al. 2007). If such epimutations affect modifier genes involved in DNA repair, a “slippery slope” of somatic and transgenerational genetic instability (i.e., a mutator phenotype) may result (Jacinto and Esteller 2007), leading not only to an increase in deleterious (purifiable) mutations (Wu et al. 2007; Morak et al. 2008) but also to occasional advantageous (positively selectable) mutations (Sniegowski et al. 2000; Cirz and Romesberg 2007) and/or speciation events (Sniegowski 1998). Selection of such “driver” beneficial mutations may lead in turn to “hitchhiking” of mutator (epi)mutations in modifier genes (Johnson 1999) as “passengers” (Frohling et al. 2007). Such mutational buffering could enhance evolvability (Wagner 2008)—consistent with the idea that error-free DNA repair may be maladaptive in mutagenic or stressful environments (Breivik and Gaudernack 2004; Ponder et al. 2005; Siegl-Cachedenier et al. 2007)—yet may also impair performance and hence robustness (Lenski et al. 2006; Frank 2007; Petrie and Roberts 2007). The “evo-devo” conundrum thus remains as to whether evolvability is indeed selectable (Hendrikse et al. 2007; Lynch 2007), and if so, by what mechanism (Colegrave and Collins 2008; Pigliucci 2008). Even if such a mechanism exists (King and Jukes 1969; Monk 1995), traditional thinking predicts that such selection may act only very weakly at a “good-for-the-species” level (Hiraiwa-Hasegawa 2000).
We have addressed this dilemma by comparing 2 classes of human genes implicated in prevention of cancer, a disease of disordered microevolution (Gatenby and Vincent 2003; Iwasa et al. 2004; Breivik 2005). Tumorigenesis is potentiated by genomic instability (Schneider and Kulesz-Martin 2004; Bielas et al. 2006) arising via multistep inactivation of so-called tumor suppressor genes (Nowak et al. 2004), which, like proto-oncogenes, have been reported to be under strong negative selection pressure (Thomas et al. 2003). These carcinogenic loss-of-function events mainly affect DNA repair—mediated by caretaker genes (CTs) such as BRCA1 and MLH1—or apoptosis, mediated by gatekeeper genes (GKs) such as TP53 and Rb (Kinzler and Vogelstein 1997). These suppressor gene subsets, as well as their disease-causing mutations (Futreal et al. 2004; University Medical Center Groningen 2006), are distinguishable using gene databases (Doctor et al. 2003; Wood et al. 2005). Although long regarded as recessive oncogenes that require 2 “hits” for disease expression (Knudson 2000), suppressor genes are increasingly recognized to exhibit clonal haploinsufficiency in tumors (Santarosa and Ashworth 2004; Smilenov 2006). Given recent evidence for the role of adaptive evolution in cancer progression (Babenko et al. 2006; Crespi and Summers 2006), the occurrence of such haploinsufficiency supports the view that gene loss and pseudogenization (“less is more”) can accelerate genome evolution in certain contexts (Olson 1999). Because deleterious mutations (those causing genetic death) are purged by negative selection, whereas nondeleterious mutations may be positively selected, systematic comparison of CT and GK evolutionary rates should clarify whether these repair and apoptosis gene subsets are subject to distinct evolutionary forces. Consistent with this possibility, comparisons of human and chimpanzee genomes have confirmed different evolutionary rates in functionally distinct gene categories related to tumorigenesis (Clark et al. 2003; Bustamante et al. 2005; Nielsen et al. 2005; Kelley et al. 2006; Voight et al. 2006), whereas adaptive evolution of the BRCA1 CT has been well documented (Huttley et al. 2000; Fleming et al. 2003; Pavlicek et al. 2004). Here, we use genomic data mining to test the hypothesis that germ line CTs are commoner targets for methylation-dependent mutational inactivation than are GKs and, hence, that repair gene dysfunction contributes both to germ line evolvability and somatic tumor progression. A male-dependent prezygotic mechanism for this process, which we have termed programmed genetic instability or PGI (Epstein and Zhao 2006a), is also presented.
Materials and Methods
Identification and Classification of CTs and GKs
We mined data to compare the structural and functional characteristics of human CTs and GKs (see supplement 1 [Supplementary Material online] for sources). Given the multigenic interdependence of DNA repair and cellular apoptosis (Wee and Aguda 2006), unambiguous identification of genes that exclusively mediate 1 of these 2 processes is not straightforward. We sought to minimize the “noise” of this functional overlap in 2 ways. First, we used a familial tumor suppressor gene database (Futreal et al. 2004; University Medical Center Groningen 2006) to restrict the choice of genes to those with major neoplastic effects (i.e., heritable cancer syndromes) when deleted in the germ line; this yielded a total of 74 tumor suppressor genes (table 1). Second, by cross-correlating the former data set with a database of DNA repair genes (Wood et al. 2005), we subclassified this familial cancer susceptibility gene subset as CTs (n = 38) and then designated the remainder—the majority of which were confirmed to mediate apoptosis (Doctor et al. 2003)—as GKs (n = 36; table 1).
Table 1.
Classification of CT and GK Suppressor Genes, Listing RefSeq, EntrezGene, and Ensembl Identifiers
| CTs | RefSeq | Entrez Gene | Ensembl | GKs | RefSeq | Entrez Gene | Ensembl | |
| 1 | ATM | NM_000051 | 472 | ENST00000278616 | APC | NM_000038 | 324 | ENST00000257430 |
| 2 | BLM | NM_000057 | 641 | ENST00000355112 | AXIN2 | NM_004655 | 8313 | ENST00000307078 |
| 3 | BRCA1 | NM_007295 | 672 | ENST00000309486 | BMPR1A | NM_004329 | 657 | ENST00000372037 |
| 4 | BRCA2 | NM_000059 | 675 | ENST00000267071 | BUB1B | NM_001211 | 701 | ENST00000287598 |
| 5 | BRIP1 | NM_032043 | 83990 | ENST00000259008 | CDC73 | NM_024529 | 79577 | ENST00000367436 |
| 6 | DDB2 | NM_000107 | 1643 | ENST00000256996 | CDH1 | NM_004360 | 999 | ENST00000268794 |
| 7 | ERCC2 | NM_000400 | 2068 | ENST00000221481 | EXT1 | NM_000127 | 2131 | ENST00000378204 |
| 8 | ERCC3 | NM_000122 | 2071 | ENST00000285398 | MEN1 | NM_130803 | 4221 | ENST00000337652 |
| 9 | ERCC4 | NM_005236 | 2072 | ENST00000311895 | NF1 | NM_000267 | 4763 | ENST00000358273 |
| 10 | ERCC5 | NM_000123 | 2073 | ENST00000375971 | NF2 | NM_000268 | 4771 | ENST00000338641 |
| 11 | ERCC6 | NM_000124 | 2074 | ENST00000355832 | PRKAR1A | NM_212472 | 5573 | ENST00000358598 |
| 12 | ERCC8 | NM_000082 | 1161 | ENST00000265038 | PTCH | NM_000264 | 5727 | ENST00000331920 |
| 13 | FANCA | NM_000135 | 2175 | ENST00000305699 | PTEN | NM_000314 | 5728 | ENST00000371953 |
| 14 | FANCB | NM_001018113 | 2187 | ENST00000340604 | RB1 | NM_000321 | 5925 | ENST00000267163 |
| 15 | FANCC | NM_000136 | 2176 | ENST00000289081 | SBDS | NM_016038 | 51119 | ENST00000246868 |
| 16 | FANCD2 | NM_033084 | 2177 | ENST00000287647 | SDHB | NM_003000 | 6390 | ENST00000375499 |
| 17 | FANCE | NM_021922 | 2178 | ENST00000229769 | SDHC | NM_003001 | 6391 | ENST00000367975 |
| 18 | FANCF | NM_022725 | 2188 | ENST00000327470 | SDHD | NM_003002 | 6392 | ENST00000375549 |
| 19 | FANCG | NM_004629 | 2189 | ENST00000378643 | SMAD4 | NM_005359 | 4089 | ENST00000342988 |
| 20 | FANCL | NM_018062 | 55120 | ENST00000233741 | SMARCB1 | NM_001007468 | 6598 | ENST00000344921 |
| 21 | FANCM | NM_020937 | 57697 | ENST00000267430 | STK11 | NM_000455 | 6794 | ENST00000326873 |
| 22 | LIG4 | NM_002312 | 3981 | ENST00000310534 | SUFU | NM_016169 | 51684 | ENST00000369902 |
| 23 | MLH1 | NM_000249 | 4292 | ENST00000231790 | TCF1 | NM_000545 | 6927 | ENST00000257555 |
| 24 | MLH3 | NM_014381 | 27030 | ENST00000238662 | TSC1 | NM_000368 | 7248 | ENST00000298552 |
| 25 | MRE11A | NM_005591 | 4361 | ENST00000323929 | TSC2 | NM_000548 | 7249 | ENST00000219476 |
| 26 | MSH2 | NM_000251 | 4436 | ENST00000233146 | TSHR | NM_000369 | 7253 | ENST00000298171 |
| 27 | MSH3 | NM_002439 | 4437 | ENST00000265081 | VHL | NM_000551 | 7428 | ENST00000256474 |
| 28 | MSH6 | NM_000179 | 2956 | ENST00000234420 | WT1 | NM_024426 | 7490 | ENST00000379079 |
| 29 | MUTYH | NM_012222 | 4595 | ENST00000372112 | CDK4 | NM_000075 | 1019 | ENST00000257904 |
| 30 | NBN | NM_001024688 | 4683 | ENST00000265433 | CHEK2 | NM_001005735 | 11200 | ENST00000382580 |
| 31 | PMS1 | NM_000534 | 5378 | ENST00000342075 | CYLD | NM_015247 | 1540 | ENST00000311559 |
| 32 | PMS2 | NM_000535 | 5395 | ENST00000265849 | EXT2 | NM_000401 | 2132 | ENST00000358681 |
| 33 | POLH | NM_006502 | 5429 | ENST00000372236 | FH | NM_000143 | 2271 | ENST00000205832 |
| 34 | RAD51 | NM_002875 | 5888 | ENST00000382643 | FLCN | NM_144997 | 201163 | ENST00000285071 |
| 35 | RECQL4 | NM_002907 | 5965 | ENST00000314748 | GPC3 | NM_004484 | 2719 | ENST00000370818 |
| 36 | WRN | NM_000553 | 7486 | ENST00000298139 | TP53 | NM_000546 | 7157 | ENST00000269305 |
| 37 | XPA | NM_000380 | 7507 | ENST00000259463 | ||||
| 38 | XPC | NM_004628 | 7508 | ENST00000285021 |
Analyses of Gene Sequences, Mutations, and Evolutionary Rate
Human and mouse reference sequences, and species gene numbers, were downloaded from NCBI Entrez Gene (http://www.ncbi.nlm.nih.gov/Entrez/Gene). Mutation data were downloaded from the Human Gene Mutation Database. K-estimator 6.1 (with window size of 33 codons and step size of 10 codons using Kimura 2-parameter [2p] method) (Comeron 1999) and PAML 3.15 with yn00 model (Yang and Nielsen 2002) were used for evolutionary rate calculations. For analysis of gene evolutionary profiles, we downloaded coding sequences from Ensembl (http://www.ensembl.org). Kendall package from R-gui (http://www.r-project.org) was used for statistical analysis. Other analyses were done using MATLAB (7.6) Statistical Toolbox (5.1) (http://www.mathworks.com) for principal component analysis and nonparametric tests, and R-gui (2.60) for χ2 or Fisher's exact test.
