Abstract
In the yeast Saccharomyces cerevisiae, meiotic recombination is initiated by transient DNA double-strand breaks (DSBs) that are repaired by interaction of the broken chromosome with its homologue. To identify a large number of DSB sites and gain insight into the control of DSB formation at both the local and the whole chromosomal levels, we have determined at high resolution the distribution of meiotic DSBs along the 340 kb of chromosome III. We have found 76 DSB regions, mostly located in intergenic promoter-containing intervals. The frequency of DSBs varies at least 50-fold from one region to another. The global distribution of DSB regions along chromosome III is nonrandom, defining large (39–105 kb) chromosomal domains, both hot and cold. The distribution of these localized DSBs indicates that they are likely to initiate most crossovers along chromosome III, but some discrepancies remain to be explained.
Keywords: recombination, transcription promoter, meiosis, chromosome organization
Meiotic recombination plays important biological roles at genomic and cellular levels: it creates the genetic diversity transmitted through the germ line and ensures the proper disjunction of homologous chromosomes during the first meiotic division that is necessary for the production of viable haploid gametes (1–3). The complexity of these fundamental processes, conserved over the course of eukaryotic evolution, is not yet fully elucidated.
In the yeast Saccharomyces cerevisiae, a series of studies, mostly performed at recombinational hot spots, has shown that meiotic recombination is initiated by transient localized DNA double-strand breaks (DSBs) occurring in early meiosis (4–8), which are then repaired by interaction with the unbroken homologous chromosome to yield recombinant products (1). The factors controlling the position and amount of the DSBs at few recombinational hot spots have been partially uncovered. (i) Most DSB sites are found located in intergenic regions containing transcriptional promoters (reviewed in ref. 1) but the occurrence of DSBs at other locations has been reported, including within the coding sequence of the HIS2 gene (9) and at uncharacterized positions in several artificial constructs (10). (ii) The occurrence of DSBs is controlled by determinants of chromatin structure, as they occur in regions that are hypersensitive to DNaseI and micrococcal nuclease (8, 10–13). (iii) DSB sites are not sequence-specific (14) and are clustered at many positions throughout regions ≈50–200 nucleotides long (15–18). (iv) As shown by pulsed field gel electrophoresis experiments, DSBs occur at numerous locations within the genome and along the chromosomes (6, 8, 19, 20). The factors controlling the overall distribution of DSBs are not known, but they are likely related to other aspects of the functional and structural organization of the chromosome as reported here. Finally, the formation of DSBs is also controlled by distant cis and trans interactions, as manifested by the reduction of DSB frequencies at one site in the presence of a strong DSB site nearby (10) or in the presence of heterozygosities between homologs (13, 17, 21).
Taking advantage of the sequencing of the first yeast chromosome (chromosome III, ref. 22), we have determined the distribution of meiotic DSBs along its 340 kb length, at a resolution of 100–500 bp. The identification of a large sample of DSB sites allows us to examine their locations and frequencies at the levels of both the single gene and the whole chromosome.
MATERIALS AND METHODS
Plasmids.
pTy5 contains the YCL075w ORF (gift from D. F. Voytas, Iowa State University, Ames); pJH153 contains HIS4 (gift from M. Lichten, National Institutes of Health, Bethesda); Yep351 contains LEU2; pYJ007 and pYJ006 contain YCR032w and YCR033w, respectively (gift from P. Slonimski, Centre de Génétique Moléculaire, Gif-sur-Yvette, France); pBUD5 and TSM1-L1 contain BUD5 and TSM1, respectively (gift from M. Jacquet, Université Paris-Sud, Orsay, France); a subclone of the PM5239 λ clone contains YCR047c and YCR048w (gift from L. Grivell, Universiteit van Amsterdam); YRP7–12 contains RAD18 (gift from F. Fabre, Institut Curie, Paris); 9189Δ13 contains YCR098c (gift from F. Sor, Institut Curie, Orsay, France). Other ORF-specific probes were generated by PCR and cloned into the pBluescriptKS+ vector.
Yeast Strains and Sporulation.