Supergene Concatenation
Orthologous gene sequences of human, mouse, rat, chimpanzee, and rhesus monkey were aligned with amino acid sequences using ClustalW (Thompson et al. 1994), then reverted to codon sequences. In-house Perl scripts (available upon request) were developed for aligned codon concatenation. All the aligned CT and GK sequences were concatenated for supergene construction—23,100 codons for the GK supergene and 32,335 for the CT supergene. The supergene tree was constructed using a neighbor-joining method, with a modified Nei–Gojobori approach, as input in MEGA4 (codon substitution number Molecular Evolutionary Genetics Analysis software, version 4.0), producing 2 types of tree: synonymous and nonsynonymous substitution trees.
Coding Sequence Feature Analysis
We used reference sequences downloaded from NCBI Entrez Gene. For multiple splicing forms, the longest coding sequence was used for analysis. Mono- and dinucleotide composition was assessed using in-house Perl scripts. Additional methodologic details are supplied in the supplements, text, and legends (Supplementary Material online).
Results
Phylogenetic Comparison of CT and GK Orthologs
As an initial assessment, we quantified the numbers of human CT and GK orthologs among species of differing biological complexity. Table 2 shows that orthologs of human CTs occur more often than those of GKs in unicellular (P = 0.047) than in multicellular organisms (P = 0.192)—suggesting that CTs are phylogenetically older, whereas GKs may be more essential to the evolution of multicellular organisms. Although this phylogenetic rise in GK ortholog frequency may reflect selection for increased developmental complexity as predicted by Cope's rule, it may also reflect the increasing importance of policing rogue elements (either intragenomic elements like meiotic drivers or intercellular outlaws like cancer cells) as multicellularity evolves and organismal cell number increases.
Table 2.
Phylogenetic and Gene Essentiality Profiles of Familial Cancer Syndrome Genes
| Parameters | CTs | GKs | P value |
| Yeast orthologs | 0.047 | ||
| Present | 30 | 20 | |
| Absent | 8 | 16 | |
| Worm orthologs | 0.192 | ||
| Present | 30 | 33 | |
| Absent | 8 | 3 | |
| Yeast essentiality | 0.63 | ||
| Deletion lethal | 3 | 2 | |
| Deletion nonlethal | 27 | 18 | |
| Worm essentiality | 0.003 | ||
| RNAi lethal | 1 | 11 | |
| RNA nonlethal | 29 | 22 | |
| Mouse essentiality | <0.00001 | ||
| Knockout lethal | 4 | 28 | |
| Normal | 21 | 8 | |
| Male infertility | 13 | 0 |
NOTE.—.For yeast phylogenetic analysis, CT analysis excludes the numerous Fanconi anemia gene orthologs in order to avoid data confounding. Gene knockout phenotypes were sourced using data mined from http://www.informatics.jax.org/. All P values were computed using Fisher's exact test (2 sided).
We next assessed differences in CT and GK gene essentiality (Liao et al. 2006) based on deletion, RNAi, and gene-targeting data (supplement 2, Supplementary Material online). This analysis shows that germ line GK ortholog disruption is more often lethal than CT knockout in multicellular (worm RNAi data and mouse knockouts; P < 0.00001, Fisher's exact test, 2 sided) but not in unicellular organisms (yeast deletions; P = 0.63; table 2). Hence, relative to GK function, germ line CT function appears selectively dispensable in multicellular organisms.
Analysis of mammalian gene-targeting phenotypes further reveals that, unlike GK knockouts, viable CT knockouts are associated with male sterility (table 2). We infer from this finding that CT dysfunction selectively permits (organism) viability at the expense of (genetic) fidelity, severe defects of which might be expected to cause sperm dysfunction or death. As discussed below, however, the possibility is raised that less profound (i.e., nondeletional or epimutational) germ line repair deficiencies could be associated with offspring fertility.
To assess further the impact of repair and apoptotic gene defects transmitted through the germ line, we compared CT and GK mutation frequencies in cancer families and somatic tumors. This shows that isolated germ line CT mutations are commoner than isolated GK mutations (P < 0.00001; table 3), reinforcing the notion that CT/repair function is significantly more dispensable for survival than is GK/apoptosis function.
Table 3.
Mutational Analysis of Familial (germ line) and Sporadic (somatic) Human Tumor Mutations of CTs and GKs (i.e., relative frequencies of familial vs. sporadic human tumor mutations in the germ line and/or somatic lineages)
| Germ line versus somatic human tumor mutations | Germ line and somatic | Germ line only | P value |
| CT (n = 38) | 6 | 32 | <0.00001 |
| GK (n = 36) | 25 | 11 |
NOTE.—Mutation frequencies were determined by mining Cancer Gene Census and PubMed with P values computed using Fisher's exact test (2 sided).
Phylogenetic Analysis of Apoptosis versus Repair
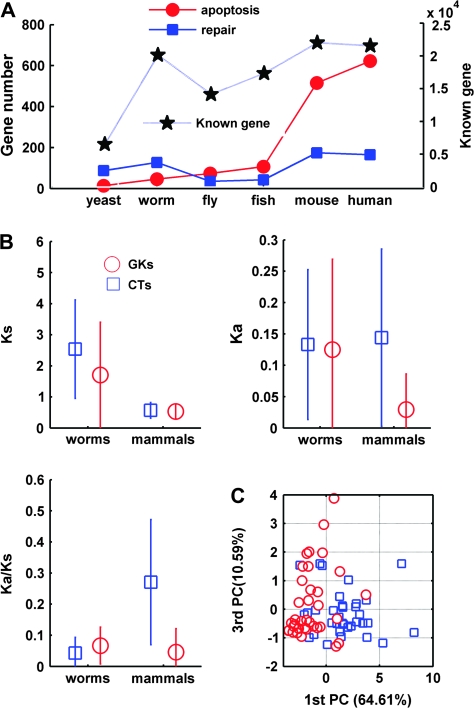
The foregoing data apply only to tumor suppressor genes. To determine whether these results can be generalized, we performed a cross-species quantitation of orthologs implicated in either apoptosis or repair (supplement 1, Supplementary Material online). Based on the assumptions that organism complexity is increasing from yeast to humans and that assignation of gene ontology includes some random effects, we performed Kendall rank test (R-gui Kendall package, www.r-project.org) for correlation analysis of DNA repair genes and apoptosis genes. As shown in figure 1A, phylogenetic differences in apoptosis gene numbers are significant (tau = 1, 2-sided P = 0.008535), whereas for DNA repair genes this is not the case (tau = 0.333, 2-sided P = 0.45237). Considered together with table 2, this difference confirms that increases in biological complexity depend more upon apoptotic than repair gene number.
FIG. 1.—
Evolutionary characteristics of human CTs and GKs. (A) Overall quantitation of apoptotic gene versus repair gene number in different phyla: human (H. sapiens), mouse (Mus musculus), fish (D. rerio), fly (D. melanogaster), worm (C. elegans), and yeast (S. pombe). Blue filled squares, DNA repair genes; red filled circles, apoptosis genes; black stars, total gene number of respective genome from Ensemble 49 (www.ensembl.org). Data were mined as detailed in the Materials and Methods. (B) Relative divergence of CT and GK genes in mammals (human–mouse; divergence time approximately 85 MYA) and worms (C. elegans–C. briggsae; divergence time approximately 100 MYA) quantified using Ka/Ks. Blue squares, CT orthologs; red circles, GK orthologs. (C) Principal component analysis with parameters of Ka/Ks of CTs and GKs. We normalized the computed 10 pairwise divergence of human, chimpanzee, rhesus, monkey, mice, and rats using Ka/Ks analysis. 1st PC, first principal component (x axis); 3rd PC, third principal component (y axis).
Comparison of CT and GK Evolutionary Rates
We then used nonparametric 2-sample Kolmogorov–Smirnov goodness of fit hypothesis testing with kstest2 function (MATLAB, http://www.mathworks.com, statistical toolbox) for Ka/Ks—a positive correlate of positive selection—ortholog analysis. This confirmed that CTs evolve more rapidly than GKs in sexually reproducing (human vs. mouse, estimated divergence time 85 Myr; P = 0.000003) but not in self-fertilizing (2 worm species, estimated divergence time 100 Myr; P = 0.2582) multicellular organisms (fig. 1B; Kruskal–Wallis test, P < 0.004); Ks was different in worms (P = 0.007) but not in mammals (P = 0.091). Consistent with our earlier finding of selective male sterility in CT knockouts, these data support the hypothesis that rapid CT evolution is related in some way to sexual reproduction. Principal component analysis based on evolutionary rate as well as variables such as GC content (see fig. 5) and gene length (see supplement 3, Supplementary Material online) provides further visual evidence that CTs and GKs are distinguishable based on genetic evolutionary parameters (fig. 1C).