The strain used in this study is the diploid ORD1181 obtained by mating NKY1000 (Matα, ho::LYS2, lys2, ura3, rad50SKI81.URA3) and NKY1001 (Mata, ho::LYS2, lys2, ura3, rad50SKI81.URA3), obtained from N. Kleckner (Harvard University, Cambridge, MA). These strains are SK1 derivatives. Complementary experiments have been performed with the ORD307 diploid (14), which is a S288C derivative. For sporulation, diploid cells were grown to a concentration of 2–5 × 107 cells/ml in yeast extract/peptone/acetate medium, washed once with water, and incubated at the same cell concentration in 1% potassium acetate at 30°C with vigorous shaking (23).
Analysis of Meiotic DSBs.
Pulsed-field gel electrophoresis shown in Fig. 1A has been made with DNA extracted in agarose plugs according to Bio-Rad recommendations. For high-resolution mapping, DNA from vegetative and meiotic cells was purified with Qiagen (Chatsworth, CA) columns. One microgram of DNA was digested with appropriate restriction enzymes. Standard gel electrophoresis and Southern blot analysis were done as described (14) and probed from the ends of the fragments of interest. The sizes of DSB fragments were measured by comparison with the λ/BstEII size marker and genomic double-digestions, which additionally allowed us to check the accuracy of the expected restriction map. Radioactive counts were quantified with a PhosphorImager (Molecular Dynamics). The frequencies of meiotic DSBs at t = 5 h or t = 6 h and t = 7 h after transfer to 1% potassium acetate medium were calculated from the ratio of individual DSB signals over the sum of parental and all DSB peaks in each lane, corrected by subtraction of the ratio observed for vegetative cells (average of yeast extract/peptone/dextrose grown cells and t = 0 h in 1% potassium acetate medium). These values, only based on standard gel electrophoresis experiments (not pulsed field) are not corrected for the efficiency of sporulation (≈80% in ORD1181). The term “cumulative DSB frequencies” refers to the sum of all DSB frequencies within a given interval.
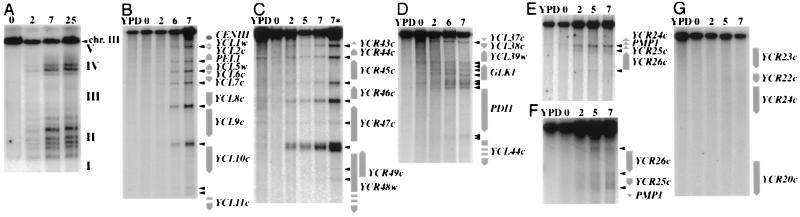
Figure 1.
Experimental detection of meiotic double-strand breaks on chromosome III. (A–G) DNA was extracted from ORD1181 grown either in rich medium (yeast extract/peptone/dextrose, YPD) or after transfer into sporulation medium for various times (0, 2, 5, 6, or 7 h). (A) Detection of chromosome III DSBs by DNA pulsed-field gel electrophoresis and Southern blot analysis and probing with YCL60c (see Fig. 2 for position). I–V represent the five DSB domains: left end DSB-poor (I), left arm DSB-rich (II), internal DSB-poor (III), right arm DSB-rich (IV), and right end DSB-poor (V) domains. Bio-Rad clamped homogeneous electric fields (CHEF) electrophoresis conditions: 180 V, 5- to 30-sec pulses, 38 h, 1.1% agarose gel in 0.5× TBE. (B–G) Detection of chromosome III DSBs by standard electrophoresis and Southern blot analysis; (B) YCL11c-CENIII interval probed with YCL11c (GBP2) (Psp5II); (C) YCR43c-YC48w interval probed with YCR48w (AseI digest). Lanes 7 and 7* correspond to DNA from two independent sporulations; (D) GLK1 region probed with YCL45c (XbaI digest); (E and F) YCR24c-YCR27c interval probed with YCR32w or YCR24c, respectively (EagI); and (G) YCR20c-YCR23c interval probed with YCR19w(PvuII). Vertical arrows indicate genes or ORFs predicted by the DNA sequence (22). Arrowheads indicate the positions of meiotic DSBs.