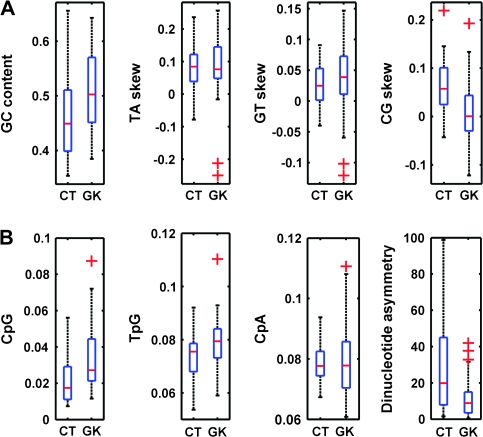
FIG. 5.—
Box plot illustration of CT- and GK-coding sequence features relating to DNA methylation and gene expression. The boxes feature lines at the lower quartile, median, and upper quartile values. The whiskers are lines extending from each end of the boxes to show the extent of the data; outliers are values beyond the ends of the whiskers. (A) Comparison of GC content and dinucleotide skew in CTs and GKs. (B) Frequency of methylation-related dinucleotides and asymmetry in CTs versus GKs. Outliers are denoted by plus signs.
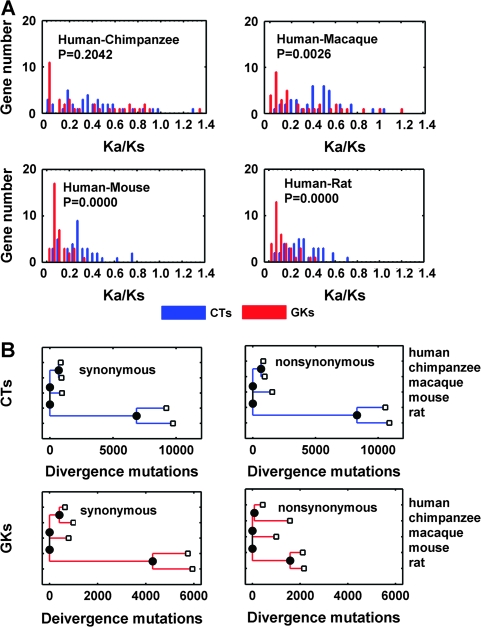
We then used neighbor-joining supergene (gene concatenation) trees (Zhang et al. 1998) to characterize CT and GK branches under selection. This methodology, which has been reported to yield more accurate phylogenetic data than multigene approaches (Gadagkar et al. 2005), confirms that accelerated evolution of CTs compared with GKs is evident in human–macaca, human–mouse, and mouse–rat comparisons, though apparently not in human–chimpanzee comparisons (fig. 2A). This difference is further illustrated by a tree diagram showing the distinct divergence parameters of CTs and GKs (fig. 2B). It is noted that the ratio of missense to silent mutations (A/S) encoded by CT single-nucleotide polymorphisms (SNPs) is no higher than in GKs (supplement 1, Supplementary Material online) and that, compared with historical data (Zhang et al. 1998; Fay et al. 2001), larger polymorphisms are present in both rare and common SNPs (P < 0.001, Pearson's χ2). We have also noted that CTs evolve more rapidly than tissue-specific genes, whereas GKs evolve more slowly than housekeeping genes (P < 0.001, Mann–Whitney U test; data not shown), emphasizing the wide class differences in evolutionary rates.
FIG. 2.—
Evolutionary rate comparison of CTs and GKs. (A) Distribution of Ka/Ks in pairwise comparisons between CTs and GKs within 4 lineages including human–chimpanzee (top left), human–macaca (top right), human–mouse (bottom left), and human–rat (bottom right). Bin size 0.05 was used for distribution computation. For this analysis—which compares the different evolutionary rates of human CTs and GKs using a variety of species comparators, as distinct from comparing gene evolutionary rates in multiple species—P values were calculated by ranked 2-sample Mann–Whitney U test using MATLAB Statistical toolbox function rank sum. (B) Lineage-specific comparison of evolutionary rate of CTs and GKs using supergene concatenation. Blue bars, CTs; red bars, GKs.
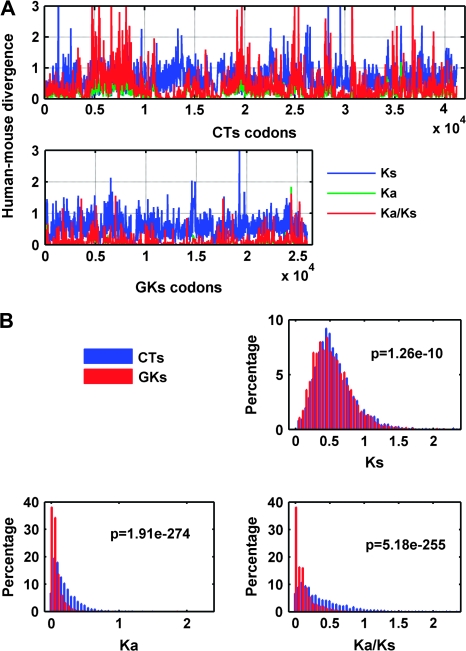
To explore possible differences of divergence between CTs and GKs, we used a sliding window analysis of concatenated CT (38 genes, 41,169 codons) and GK (36 genes, 25,968 codons) supergenes. A window size of 33 codons and step size of 10 codons was used in conjunction with K-estimator (calculation of the number of nucleotide substitutions per site and the confidence intervals) 6.1, Kimura 2p model. As shown in figure 3A, the Ka/Ks (red), but neither Ka nor Ks (green and blue, respectively), is overrepresented in CTs compared with GKs. We further tested the distribution of these 3 parameters by using the nonparametric 2-sample Kolmogorov–Smirnov test. Figure 3B confirms that only Ka/Ks is significant (P = 0.0000079) but neither Ka nor Ks, which correlate with deleterious mutation and neutral mutation rate, respectively (for further details, see supplement [Supplementary Material online] and fig. 3).
FIG. 3.—
Sliding window analysis of human–mouse CT and GK divergence. (A) Evolutionary rates of CTs (top) and GKs (bottom) using human–mouse alignments (window size 33 codons, step size 10 codons). Red line, Ka/Ks; blue line, Ks; green line, Ka. Most regions with Ka/Ks >1.5, with P value <0.05 (bootstrap threshold computed with K-estimator 6.1). (B) Distribution of Ka, Ks, Ka/Ks in CTs and GKs using bin size 0.05 and nonparametric Kolmogorov–Smirnov test, revealing a significant difference of Ka/Ks (P = 0.0000079), thus confirming more rapid evolution of CTs than GKs.
To check whether human–chimpanzee divergences of CTs and GKs are in fact similar, as suggested by the findings in figure 2, we used the McDonald and Kreitman (1991) test for evolutionary rate analysis. Data were mined as described (Bustamante et al. 2005) using a Poisson random field model—a variant of the McDonald–Kreitman test—for divergence versus diversity comparison. Data were obtained from Celera Genomics, which applied exon-specific polymerase chain reaction amplification to 20,362 loci in 39 humans and 1 male chimpanzee. We note, however, that the following genes were not found in this data set: 9 CTs (BRCA2, BRIP1, FANCD2, FANCM, MLH1, MLH3, NBN, PMS2, and RECQL4) and 10 GKs (CDC73, EXT1, NF1, PTCH, SBDS, SDHD, TSHR, WT1, CHEK2, and FH). The following parameters were used: synonymous divergence, synonymous polymorphism, nonsynonymous divergence (DN), nonsynonymous polymorphism (PN), and γ (Poisson random field model parameters). All mined data are supplied (supplement [Supplementary Material online] and fig. 4). In contrast to figure 2, these results—which show a higher frequency of GKs lacking both PN and DN sites relative to CTs (Pearson's χ2, P = 0.0044)—confirm that the rapid evolution of CTs relative to GKs persisted during the human–chimpanzee divergence approximately 10 MYA (fig. 4A–D). Hence, having shown that CTs evolved faster than GKs both during primate–rodent (figs. 1–3) and human–chimpanzee divergence (fig. 4), we conclude that the rapid evolution of CTs is likely independent of timescale.
FIG. 4.—
McDonald–Kreitman testing of CT evolutionary rate relative to GKs during human–chimpanzee divergence. (A) Distributions of synonymous divergence (DS), synonymous polymorphism (PS), nonsynonymous divergence (DN), and nonsynonymous polymorphism (PN) of CTs and GKs using bin size 1 and nonparametric tests (median of 2 unpaired samples, using the MATLAB 7.6 statistical toolbox rank-sum function). This shows that DN is the most significant parameter (P = 0.00016), followed by PN (P = 0.004), with DS and PS not significant, thus confirming rapid CT evolution. (B) R-gui function χ2 test function in terms of the summarized codon changes of CTs and GKs. This also shows that the evolutionary rates of CTs and GKs are different (P = 0.0000037). (C) Poisson random field parametric test. The selection parameters are illustrated using a 95% confidence interval and show no significant difference (P = 0.1913, by nonparametric test of equal distribution of 2 samples using function kstest2), presumably reflecting numerous nonchange genes in terms of the parameters. (D) Parametric retesting of CT and GKs in terms of DS, PS, DN, and PN, confirming that CTs evolved more rapidly than GKs during human–chimpanzee divergence.
Sequence-Based Evidence for Evolutionary CT Gene Methylation
Our previous studies implicated nonrandom CG → TA transitional mutations (CpG decay) as a correlate of adaptive evolution in less transcribed (Tang and Epstein 2007) and/or less essential coding sequences (Epstein et al. 2000); conversely, we have implicated CpG conservation as a correlate of negative selection in more transcribed (Tang et al. 2006) and/or more essential sequences (Lin et al. 2003). These results suggested an evolutionary paradigm in which concomitant promoter and coding sequence methylation accelerate (epi)mutational functional loss of nonessential coding sequences. To test whether any such methylation-dependent signatures distinguish CTs and GKs, we computed the sequence component of the relevant transcribed coding strand and applied a nonparametric 2-sample Kolmogorov–Smirnov goodness of fit hypothesis test with kstest2 function of MATLAB (http://www.mathworks.com) statistical tool box for all parameters compared. Meaningful CT and GK gene expression data were unable to be derived from GNF Expression Atlas 2 based on U133A and GNF1H Chips, perhaps reflecting parametric uncertainty (Su et al. 2004). However, for gene expression-related sequence features relating to CpG mutation and asymmetric transcription-related repair (Tang et al. 2006), we compared GC content ((G + C)/(G + C + A + T), P = 0.0106), TA skew ((T − A)/(T + A), P = 0.5242), GC skew ((G − C)/(G + C), P = 0.0028), and B factor ((G + T − A − C)/(A + T + G + C), P = 0.0860) (Majewski 2003) (fig. 5A). These differences suggest greater methylation-dependent mutation of CTs relative to GKs during recent human evolution. We then compared the methylation-related sequence features of CT/GK transcribed strands, revealing differences in CpG content (CpG count/total dinucleotide count, P = 0.0011), CpA content (CpA count/total dinucleotide count, P = 0.2555), TpG content (TpG count/total dinucleotide count, P = 0.0174), and DNA methylation-related dinucleotide asymmetry (TpG × CpA)/(CpG × CpG), P = 0.00105) (fig. 5B). These results indicate less transcription-related repair of methylated CTs relative to GKs in germ line–coding sequences. Considered together with the gene essentiality differences presented in table 2, the rapid evolution of CTs relative to GKs suggests an epimutational mechanism for CT functional inhibition that escapes purification while accelerating mutation.
Discussion
The central finding of this study is that CTs are evolving more rapidly than GKs and that this process—which appears likely to be mediated by methylation-dependent mutation—is confined to higher sexually reproducing species. These CT–GK distinctions are consistent with earlier work showing that apoptosis-regulatory genes are essential for development of higher organisms (Aravind et al. 2001), whereas germ line mutation of DNA repair genes drives species evolution (Fay et al. 2001). However, because loss of DNA repair capacity also contributes to cancer development (Cleaver et al. 1995; Breivik and Gaudernack 2004), it is also reasonable to expect tumor suppressor genes in general to be under strong purifying selection (Thomas et al. 2003). Here, we propose that this apparent discrepancy arises from a complex mix of dualistic variables: 1) the bifunctional evolutionary role of repair genes in either conserving genetic information or permitting genetic variation, depending on selection pressure; 2) the bifunctional ability of sexual reproduction either to promote (prezygotically) variation through repair inhibition and intra- or intermale sperm competition or to conserve (postzygotically) genetic fidelity through repair activation, apoptosis, and miscarriage; and 3) the bifunctional role of CpG dinucleotides and promoter CpG islands in either enhancing transcription and repair (when demethylated) or in repressing transcription and predisposing to mutation (when methylated).
Epigenetic reprogramming occurs not only in the early embryo, where somatic patterns of gene expression are set, but also during germ cell development (Morgan et al. 2005)—which changes can be heritable for at least 2 generations (Anway et al. 2008). The methylation dynamics of sperm/testis DNA are unique (Oakes et al. 2007a); unlike oocyte DNA, prezygotic protamine-compacted male germ cell DNA tends to be methylated in nonpromoter regions (Oakes et al. 2007b) that undergo rapid demethylation following fertilization (Haaf 2006). Such sex-specific DNA methylation appears necessary but not sufficient (El-Maarri et al. 1998) to explain the higher mutation rate of mammalian male germ cells (Agulnik et al. 1997; Hurst and Ellegren 1998; Wyckoff et al. 2000; Makova and Li 2002)—suggesting in turn that exposures of the (external) testes to heat (Nikolopoulos et al. 2007), DNA damage (Barber et al. 2006), or other insults (Hara et al. 1999) could well play an evolutionarily programmed epimutagenic role, consistent with the postnatal timing of male germ cell promoter methylation (Driscoll and Migeon 1990). Notably, this hypothesis differs from the standard view of male-driven evolution reflecting a simple excess of male germ cell divisions, giving rise in turn to more replicative mutations (Drost and Lee 1995).
In earlier work, we showed that rarely transcribed genes with promoter CpG islands are hot spots for adaptive evolution (Tang and Epstein 2007), raising the possibility that promoter methylation of sperm target gene classes such as CTs could be a mechanistic “missing link” between environmental selection for specific transcriptomes (Su et al. 2004) and/or coding sequence CpG mutations that permit transgenerational propagation of genetic instability (Wu et al. 2007; Morak et al. 2008). Consistent with this, CTs more often contain promoter CpG islands than do GKs (Zhao Y, unpublished data): classic GKs such as TP53 do not contain promoter CpG islands, whereas canonical CTs such as MLH1 and BRCA1 do. Instructively, the latter gene is mutated and not methylated in familial cancer syndromes (Chen et al. 2006), yet is methylated and not mutated in chemotherapy-induced second malignancies (Scardocci et al. 2006); similar exclusivity between repair gene methylation events and oncogenic indels or point mutations in repair-deficient tumors (Esteller et al. 2001; Toyooka et al. 2006) suggests the fluidity of such epimutations. Although in this model extrinsic damage is the major regulator of prezygotic male germ cell CT methylation—as well as being both a cause and effect of progressive suppressor gene repression in precancerous adult somatic tissues (Neri et al. 2007)—age may be as important as damage in the latter process (Kim et al. 2005), with parental (especially paternal) age playing a synergistic role in the former (Oakes et al. 2003).
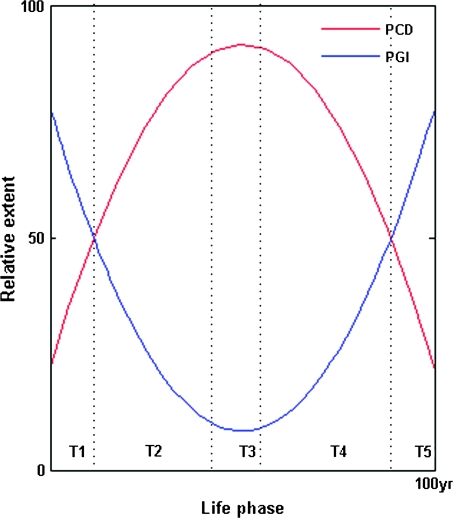
We define this model of a “methylation clock” regulating the epigenetic inactivation of CT/repair genes in the male germ line and adult somatic tissues—commensurate with extrinsic damage/stress or intrinsic ageing/senescence—as PGI; just as programmed cell death (PCD) is the mechanism of negative selection, so is PGI proposed to be the mechanism of positive selection (fig. 6). We suggest that PGI intensifies genetic divergence during sexual reproduction at the level of sperm–egg fusion, in contrast to PCD which eliminates both oocyte (Suh et al. 2006) and sperm DNA defects after syngamy (Fatehi et al. 2006). This sequence (sperm → ovum, zygote → embryo) of positive followed by negative selection fits with the notion of sexual conflict (Partridge and Hurst 1998), which should enhance biological system robustness and evolvability (Kitano 2004). We also note that heritable, though not necessarily familial, predisposition to carcinogenesis could also be propagated transgenerationally by PGI (Epstein and Zhao 2006b).
FIG. 6.—
Model of changing human male CT and GK function from germ line to somatic contexts. The x axis is divided into the following time frames: T1, prezygotic spermatogonia and spermatocytes; T2, postzygotic embryonic development; T3, prereproductive infancy and childhood; T4, reproductive adult life; and T5, postreproductive senescence. The y axis models the relative extent of either GK functionality (PCD) or CT dysfunctionality (PGI) during these time frames. Damage/stress-inducible prezygotic promoter (CpG island) methylation of spermatogonial/spermatocyte CTs and GKs, respectively, increases PGI while reducing PCD—thus maximizing genetic variability in response to changing environmental selection pressures. Postfertilization male germ line gene demethylation has the opposite effect, causing a sustained decline in PGI and a rise in PCD. During postreproductive adult life, an age-dependent (as well as damage-inducible) methylation clock induces somatic CT/GK gene inactivation, leading in turn to a senescent rise in PGI and decline of PCD that predisposes to sporadic tumor outgrowth.
What evidence do we have for PGI operating through the male germ line? The original hypothesis arose from prior knowledge, namely, 1) evidence for male-driven evolution from other groups (Agulnik et al. 1997; Hurst and Ellegren 1998; Wyckoff et al. 2000; Makova and Li 2002); 2) evidence for repair genes being speciation genes, implying a permissive role in rapid evolution (Radman and Wagner 1993; Cleaver et al. 1995; Sniegowski 1998); and 3) the external anatomic location of the testis, making male germ cells uniquely vulnerable to mutagenic, yet potentially reparable, environmental damage (Roest et al. 1996; Hsia et al. 2003; Feitsma et al. 2007). Against this background, we needed to integrate the following new data: 1) CT/repair genes are evolving with unexpected rapidity; 2) CT knockouts are relatively dispensable for survival, resulting only in reduced male fertility; and 3) abnormally low CT/repair gene expression is characteristic of teratozoospermia (Zhao Y, unpublished data), a condition of abnormal sperm morphology that does not significantly reduce in vitro fertilization success rates (Keegan et al. 2007). These considerations invited the hypothesis: could the rapid evolution of CT/repair genes reflect selection for a permissive (i.e., causal loss-of-function) role in male-driven evolution? This latter possibility is supported by the finding that heterozygous males and homozygous females can remain fertile when homozygous male repair gene knockouts are sterile (Roest 1996) and that even minimal repair transgene expression suffices to rescue fertility in repair-knockout males (Hsia 2003). Moreover, homozygous repair-deficient males may survive—albeit at the cost of tumor susceptibility—in genetic backgrounds where homozygous females die (Cranston et al. 1997). This striking gender-specific difference strongly suggests a male-specific significance for accelerated CT evolution, which involves interaction of ambient damage with selection for subtle (e.g., epimutational) male germ cell repair defects.