RESULTS
To examine meiotic DSBs on chromosome III, we have used the efficiently sporulating ORD1181 diploid strain (SK1 background), homozygous for the rad50S mutation (5). This mutation leads to the accumulation of unrepaired meiotic DSBs, which thereby greatly facilitates their mapping and quantification by Southern blot analysis. The locations of DSBs were determined by dividing chromosome III into 80 overlapping restriction fragments (10–20 kb each) and probing them from both ends. This allowed for the precise mapping and quantification of DSBs with respect to the nucleotide sequence (22), taking into account the sequence differences of ORD1181 as described in the legend to Fig. 2. Each segment of chromosome III was examined at least twice, except for the chromosomal end regions that cannot be uniquely probed with telomeric sequences, since these are repeated on other chromosomes (24). Fig. 1 illustrates the experimental detection of meiotic DSBs in representative regions of the chromosome.
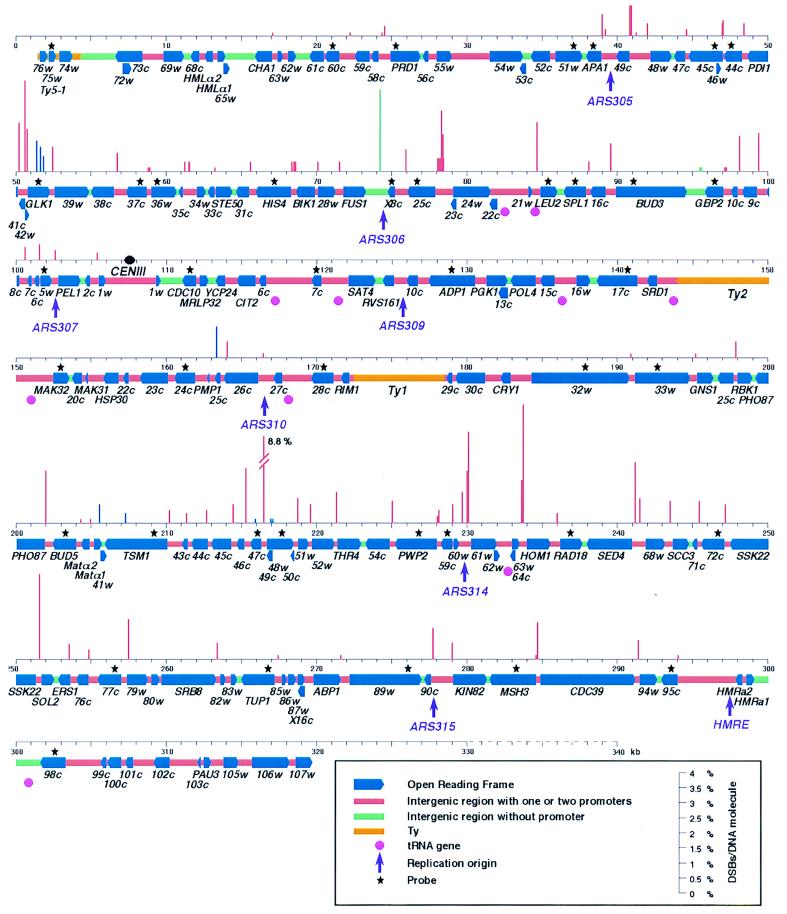
Figure 2.
Location and amount of meiotic DSBs on chromosome III. For simplification, the ORF numbers (22) are indicated without YCL or YCR. The physical map is drawn to scale from the sequence (coordinates are in kb) and is corrected for the major differences found in ORD1181 derived from SK1 that lead chromosome III to a length of 340 kb instead of 315 kb in the published sequence (22): Ty2–17 near LEU2 is absent, as is the delta LTR between YCR6c and YCR7c; a Ty2 identified by PCR amplification and restriction analysis is inserted at coordinate 151,000; a Ty1 is inserted between RIM1 and YCR29c (25); additional 396 bp are in YCR89w; the right chromosomal end is 15–25 kb longer and of unknown sequence. Bar height represents the percent of meiotic DSBs per DNA molecule. Generally, two measurements were made at each region, with different restriction digests and probes. Standard errors between different measurements of the same DSB sites depend on the DSB frequencies: 0.05–0.2% for DSB ≤ 0.5%, 0.2–0.5% for DSB between 0.5% and 1.5%, and 0.5–1% for DSB > 1.5%. Red, green, and blue bars represent DSBs in intergenic-promoter, intergenic terminators, and coding regions, respectively. Other information is provided in the legend box. The probes (ORF name, coordinates on chromosome III sequence) and the various single restriction digests used are as follow: Ty5–1 (2,044–2,881), SmaI, PpuMI; L60c (20,905–21,308), BglII, Psp5II, FspI, NsiI; L57w (25,667–26,104), BamHI, Psp5II, StuI; L51w (37,222–37,603), NruI, DraIII, AvrII; L50c (37,870–38,427), Psp5II; L45c (46,155–46,731), SalI, XbaI; L44c (47,229–47,639), PstI; L37c (58,303–58,728), SalI; L36w (59,350–59,918), XbaI, BamHI; HIS4 (64,862–65,971), SalI, XbaI; HIS4 (66,402–67,581), HpaI; LX8c (74,841–75,300), BspHI; L25c (77,036–77,612), XbaI, Bsu36I; LEU2 (91,646–92,260), BstEII, PstI; SPL1 (93,205–93,771), PstI; BUD3 (96,872–97,621), DraIII; GBP2 (102,136–102,809), HindIII, Psp5II;L5w (107,950–108,386), NdeI, BclI; CDC10 (117,167–117,837), XhoI, EcoRV; R7c (125,703–126,220), Tth111I, XhoI; ADP1 (135,227–135,681), PvuI, BglI; R17c (145,930–146,751), ApaLI, BlpI; MAK32 (159,990–153,448), EcoNI, PvuII; R24c (161,088–161,452), EcoRI, NsiI, ScaI, EagI; FEN2 (170,327–170,967), EagI, XhoI; R32w (179,254–180,256), PstI, SacI, BamHI; R33w (186,230–187,326), XhoI, SacI; BUD5 (196,447–197,353), AatII, PmlI; TSM1 (202,617–203,464), BclI, KpnI; R47c (209,451–210,119), XbaI; ARE1 (210,981–212,598), AseI; CTR86 (217,182–217,837) XbaI; R59c (222,343–222,825), ApaLI, EcoNI; RAD18 (230,349–230,967), SalI, Bsu36I; R72c (239,598–240,466), BamHI, PacI; R77c (250,130–250,608), EcoNI, AatII; TUP1 (259,636–260,424), BamHI, EcoRI; R89w (269,902–270,364), KpnI, DraIII; MSH3 (277,837–278,335), AvaI, Bsu36I; R95c (287,179–287,644), SphI, AvrII, BspEI; R98c (296,513–297,075), SalI, BamHI, HindIII, MluI.
Meiotic DSBs Occur Almost Exclusively in Intergenic Promoter-Containing Intervals.
Along the 340 kb of chromosome III, we have detected and mapped 76 regions where DSBs form, confirming that they occur in multiple regions as indicated by lower resolution studies based on pulsed-field gel electrophoresis (6, 19) and Fig. 1A) and standard gel electrophoresis for three regions of chromosome III and VIII that comprise about 38 kb (8). Fig. 2 shows our high resolution (100–500 bp) mapping of DSB regions with respect both to the physical map of the chromosome and to the distribution of ORFs (22). Strikingly, the vast majority (68 of 76) of these DSB regions is located in intergenic promoter-containing regions. Intergenic regions containing either one promoter/one terminator combination or two divergent promoters are equally subject to DSBs (50% and 56% of them have DSBs, respectively). Both cases are illustrated in Fig. 1B. Only two intergenic convergent-terminator regions, FUS1-YCLX8c and YCL12w-GBP2, have DSBs. In seven cases, DSBs are detected in coding regions: two of these are hypothetical ORFs (YCR25c and YCR41w), three flank the strongest DSB region (one in YCR47c, and two in YCR48w, one of which occurs in the putative promoter region of the overlapping YCR49c ORF, Fig. 1C) and two are in TSM1 and GLK1, where multiple DSBs occur in both coding and 5′ sequences (Fig. 1D). Along chromosome III, among a total of 128 intergenic promoter-containing regions (22) that are expected to be located upstream of a gene transcribed by RNA polymerase II, 53% have DSBs. In contrast, among the 10 tRNA genes present on chromosome III, only the tRNALeu located 5′ of LEU2 is near a detectable DSB region, but our mapping resolution was not sufficient to determine whether these DSBs are upstream or downstream of this tRNA gene (Fig. 2). These results suggest that RNA polymerase III promoters of tRNAs are less prone and even perhaps totally refractory to DSB formation.