Our gene targeting and familial cancer data confirm that germ line CT defects are less lethal than GK defects (tables 2 and 3). The effects of repair gene targeting depend critically on the extent of functional inhibition and the presence or otherwise of associated mutations; in general, mild (e.g., haploinsufficient) CT/repair defects tend to increase mutation while decreasing apoptosis (Frank et al. 2005; Smilenov et al. 2005) and may even enhance fitness and lifespan (Siegl-Cachedenier et al. 2007), whereas severe repair defects tend to cause increased apoptosis and premature lethality (Henrie et al. 2003). Hence, because low-level sperm DNA damage can be repaired after fertilization of repair-proficient oocytes (Menezo 2006; Marchetti et al. 2007; Fernandez-Gonzalez et al. 2008)—particularly in the setting of prior DNA damage “conditioning” of such oocytes (Agrawal and Wang 2008)—mild nondeletional prezygotic male germ line repair defects could plausibly enhance sperm divergence with minimal fertility compromise, thereby safeguarding species’ evolvability while offsetting transgenerational risks of paternally transmitted birth defects (Marchetti and Wyrobek 2008) or cancer (Zenzes et al. 1999; Yauk et al. 2007). This conclusion is further supported by the finding that chronic exposure of spermatogonia to low-dose damage greatly reduces genetic damage induced by an acute second hit (Cai et al. 1993; Koana et al. 2007). Our findings therefore suggest the evolution of an environmentally interactive genetic program for promoting divergence in the germ line. We note that the related age-dependent predisposition to cancer is not wholly disadvantageous to the species as such mortality may promote redistribution of environmental resources to younger and more fertile individuals.
As an extension of the “immortal strand” hypothesis of stem cell fidelity (Cairns 2006), our findings raise the notion that the transcribed (well repaired, CpG-demethylated) DNA strand represents the “immovable object” of negative selection, whereas the untranscribed (poorly repaired, CpG-methylated) strand provides the “irresistible force” of positive selection. Furthermore, consistent with the notion that sexual conflict selects for distinct gender phenotypes (Hosken et al. 2001), the evolutionary paradigm presented here implies that the usual targets of positive selection—choice and specialization (Barkman 2003; Hamm et al. 2007), discrimination and success (Swanson et al. 2003), and beauty (Morris RD and Morris JA 2004)—are intrinsically male (sperm/PGI dependent) in origin, whereas the targets of negative selection (normality, utility, longevity, and fidelity) are genetically female or oocyte/PCD dependent. Both positively and negatively selected traits are vital for fitness in sexually reproducing species: for example, the reproductive success of butterflies, birds, and peacocks depends on highly variegated yet symmetric surface patterns (Gould JL and Gould CG 1997) that predict a low underlying frequency of deleterious functional mutations (Morris RD and Morris JA 2004). In our model, PGI is driven by sexual selection pressure on the male germ line to optimize as opposed to normalize whatever phenotypes can be optimized—such as sperm velocity (Gage et al. 2004) or ion channel function regulating motility (Podlaha et al. 2005)—while also enhancing variation in species-discriminatory phenotypes such as egg-binding proteins (Moller and Cuervo 2003). Hence, the basis for sexual conflict in our model is that “average” can never be “best” and that “health” may not guarantee “popularity” (Zaidel et al. 2005)—whether with respect to beauty (DeBruine et al. 2007) or to sperm competition (Neff and Pitcher 2005)—thus helping to explain the paradox that mutation rates tend to be reduced by natural selection yet increased by sexual selection (Moller and Cuervo 2003).
In conclusion, we have shown that CTs are less essential for germ line viability and hence more rapidly evolving than GKs. The resulting model of PGI implicates sexual reproduction as an evolutionary masterstroke: uniquely, sex plays off prezygotic positive selection (in which epimutational divergence is accelerated by environmental stress and damage, and efficiently selected and fixed by intra- and/or intermale sperm competition) against the evolutionary safety valves of postzygotic repair and negative selection (Menezo 2006). Via this mechanism, we propose that environmental stressors succeed in the otherwise oxymoronic task of “selecting for divergence” via epigenetic inhibition of DNA repair, thus helping to settle the “4-billion-year struggle of selfish genes to balance the need for variation with the equally important goal of conserving success” (Gould JL and Gould CG 1997). Moreover, if tumors do indeed arise in part as a side effect of PGI, cancer susceptibility may be most accurately viewed as the tumor-permissive “price” paid by multicellular organisms for genetic plasticity, with the species reaping the ultimate evolutionary “reward” of a delayed time to extinction.
Supplementary Material
Supplement 1, 2, and 3 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
We thank both anonymous reviewers for constructive scrutiny of the manuscript and the Hong Kong Jockey Club for support of the Clinical Research Centre.
References
- Agrawal AF, Wang AD. Increased transmission of mutations by low-condition females: evidence for condition-dependent DNA repair. PLoS Biol. 2008;6:e30. doi: 10.1371/journal.pbio.0060030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agulnik AI, Bishop CE, Lerner JL, Agulnik SI, Solovyev VV. Analysis of mutation rates in the SMCY/SMCX genes shows that mammalian evolution is male driven. Mamm Genome. 1997;8:134–138. doi: 10.1007/s003359900372. [DOI] [PubMed] [Google Scholar]
- Andrews JD, Mancini DN, Singh SM, Rodenhiser DI. Site and sequence specific DNA methylation in the neurofibromatosis (NF1) gene includes C5839T: the site of the recurrent substitution mutation in exon 31. Hum Mol Genet. 1996;5:503–507. doi: 10.1093/hmg/5.4.503. [DOI] [PubMed] [Google Scholar]
- Anway MD, Rekow SS, Skinner MK. Transgenerational epigenetic programming of the embryonic testis transcriptome. Genomics. 2008;91:30–40. doi: 10.1016/j.ygeno.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Dixit VM, Koonin EV. Apoptotic molecular machinery: vastly increased complexity in vertebrates revealed by genome comparisons. Science. 2001;291:1279–1284. doi: 10.1126/science.291.5507.1279. [DOI] [PubMed] [Google Scholar]
- Babenko VN, Basu MK, Kondrashov FA, Rogozin IB, Koonin EV. Signs of positive selection of somatic mutations in human cancers detected by EST sequence analysis. BMC Cancer. 2006;6:36. doi: 10.1186/1471-2407-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber RC, Hickenbotham P, Hatch T, et al. (11 co-authors) Radiation-induced transgenerational alterations in genome stability and DNA damage. Oncogene. 2006;25:7336–7342. doi: 10.1038/sj.onc.1209723. [DOI] [PubMed] [Google Scholar]
- Barkman TJ. Evidence for positive selection on the floral scent gene isoeugenol-O-methyltransferase. Mol Biol Evol. 2003;20:168–172. doi: 10.1093/molbev/msg030. [DOI] [PubMed] [Google Scholar]
- Bielas JH, Loeb KR, Rubin BP, True LD, Loeb LA. Human cancers express a mutator phenotype. Proc Natl Acad Sci USA. 2006;103:18238–18242. doi: 10.1073/pnas.0607057103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco D, Vicent S, Fraga MF, et al. (11 co-authors) Molecular analysis of a multistep lung cancer model induced by chronic inflammation reveals epigenetic regulation of p16 and activation of the DNA damage response pathway. Neoplasia. 2007;9:840–852. doi: 10.1593/neo.07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breivik J. The evolutionary origin of genetic instability in cancer development. Semin Cancer Biol. 2005;15:51–60. doi: 10.1016/j.semcancer.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Breivik J, Gaudernack G. Resolving the evolutionary paradox of genetic instability: a cost-benefit analysis of DNA repair in changing environments. FEBS Lett. 2004;563:7–12. doi: 10.1016/S0014-5793(04)00282-0. [DOI] [PubMed] [Google Scholar]
- Bustamante CD, Fledel-Alon A, Williamson S, et al. (14 co-authors) Natural selection on protein-coding genes in the human genome. Nature. 2005;437:1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- Cai L, Jiang J, Wang B, Yao H, Wang X. Induction of an adaptive response to dominant lethality and to chromosome damage of mouse germ cells by low dose radiation. Mutat Res. 1993;303:157–161. doi: 10.1016/0165-7992(93)90017-p. [DOI] [PubMed] [Google Scholar]
- Cairns J. Cancer and the immortal strand hypothesis. Genetics. 2006;174:1069–1072. doi: 10.1534/genetics.104.66886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerda S, Weitzman SA. Influence of oxygen radical injury on DNA methylation. Mutat Res. 1997;386:141–152. doi: 10.1016/s1383-5742(96)00050-6. [DOI] [PubMed] [Google Scholar]
- Chen Y, Toland AE, McLennan J, Fridlyand J, Crawford B, Costello JF, Ziegler JL. Lack of germ-line promoter methylation in BRCA1-negative families with familial breast cancer. Genet Test. 2006;10:281–284. doi: 10.1089/gte.2006.10.281. [DOI] [PubMed] [Google Scholar]