DSB Frequency Is Extremely Variable from One Region to Another.
The frequency of DSBs per DNA molecule ranges from 0.2% (the level of detection) to 8.8% per DNA molecule at the previously identified YCR47c-YCR48w hotspot (6, 8). Notably, four novel hotspot regions with a DSB frequency of >5% have been precisely mapped: GLK1 (6%), YCL25c-YCL24w (5%), YCR61w (5.7%), and HOM1 (5.3%) (Fig. 2). Nevertheless, three quarters of DSB regions have a DSB frequency of <1%. There is no obvious relationship between the frequency of DSB formation and the length of the intergenic region, although we observe that the pattern of DSB signals often appears to be more diffuse or to be split into several subregions in the long intergenic intervals.
DSBs Are Clustered in Chromosomal Domains.
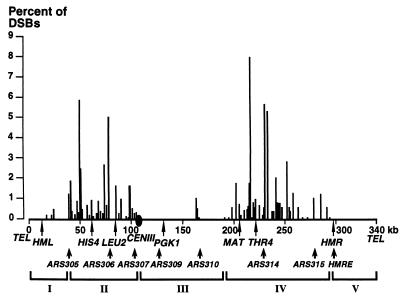
The distribution of DSBs along chromosome III is heterogeneous and defines five chromosomal domains (Figs. 1A and 3; domains I–V). Indeed, 70 of 76 DSB regions are clustered in two main regions, one on each arm. These domains are 68 kb (domain II: coordinates 39,000–107,000) and 105 kb long (domain IV: coordinates 190,000–295,000) with DSBs occurring in almost every intergenic promoter-containing interval (29/32 and 36/43 in the left and right arm clusters, respectively). The cumulative DSB frequencies per molecule in domains II and IV are 0.32 and 0.44, respectively. In contrast, the regions near the chromosomal ends contain few and only weakly detectable DSBs (domains I and V, coordinates 1–39,000 and 295,000–340,000, respectively), as does the 84-kb region between CENIII and YCR32w (domain III: coordinates 107,000–190,000) (Fig. 1 A, E–G). In this latter cold domain, the occurrence of DSBs at YCR25c represents a notable exception (Fig. 1 E–F) but its frequency of 1.5% over 800 bp remains relatively low. The clustering of DSBs in subregions of the chromosome suggests that DSB formation is not only controlled by local intergenic-promoter elements but also by large-scale chromosomal features.
Figure 3.
Chromosomal organization of DSB regions and other chromosomal features. Summary map of the location and amount of DSBs along the chromosome III. The height of the vertical bars corresponds to the sum of the frequencies of DSBs per intergenic or coding region. The arrows indicate the location of well known genes and functional replication origins (ARS and HMRE) (refs. 25 and 26; C. Newlon, personal communication).