- Cirz RT, Romesberg FE. Controlling mutation: intervening in evolution as a therapeutic strategy. Crit Rev Biochem Mol Biol. 2007;42:341–354. doi: 10.1080/10409230701597741. [DOI] [PubMed] [Google Scholar]
- Clark AG, Glanowski S, Nielsen R, et al. (17 co-authors) Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science. 2003;302:1960–1963. doi: 10.1126/science.1088821. [DOI] [PubMed] [Google Scholar]
- Cleaver JE, Speakman JR, Volpe JP. Nucleotide excision repair: variations associated with cancer development and speciation. Cancer Surv. 1995;25:125–142. [PubMed] [Google Scholar]
- Colegrave N, Collins S. Experimental evolution: experimental evolution and evolvability. Heredity. 2008;100:464–470. doi: 10.1038/sj.hdy.6801095. [DOI] [PubMed] [Google Scholar]
- Comeron JM. K-Estimator: calculation of the number of nucleotide substitutions per site and the confidence intervals. Bioinformatics. 1999;15:763–764. doi: 10.1093/bioinformatics/15.9.763. [DOI] [PubMed] [Google Scholar]
- Cooper DN, Krawczak M. Cytosine methylation and the fate of CpG dinucleotides in vertebrate genomes. Hum Genet. 1989;83:181–188. doi: 10.1007/BF00286715. [DOI] [PubMed] [Google Scholar]
- Cranston A, Bocker T, Reitmair A, Palazzo J, Wilson T, Mak T, Fishel R. Female embryonic lethality in mice nullizygous for both Msh2 and p53. Nat Genet. 1997;17:114–118. doi: 10.1038/ng0997-114. [DOI] [PubMed] [Google Scholar]
- Crespi BJ, Summers K. Positive selection in the evolution of cancer. Biol Rev Camb Philos Soc. 2006;81:407–424. doi: 10.1017/S1464793106007056. [DOI] [PubMed] [Google Scholar]
- DeBruine LM, Jones BC, Unger L, Little AC, Feinberg DR. Dissociating averageness and attractiveness: attractive faces are not always average. J Exp Psychol Hum Percept Perform. 2007;33:1420–1430. doi: 10.1037/0096-1523.33.6.1420. [DOI] [PubMed] [Google Scholar]
- Doctor KS, Reed JC, Godzik A, Bourne PE. The apoptosis database. Cell Death Differ. 2003;10:621–633. doi: 10.1038/sj.cdd.4401230. [DOI] [PubMed] [Google Scholar]
- Dolinoy DC, Jirtle RL. Environmental epigenomics in human health and disease. Environ Mol Mutagen. 2008;49:4–8. doi: 10.1002/em.20366. [DOI] [PubMed] [Google Scholar]
- Driscoll DJ, Migeon BR. Sex difference in methylation of single-copy genes in human meiotic germ cells: implications for X chromosome inactivation, parental imprinting, and origin of CpG mutations. Somat Cell Mol Genet. 1990;16:267–282. doi: 10.1007/BF01233363. [DOI] [PubMed] [Google Scholar]
- Drost JB, Lee WR. Biological basis of germline mutation: comparisons of spontaneous germline mutation rates among drosophila, mouse, and human. Environ Mol Mutagen. 1995;25(Suppl. 26):48–64. doi: 10.1002/em.2850250609. [DOI] [PubMed] [Google Scholar]
- Elena SF, de Visser JA. Environmental stress and the effects of mutation. J Biol. 2003;2:12. doi: 10.1186/1475-4924-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Maarri O, Olek A, Balaban B, Montag M, van der Ven H, Urman B, Olek K, Caglayan SH, Walter J, Oldenburg J. Methylation levels at selected CpG sites in the factor VIII and FGFR3 genes, in mature female and male germ cells: implications for male-driven evolution. Am J Hum Genet. 1998;63:1001–1008. doi: 10.1086/302065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein RJ, Lin K, Tan TW. A functional significance for codon third bases. Gene. 2000;245:291–298. doi: 10.1016/s0378-1119(00)00042-1. [DOI] [PubMed] [Google Scholar]
- Epstein RJ, Zhao Y. Programmed genetic instability revealed by adaptive evolution of caretaker tumor suppressor genes. AACR Meeting Abstracts. 2006a [Google Scholar]
- Epstein RJ, Zhao Y. Racial differences in lung cancer. N Engl J Med. 2006b;354:1951–1953;. doi: 10.1056/NEJMc060512. author reply 1951–1953. [DOI] [PubMed] [Google Scholar]
- Esteller M, Fraga MF, Guo M, et al. (24 co-authors) DNA methylation patterns in hereditary human cancers mimic sporadic tumorigenesis. Hum Mol Genet. 2001;10:3001–3007. doi: 10.1093/hmg/10.26.3001. [DOI] [PubMed] [Google Scholar]
- Fatehi AN, Bevers MM, Schoevers E, Roelen BA, Colenbrander B, Gadella BM. DNA damage in bovine sperm does not block fertilization and early embryonic development but induces apoptosis after the first cleavages. J Androl. 2006;27:176–188. doi: 10.2164/jandrol.04152. [DOI] [PubMed] [Google Scholar]
- Fay JC, Wyckoff GJ, Wu CI. Positive and negative selection on the human genome. Genetics. 2001;158:1227–1234. doi: 10.1093/genetics/158.3.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feitsma H, Leal MC, Moens PB, Cuppen E, Schulz RW. Mlh1 deficiency in zebrafish results in male sterility and aneuploid as well as triploid progeny in females. Genetics. 2007;175:1561–1569. doi: 10.1534/genetics.106.068171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Gonzalez R, Moreira PN, Perez-Crespo M, et al. (11 co-authors) Long-term effects of mouse intracytoplasmic sperm injection with DNA-fragmented sperm on health and behavior of adult offspring. Biol Reprod. 2008;78:761–772. doi: 10.1095/biolreprod.107.065623. [DOI] [PubMed] [Google Scholar]
- Fleming MA, Potter JD, Ramirez CJ, Ostrander GK, Ostrander EA. Understanding missense mutations in the BRCA1 gene: an evolutionary approach. Proc Natl Acad Sci USA. 2003;100:1151–1156. doi: 10.1073/pnas.0237285100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SA. Maladaptation and the paradox of robustness in evolution. PLoS ONE. 2007;2:e1021. doi: 10.1371/journal.pone.0001021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SA, Chen PC, Lipkin SM. Kinetics of cancer: a method to test hypotheses of genetic causation. BMC Cancer. 2005;5:163. doi: 10.1186/1471-2407-5-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohling S, Scholl C, Levine RL, et al. (21 co-authors) Identification of driver and passenger mutations of FLT3 by high-throughput DNA sequence analysis and functional assessment of candidate alleles. Cancer Cell. 2007;12:501–513. doi: 10.1016/j.ccr.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadagkar SR, Rosenberg MS, Kumar S. Inferring species phylogenies from multiple genes: concatenated sequence tree versus consensus gene tree. J Exp Zoolog B Mol Dev Evol. 2005;304:64–74. doi: 10.1002/jez.b.21026. [DOI] [PubMed] [Google Scholar]
- Gage MJ, Macfarlane CP, Yeates S, Ward RG, Searle JB, Parker GA. Spermatozoal traits and sperm competition in Atlantic salmon: relative sperm velocity is the primary determinant of fertilization success. Curr Biol. 2004;14:44–47. [PubMed] [Google Scholar]
- Galhardo RS, Hastings PJ, Rosenberg SM. Mutation as a stress response and the regulation of evolvability. Crit Rev Biochem Mol Biol. 2007;42:399–435. doi: 10.1080/10409230701648502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway LF, Etterson JR. Transgenerational plasticity is adaptive in the wild. Science. 2007;318:1134–1136. doi: 10.1126/science.1148766. [DOI] [PubMed] [Google Scholar]
- Gatenby RA, Vincent TL. An evolutionary model of carcinogenesis. Cancer Res. 2003;63:6212–6220. [PubMed] [Google Scholar]
- Gould JL, Gould CG. Sexual selection: mate choice and courtship in nature. Basingstoke (UK): Macmillan; 1997. [Google Scholar]
- Gulick JT. Divergent evolution through cumulative segregation. Westminster (MD): Smithsonian Institute; 1893. [Google Scholar]
- Haaf T. Methylation dynamics in the early mammalian embryo: implications of genome reprogramming defects for development. Curr Top Microbiol Immunol. 2006;310:13–22. doi: 10.1007/3-540-31181-5_2. [DOI] [PubMed] [Google Scholar]
- Hamm D, Mautz BS, Wolfner MF, Aquadro CF, Swanson WJ. Evidence of amino acid diversity-enhancing selection within humans and among primates at the candidate sperm-receptor gene PKDREJ. Am J Hum Genet. 2007;81:44–52. doi: 10.1086/518695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Sui H, Kawakami K, Shimada Y, Shibuya T. Partial hepatectomy strongly increased the mutagenicity of N-ethyl-N-nitrosourea in MutaMouse liver. Environ Mol Mutagen. 1999;34:121–123. [PubMed] [Google Scholar]
- Hastings PJ, Slack A, Petrosino JF, Rosenberg SM. Adaptive amplification and point mutation are independent mechanisms: evidence for various stress-inducible mutation mechanisms. PLoS Biol. 2004;2:e399. doi: 10.1371/journal.pbio.0020399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrikse JL, Parsons TE, Hallgrimsson B. Evolvability as the proper focus of evolutionary developmental biology. Evol Dev. 2007;9:393–401. doi: 10.1111/j.1525-142X.2007.00176.x. [DOI] [PubMed] [Google Scholar]
- Henrie MS, Kurimasa A, Burma S, Menissier-de Murcia J, de Murcia G, Li GC, Chen DJ. Lethality in PARP-1/Ku80 double mutant mice reveals physiological synergy during early embryogenesis. DNA Repair (Amst) 2003;2:151–158. doi: 10.1016/s1568-7864(02)00199-4. [DOI] [PubMed] [Google Scholar]
- Hiraiwa-Hasegawa M. The sight of the peacock's tail makes me sick: the early arguments on sexual selection. J Biosci. 2000;25:11–18. doi: 10.1007/BF02985176. [DOI] [PubMed] [Google Scholar]
- Hone DW, Benton MJ. The evolution of large size: how does Cope's rule work? Trends Ecol Evol. 2005;20:4–6. doi: 10.1016/j.tree.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Horsthemke B. Epimutations in human disease. Curr Top Microbiol Immunol. 2006;310:45–59. doi: 10.1007/3-540-31181-5_4. [DOI] [PubMed] [Google Scholar]
- Hosken DJ, Garner TW, Ward PI. Sexual conflict selects for male and female reproductive characters. Curr Biol. 2001;11:489–493. doi: 10.1016/s0960-9822(01)00146-4. [DOI] [PubMed] [Google Scholar]
- Hsia KT, Millar MR, King S, Selfridge J, Redhead NJ, Melton DW, Saunders PT. DNA repair gene Ercc1 is essential for normal spermatogenesis and oogenesis and for functional integrity of germ cell DNA in the mouse. Development. 2003;130:369–378. doi: 10.1242/dev.00221. [DOI] [PubMed] [Google Scholar]
- Hurst LD, Ellegren H. Sex biases in the mutation rate. Trends Genet. 1998;14:446–452. doi: 10.1016/s0168-9525(98)01577-7. [DOI] [PubMed] [Google Scholar]
- Huttley GA, Easteal S, Southey MC, Tesoriero A, Giles GG, McCredie MR, Hopper JL, Venter DJ. Adaptive evolution of the tumour suppressor BRCA1 in humans and chimpanzees. Australian Breast Cancer Family Study. Nat Genet. 2000;25:410–413. doi: 10.1038/78092. [DOI] [PubMed] [Google Scholar]
- Iwasa Y, Michor F, Nowak MA. Stochastic tunnels in evolutionary dynamics. Genetics. 2004;166:1571–1579. doi: 10.1534/genetics.166.3.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto FV, Esteller M. Mutator pathways unleashed by epigenetic silencing in human cancer. Mutagenesis. 2007;22:247–253. doi: 10.1093/mutage/gem009. [DOI] [PubMed] [Google Scholar]
- Johnson T. Beneficial mutations, hitchhiking and the evolution of mutation rates in sexual populations. Genetics. 1999;151:1621–1631. doi: 10.1093/genetics/151.4.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan BR, Barton S, Sanchez X, Berkeley AS, Krey LC, Grifo J. Isolated teratozoospermia does not affect in vitro fertilization outcome and is not an indication for intracytoplasmic sperm injection. Fertil Steril. 2007;88:1583–1588. doi: 10.1016/j.fertnstert.2007.01.057. [DOI] [PubMed] [Google Scholar]
- Kelley JL, Madeoy J, Calhoun JC, Swanson W, Akey JM. Genomic signatures of positive selection in humans and the limits of outlier approaches. Genome Res. 2006;16:980–989. doi: 10.1101/gr.5157306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Beart RW, Shibata D. Stability of colon stem cell methylation after neo-adjuvant therapy in a patient with attenuated familial adenomatous polyposis. BMC Gastroenterol. 2005;5:19. doi: 10.1186/1471-230X-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. Evolutionary rate at the molecular level. Nature. 1968;217:624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- King JL, Jukes TH. Non-Darwinian evolution. Science. 1969;164:788–798. doi: 10.1126/science.164.3881.788. [DOI] [PubMed] [Google Scholar]
- Kingsolver JG, Pfennig DW. Individual-level selection as a cause of Cope's rule of phyletic size increase. Evolution Int J Org Evolution. 2004;58:1608–1612. doi: 10.1111/j.0014-3820.2004.tb01740.x. [DOI] [PubMed] [Google Scholar]
- Kinzler KW, Vogelstein B. Cancer-susceptibility genes. Gatekeepers and caretakers. Nature. 1997;386:761–763. doi: 10.1038/386761a0. [DOI] [PubMed] [Google Scholar]
- Kitano H. Biological robustness. Nat Rev Genet. 2004;5:826–837. doi: 10.1038/nrg1471. [DOI] [PubMed] [Google Scholar]
- Knudson AG. Chasing the cancer demon. Annu Rev Genet. 2000;34:1–19. doi: 10.1146/annurev.genet.34.1.1. [DOI] [PubMed] [Google Scholar]
- Koana T, Okada MO, Ogura K, Tsujimura H, Sakai K. Reduction of background mutations by low-dose X irradiation of Drosophila spermatocytes at a low dose rate. Radiat Res. 2007;167:217–221. doi: 10.1667/rr0705.1. [DOI] [PubMed] [Google Scholar]
- Lavon I, Fuchs D, Zrihan D, Efroni G, Zelikovitch B, Fellig Y, Siegal T. Novel mechanism whereby nuclear factor kappaB mediates DNA damage repair through regulation of O(6)-methylguanine-DNA-methyltransferase. Cancer Res. 2007;67:8952–8959. doi: 10.1158/0008-5472.CAN-06-3820. [DOI] [PubMed] [Google Scholar]
- Lenski RE, Barrick JE, Ofria C. Balancing robustness and evolvability. PLoS Biol. 2006;4:e428. doi: 10.1371/journal.pbio.0040428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levasseur A, Orlando L, Bailly X, Milinkovitch MC, Danchin EG, Pontarotti P. Conceptual bases for quantifying the role of the environment on gene evolution: the participation of positive selection and neutral evolution. Biol Rev Camb Philos Soc. 2007;82:551–572. doi: 10.1111/j.1469-185X.2007.00024.x. [DOI] [PubMed] [Google Scholar]
- Liao BY, Scott NM, Zhang J. Impacts of gene essentiality, expression pattern, and gene compactness on the evolutionary rate of mammalian proteins. Mol Biol Evol. 2006;23:2072–2080. doi: 10.1093/molbev/msl076. [DOI] [PubMed] [Google Scholar]
- Lin K, Tan SB, Kolatkar PR, Epstein RJ. Nonrandom intragenic variations in patterns of codon bias implicate a sequential interplay between transitional genetic drift and functional amino acid selection. J Mol Evol. 2003;57:538–545. doi: 10.1007/s00239-003-2507-5. [DOI] [PubMed] [Google Scholar]
- Lynch M. The evolution of genetic networks by non-adaptive processes. Nat Rev Genet. 2007;8:803–813. doi: 10.1038/nrg2192. [DOI] [PubMed] [Google Scholar]
- Majewski J. Dependence of mutational asymmetry on gene-expression levels in the human genome. Am J Hum Genet. 2003;73:688–692. doi: 10.1086/378134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makova KD, Li WH. Strong male-driven evolution of DNA sequences in humans and apes. Nature. 2002;416:624–626. doi: 10.1038/416624a. [DOI] [PubMed] [Google Scholar]
- Marchetti F, Essers J, Kanaar R, Wyrobek AJ. Disruption of maternal DNA repair increases sperm-derived chromosomal aberrations. Proc Natl Acad Sci USA. 2007;104:17725–17729. doi: 10.1073/pnas.0705257104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti F, Wyrobek AJ. DNA repair decline during mouse spermiogenesis results in the accumulation of heritable DNA damage. DNA Repair (Amst) 2008;7:572–581. doi: 10.1016/j.dnarep.2007.12.011. [DOI] [PubMed] [Google Scholar]
- Maresca B, Schwartz JH. Sudden origins: a general mechanism of evolution based on stress protein concentration and rapid environmental change. Anat Rec B New Anat. 2006;289:38–46. doi: 10.1002/ar.b.20089. [DOI] [PubMed] [Google Scholar]
- Markov AV. The return of the Red Queen, or the law of the growth in the mean duration of the existence of genera during evolution. Zh Obshch Biol. 2000;61:357–370. [PubMed] [Google Scholar]
- Martin DI, Ward R, Suter CM. Germline epimutation: a basis for epigenetic disease in humans. Ann N Y Acad Sci. 2005;1054:68–77. doi: 10.1196/annals.1345.009. [DOI] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Menezo Y. Oocyte capacity to repair DNA damage induced in sperm. J Gynecol Obstet Biol Reprod (Paris) 2006;35:2S19–2S23. [PubMed] [Google Scholar]
- Moller AP, Cuervo JJ. Sexual selection, germline mutation rate and sperm competition. BMC Evol Biol. 2003;3:6. doi: 10.1186/1471-2148-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk M. Epigenetic programming of differential gene expression in development and evolution. Dev Genet. 1995;17:188–197. doi: 10.1002/dvg.1020170303. [DOI] [PubMed] [Google Scholar]
- Morak M, Schackert HK, Rahner N, et al. (14 co-authors) Further evidence for heritability of an epimutation in one of 12 cases with MLH1 promoter methylation in blood cells clinically displaying HNPCC. Eur J Hum Genet. 2008 doi: 10.1038/ejhg.2008.25. [DOI] [PubMed] [Google Scholar]
- Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet. 14 Spec No. 2005;1:R47–R58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- Morris RD, Morris JA. Sexual selection, redundancy and survival of the most beautiful. J Biosci. 2004;29:359–366. doi: 10.1007/BF02702619. [DOI] [PubMed] [Google Scholar]
- Neff BD, Pitcher TE. Genetic quality and sexual selection: an integrated framework for good genes and compatible genes. Mol Ecol. 2005;14:19–38. doi: 10.1111/j.1365-294X.2004.02395.x. [DOI] [PubMed] [Google Scholar]
- Nei M. Selectionism and neutralism in molecular evolution. Mol Biol Evol. 2005;22:2318–2342. doi: 10.1093/molbev/msi242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri S, Pawelec G, Facchini A, Mariani E. Microsatellite instability and compromised mismatch repair gene expression during in vitro passaging of monoclonal human T lymphocytes. Rejuvenation Res. 2007;10:145–156. doi: 10.1089/rej.2006.0510. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Bustamante C, Clark AG, et al. (13 co-authors) A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3:e170. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolopoulos G, Pyrpassopoulos S, Thanassoulas A, Klimentzou P, Zikos C, Vlassi M, Vorgias CE, Yannoukakos D, Nounesis G. Thermal unfolding of human BRCA1 BRCT-domain variants. Biochim Biophys Acta. 2007;1774:772–780. doi: 10.1016/j.bbapap.2007.03.018. [DOI] [PubMed] [Google Scholar]
- Nowak MA, Michor F, Komarova NL, Iwasa Y. Evolutionary dynamics of tumor suppressor gene inactivation. Proc Natl Acad Sci USA. 2004;101:10635–10638. doi: 10.1073/pnas.0400747101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes CC, La Salle S, Smiraglia DJ, Robaire B, Trasler JM. A unique configuration of genome-wide DNA methylation patterns in the testis. Proc Natl Acad Sci USA. 2007a;104:228–233. doi: 10.1073/pnas.0607521104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes CC, La Salle S, Smiraglia DJ, Robaire B, Trasler JM. Developmental acquisition of genome-wide DNA methylation occurs prior to meiosis in male germ cells. Dev Biol. 2007b;307:368–379. doi: 10.1016/j.ydbio.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Oakes CC, Smiraglia DJ, Plass C, Trasler JM, Robaire B. Aging results in hypermethylation of ribosomal DNA in sperm and liver of male rats. Proc Natl Acad Sci USA. 2003;100:1775–1780. doi: 10.1073/pnas.0437971100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson R, Cui H, He L, Pfeifer S, Malmikumpu H, Jiang S, Feinberg AP, Hedborg F. Mosaic allelic insulin-like growth factor 2 expression patterns reveal a link between Wilms’ tumorigenesis and epigenetic heterogeneity. Cancer Res. 1999;59:3889–3892. [PubMed] [Google Scholar]