The cold domain III contains two transposable elements in the strain ORD1181 (a Ty2 and a Ty1; see Fig. 2). To determine whether the absence of DSBs in this domain is related to the presence of Ty, we have mapped the distribution of DSBs in the strain ORD307 (derived from S288C background), which differs from ORD1181 by the presence of a Ty2 (Ty2–17) upstream of LEU2 and the absence of Ty2 near coordinate 150,000 (22). The two strains show a similar DSB distribution in these regions, with DSBs upstream of YCL25c (6 kb upstream of the Ty2–17 locus) and LEU2 (immediately downstream of the Ty2–17 locus), but no DSBs around coordinate 150,000 (data not shown). Thus the presence of Ty does not seem to correlate with the absence of DSBs in regions in which they are inserted (see Discussion).
DISCUSSION
The main results of this report are: (i) DSBs occur during prophase I of meiosis in 76 regions along chromosome III; (ii) most of the DSB regions (89%) are localized in intergenic promoter-containing intervals; (iii) DSB regions are strongly clustered in two domains, one per arm, where almost all intergenic promoter-containing intervals have DSBs with variable frequencies (0.2–8.8% per molecule); in contrast, DSBs are rare near the chromosomal ends and in the internal region between CENIII and CRY1; and (iv) the cumulative frequency of DSBs for the whole chromosome is at least 0.81. The implications of these findings for the local control of DSB formation and how the distribution of DSBs relates to overall chromosomal structure are discussed below.
Local Control of DSB Formation.
In agreement with information obtained from previously studied recombination hotspots (1) and from the DSB regions mapped by Wu and Lichten (8) over 30 kb of chromosome III, the present data demonstrate that almost all DSBs along chromosome III occur in promoter-containing intergenic intervals. What is the reason for this specificity? So far, no consensus sequences have been yet identified by DNA sequence comparison of intergenic regions proficient for DSB formation. One common feature of DSB regions is their preferential chromatin accessibility. Indeed, all natural and artificial DSB regions studied are located in regions of chromatin that are hypersensitive to DNaseI and micrococcal nuclease (8, 11–13). Thus, DSBs may occur where nucleosomes are absent or disrupted and the underlying DNA is exposed to the DSB-forming nuclease activity. Additionally, experiments with micrococcal nuclease have revealed an increase of accessibility of the DSB regions early in meiosis (11) but the factors that change the meiotic state of chromatin are not known. The deletion of cis-acting transcriptional elements of the ARG4 and HIS4 genes has been found to affect the amount and location of DSBs (14, 27, 28), but the relationship among transcriptional activity, DSB formation, and chromatin accessibility remains to be clarified (reviewed in ref. 1).
The rare cases when DSBs occur in non intergenic-promoter regions may also be explained in terms of chromatin accessibility, either due to the presence of an unidentified promoter (or promoter-like elements) or to the formation of a promoter-independent open chromatin structure. Indeed, DSBs in the YCR48w coding region might reflect the existence of an internal promoter, since there is a putative ORF (YCR49c) encoded on the opposite strand. Multiple DSBs also occur in the coding sequence of the GLK1 gene, which has a high transcript level in both vegetative and meiotic cells (data not shown). At the ARG4 locus, however, DSB formation is inhibited by overlapping transcription (29). The cases of DSBs occurring in the coding regions of GLK1 and HIS2 genes (9, 20) show that DSB inhibition by transcription through a gene can be overcome. Still to be elucidated is whether the formation of DSBs within coding regions is controlled by internal specific sequences or by distant cis-acting elements. Additionally, the intergenic terminator-convergent region between FUS1 and YCLX8c, where DSBs occur at a relatively high frequency (2.7%), contains an origin of replication (ARS306). Origins of replication are bound by the origin recognition complex proteins and have an easily unwound domain that is required for their function (reviewed in ref. 30). Thus, the presence of ARS306 might create an accessible chromatin structure competent to form DSBs. The occurrence of DSBs in other replication origins that are in DSB-rich domains does not contradict this hypothesis, but may be explained solely by their location in promoter-containing intervals.
Why Are DSBs Clustered in Chromosomal Domains?