- Ohta T. Evolution by nearly-neutral mutations. Genetica. 1998;102–103:83–90. [PubMed] [Google Scholar]
- Olson MV. When less is more: gene loss as an engine of evolutionary change. Am J Hum Genet. 1999;64:18–23. doi: 10.1086/302219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L, Hurst LD. Sex and conflict. Science. 1998;281:2003–2008. doi: 10.1126/science.281.5385.2003. [DOI] [PubMed] [Google Scholar]
- Pavlicek A, Noskov VN, Kouprina N, Barrett JC, Jurka J, Larionov V. Evolution of the tumor suppressor BRCA1 locus in primates: implications for cancer predisposition. Hum Mol Genet. 2004;13:2737–2751. doi: 10.1093/hmg/ddh301. [DOI] [PubMed] [Google Scholar]
- Perrin MC, Brown AS, Malaspina D. Aberrant epigenetic regulation could explain the relationship of paternal age to schizophrenia. Schizophr Bull. 2007;33:1270–1273. doi: 10.1093/schbul/sbm093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie M, Roberts G. Sexual selection and the evolution of evolvability. Heredity. 2007;98:198–205. doi: 10.1038/sj.hdy.6800921. [DOI] [PubMed] [Google Scholar]
- Pigliucci M. Is evolvability evolvable? Nat Rev Genet. 2008;9:75–82. doi: 10.1038/nrg2278. [DOI] [PubMed] [Google Scholar]
- Podlaha O, Webb DM, Tucker PK, Zhang J. Positive selection for indel substitutions in the rodent sperm protein catsper1. Mol Biol Evol. 2005;22:1845–1852. doi: 10.1093/molbev/msi178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponder RG, Fonville NC, Rosenberg SM. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol Cell. 2005;19:791–804. doi: 10.1016/j.molcel.2005.07.025. [DOI] [PubMed] [Google Scholar]
- Ponting CP, Lunter G. Signatures of adaptive evolution within human non-coding sequence. Hum Mol Genet. 15 Spec No. 2006;2:R170–R175. doi: 10.1093/hmg/ddl182. [DOI] [PubMed] [Google Scholar]
- Radman M, Wagner R. Mismatch recognition in chromosomal interactions and speciation. Chromosoma. 1993;102:369–373. doi: 10.1007/BF00360400. [DOI] [PubMed] [Google Scholar]
- Roest HP, van Klaveren J, de Wit J, et al. (13 co-authors) Inactivation of the HR6B ubiquitin-conjugating DNA repair enzyme in mice causes male sterility associated with chromatin modification. Cell. 1996;86:799–810. doi: 10.1016/s0092-8674(00)80154-3. [DOI] [PubMed] [Google Scholar]
- Salnikow K, Zhitkovich A. Genetic and epigenetic mechanisms in metal carcinogenesis and cocarcinogenesis: nickel, arsenic, and chromium. Chem Res Toxicol. 2008;21:28–44. doi: 10.1021/tx700198a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarosa M, Ashworth A. Haploinsufficiency for tumour suppressor genes: when you don't need to go all the way. Biochim Biophys Acta. 2004;1654:105–122. doi: 10.1016/j.bbcan.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Scardocci A, Guidi F, D'Alo F, et al. (12 co-authors) Reduced BRCA1 expression due to promoter hypermethylation in therapy-related acute myeloid leukaemia. Br J Cancer. 2006;95:1108–1113. doi: 10.1038/sj.bjc.6603392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider BL, Kulesz-Martin M. Destructive cycles: the role of genomic instability and adaptation in carcinogenesis. Carcinogenesis. 2004;25:2033–2044. doi: 10.1093/carcin/bgh204. [DOI] [PubMed] [Google Scholar]
- Siegl-Cachedenier I, Munoz P, Flores JM, Klatt P, Blasco MA. Deficient mismatch repair improves organismal fitness and survival of mice with dysfunctional telomeres. Genes Dev. 2007;21:2234–2247. doi: 10.1101/gad.430107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smilenov LB. Tumor development: haploinsufficiency and local network assembly. Cancer Lett. 2006;240:17–28. doi: 10.1016/j.canlet.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Smilenov LB, Lieberman HB, Mitchell SA, Baker RA, Hopkins KM, Hall EJ. Combined haploinsufficiency for ATM and RAD9 as a factor in cell transformation, apoptosis, and DNA lesion repair dynamics. Cancer Res. 2005;65:933–938. [PubMed] [Google Scholar]
- Smith NG, Hurst LD. Molecular evolution of an imprinted gene: repeatability of patterns of evolution within the mammalian insulin-like growth factor type II receptor. Genetics. 1998;150:823–833. doi: 10.1093/genetics/150.2.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniegowski P. Mismatch repair: origin of species? Curr Biol. 1998;8:R59–R61. doi: 10.1016/s0960-9822(98)70035-1. [DOI] [PubMed] [Google Scholar]
- Sniegowski PD, Gerrish PJ, Johnson T, Shaver A. The evolution of mutation rates: separating causes from consequences. Bioessays. 2000;22:1057–1066. doi: 10.1002/1521-1878(200012)22:12<1057::AID-BIES3>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, et al. (13 co-authors) A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh EK, Yang A, Kettenbach A, Bamberger C, Michaelis AH, Zhu Z, Elvin JA, Bronson RT, Crum CP, McKeon F. p63 protects the female germ line during meiotic arrest. Nature. 2006;444:624–628. doi: 10.1038/nature05337. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Nielsen R, Yang Q. Pervasive adaptive evolution in mammalian fertilization proteins. Mol Biol Evol. 2003;20:18–20. doi: 10.1093/oxfordjournals.molbev.a004233. [DOI] [PubMed] [Google Scholar]
- Tang CS, Epstein RJ. A structural split in the human genome. PLoS ONE. 2007;2:e603. doi: 10.1371/journal.pone.0000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang CS, Zhao YZ, Smith DK, Epstein RJ. Intron length and accelerated 3′ gene evolution. Genomics. 2006;88:682–689. doi: 10.1016/j.ygeno.2006.06.017. [DOI] [PubMed] [Google Scholar]
- Thomas MA, Weston B, Joseph M, Wu W, Nekrutenko A, Tonellato PJ. Evolutionary dynamics of oncogenes and tumor suppressor genes: higher intensities of purifying selection than other genes. Mol Biol Evol. 2003;20:964–968. doi: 10.1093/molbev/msg110. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka S, Tokumo M, Shigematsu H, et al. (12 co-authors) Mutational and epigenetic evidence for independent pathways for lung adenocarcinomas arising in smokers and never smokers. Cancer Res. 2006;66:1371–1375. doi: 10.1158/0008-5472.CAN-05-2625. [DOI] [PubMed] [Google Scholar]
- University Medical Center Groningen. Familial Cancer Database. UICC. 2006 http://www.familialcancerdatabase.nl/ [Google Scholar]
- Van Valkenburgh B, Wang X, Damuth J. Cope's rule, hypercarnivory, and extinction in North American canids. Science. 2004;306:101–104. doi: 10.1126/science.1102417. [DOI] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. Robustness and evolvability: a paradox resolved. Proc Biol Sci. 2008;275:91–100. doi: 10.1098/rspb.2007.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee KB, Aguda BD. Akt versus p53 in a network of oncogenes and tumor suppressor genes regulating cell survival and death. Biophys J. 2006;91:857–865. doi: 10.1529/biophysj.105.077693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood RD, Mitchell M, Lindahl T. Human DNA repair genes, 2005. Mutat Res. 2005;577:275–283. doi: 10.1016/j.mrfmmm.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Wu YH, Tsai Chang JH, Cheng YW, Wu TC, Chen CY, Lee H. Xeroderma pigmentosum group C gene expression is predominantly regulated by promoter hypermethylation and contributes to p53 mutation in lung cancers. Oncogene. 2007;26:4761–4773. doi: 10.1038/sj.onc.1210284. [DOI] [PubMed] [Google Scholar]
- Wyckoff GJ, Wang W, Wu CI. Rapid evolution of male reproductive genes in the descent of man. Nature. 2000;403:304–309. doi: 10.1038/35002070. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol. 2002;19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- Yauk C, Polyzos A, Rowan-Carroll A, et al. (12 co-authors) Germ-line mutations, DNA damage, and global hypermethylation in mice exposed to particulate air pollution in an urban/industrial location. Proc Natl Acad Sci USA. 2008;105:605–610. doi: 10.1073/pnas.0705896105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauk CL, Berndt ML, Williams A, Rowan-Carroll A, Douglas GR, Stampfli MR. Mainstream tobacco smoke causes paternal germ-line DNA mutation. Cancer Res. 2007;67:5103–5106. doi: 10.1158/0008-5472.CAN-07-0279. [DOI] [PubMed] [Google Scholar]
- Zaidel DW, Aarde SM, Baig K. Appearance of symmetry, beauty, and health in human faces. Brain Cogn. 2005;57:261–263. doi: 10.1016/j.bandc.2004.08.056. [DOI] [PubMed] [Google Scholar]
- Zenzes MT, Puy LA, Bielecki R, Reed TE. Detection of benzo[a]pyrene diol epoxide-DNA adducts in embryos from smoking couples: evidence for transmission by spermatozoa. Mol Hum Reprod. 1999;5:125–131. doi: 10.1093/molehr/5.2.125. [DOI] [PubMed] [Google Scholar]
- Zhang J, Rosenberg HF, Nei M. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc Natl Acad Sci USA. 1998;95:3708–3713. doi: 10.1073/pnas.95.7.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.