One hypothesis is that the clustering of DSB regions may simply reflect a serendipitous arrangement of DSB-proficient genes, each individually controlled. In this direction, we have examined the relationships between overall gene distribution or transcription patterns and DSB domains. All domains contain numerous genes (Fig. 2) and therefore the absence of DSBs in the cold domains is not due to an unusually low gene density. The levels of chromosome III transcripts have been studied systematically in vegetative cells and ranked from level 1–5 (most expressed) (31). We observe that the transcript levels in both telomeric domains (I and V) are biased toward the low levels (rank 1–2) but the internal domains with DSBs (II, IV) and without DSBs (III) have a similar pattern (ranking 2–4). Therefore, unless the global gene transcription pattern in meiosis is region-specific and not regulated at the single gene level as in vegetative cells, it is unlikely that the DSB clustering reflects major differences in transcriptional activities.
Alternatively, the clustering of DSBs may be caused by a novel type of long distance control that is superimposed on individual gene controls. A strong argument in favor of a large scale control is the general observation that the displacement of DNA fragments potentially able to form DSBs from one location to another in the genome is frequently associated with a change in their behavior (10). Interestingly, on chromosome III, the level of DSBs in a fragment containing the ARG4 gene and pBR322 sequences is maximal upon insertion at HIS4 and LEU2 (in DSB-rich domain II), intermediate at MAT (at the flank of DSB-rich domain IV) and minimal at CHA1 (in DSB-poor domain I), in good correlation with the hot and cold chromosomal domains we have identified here. Such a control may be related to the DNA replication process, because DSBs occur after premeiotic DNA replication (23); it may also involve a high order arrangement of chromatin structure; finally, it may be mediated by the overall chromosome structure, which could be controlled by either some cis-acting short sequences or long-range variations of the primary DNA sequence along the chromosome. These different proposals are discussed below.
Origins of replication and centromeres are known to overlap nuclear scaffold attachment sites, which are involved in higher-order organization of the chromatin fiber, defining units of chromosome compaction (32). CENIII is also a replication fork pause site in both vegetative and meiotic S phases (25). These elements that participate in the architecture of the chromosome might determine the structural domains. To test this view, we have compared the DSB domains to the distribution of chromosome III replication origins, which are all known and used during both vegetative growth and meiosis (refs. 25 and 26; C. Newlon, personal communication). It is noteworthy that there are replication origins at three of the edges of the two DSB-rich domains (ARS305, ARS307, and HMRE), and that CENIII is, near ARS307, at the right limit of domain II. In support of a role for the replication origins, it has been shown that ARS307 enhances both gene conversions and crossovers (33). The presence of DSBs near ARS310 in domain III might reflect a similar situation (Fig. 1 A, E, and F and Fig. 2). Nevertheless, even if replication origins participate in the DSB domain control, they do not dictate their establishment, as indicated by the presence of replication origins in both the DSB-rich domains II and IV (ARS306, ARS314, and ARS315), at their flanks (ARS305, ARS307, and HMRE) and in the DSB-poor internal domain III (ARS309 and ARS310). The late replication of yeast telomere regions observed in vegetative cells, including chromosome III (34), might contribute, if it is also the case for premeiotic replication, to inhibit DSB formation in these regions that are free of DSBs over at least 25 kb (20).
One might also envisage that higher-order arrangement of chromatin structure, independent of transcription or DNA replication, may lead to activation or repression for DNA accessibility. A well characterized example of a chromosomal domain effect in S. cerevisiae is transcriptional silencing in the vicinity of telomeres. The machinery responsible for this phenomenon also prevents both transcription and HO endonuclease cleavage of the HML and HMR silent mating-type cassettes on chromosome III (reviewed in ref. 35). These silencing effects are thought to be mediated by the formation of a “heterochromatin-like” structure, involving numerous proteins including Sir3, Sir4, and Rap1 that localize in the spaces where telomeres are clustered, near the nuclear membrane (35–37). To test whether this silencing mechanism is involved in DSB domain formation, we examined the distribution of meiotic DSBs in a sir4 mutant. As in the wild-type, we observed no DSBs in the left telomere region (data not shown), indicating that the low frequency of DSBs in this region is not due to the spreading in meiosis of this SIR silencing mechanism. It can be noted that we have never detected DSBs within or immediately upstream of a Ty sequence, that may be related to their poor ability to recombine (38). By comparing two strains differing with respect to Ty insertions, we found that the presence of Ty does not change the DSB distribution in the region where it is inserted, suggesting that whatever the mechanism preventing DSB formation at Ty promoters, it does not affect adjacent sequences.
Another interesting example of a chromosomal domain control concerns the choice of the donor locus on chromosome III during mating-type switching (HMLα or HMRa), which is strongly position-dependent (39). In MATa cells, a donor (i.e., mating-type copy) located on the left arm of chromosome III is highly preferred. This preference depends on the presence of a 700-bp recombinational enhancer (39). In cells where α2 is expressed (MATα cells) or in which the recombinational enhancer is lacking, the left part of chromosome III is specifically repressed and does not act as a donor in mating-type switching. How this donor preference is related to chromosomal architecture and dynamics is not known, but it clearly demonstrates that a short cis-acting sequence can act to control chromosomal interactions over long distances.
Finally, we may also consider features of the DNA primary sequence, of which variations along chromosomes are suspected to reflect some aspects of chromosome organization. Computer analysis of the GC content along yeast chromosome XI shows a heterogeneous distribution with peaks separated from each other by roughly 90–100 kb (40). On chromosome III, there are two GC-rich regions located within coordinates 40,000–80,000 and 205,000–265,000, which broadly coincide with the DSB-rich domains (41), suggesting a general relationship that remains to be interpreted.
Do DSBs Initiate All Recombination Events?
Previous studies of a few meiotic recombinational hot-spots in yeast (1) and of the 20 kb chromosome III region between LEU2 and CEN3 (8) suggested that meiotic DSBs initiate a large fraction of both noncrossover (gene conversion) and crossover events. The precise determination of the DSB distribution along chromosome III allows for an examination of the relationship between DSB formation and recombination over the whole chromosome.
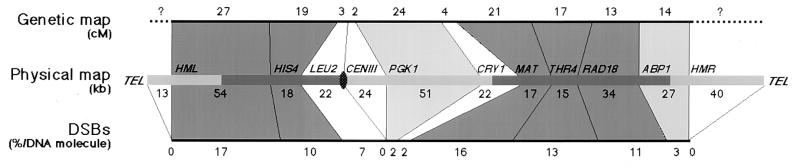
The first question is the relationship between the amount of DSBs and the genetic distances. For comparison, Fig. 4 shows chromosome III subdivided into 12 intervals between the HML and HMR loci (280 kb), and indicates genetic distances measured in cM (42), physical distances (kb) (22), and the cumulative frequencies of DSBs per interval (data from Fig. 2). In yeast as in other organisms, the amount of cM/kb, which reflects the density of crossovers per unit of physical distance, is extremely variable from one chromosomal region to another. Along chromosome III, it varies >10-fold, from 1.24 cM/kb in the hot MAT-THR4 interval to 0.08 cM/kb in the extremely cold CENIII-PGK1 interval. Therefore, taking into account the ratio of cM/kb and DSB frequencies, chromosome III seems to reflect three types of situations. (i) There are two hot or average regions (Fig. 4, ▪), one per arm, corresponding to the HML-LEU2 and MAT-ABP1 intervals with an average of 0.64 and 0.77 cM/kb respectively, correlating well with a high level of DSB formation (0.40 and 0.61% DSB/kb, respectively), which strongly suggests that most if not all recombination events in these regions are initiated by DSB lesions. (ii) This situation corresponds to cold regions (Fig. 4, □), where recombination is rare due to infrequent DSB formation (LEU2-PGK1 and CRY1-MAT intervals). (iii) Exemplified by the PGK1-CRY1 interval and to a lesser extent the ABP1-HMR interval (Fig. 4, ░⃞), DSBs are rare but recombination occurs substantially. Several hypotheses may explain this unexpected last situation: (i) an overestimate of the genetic distances that are taken from the published map compiled from various strains (42) but not measured in the SK1 strain used for this DSB study—differences between strains are known to occur as reported here for the physical distance (see legend for Fig. 2); (ii) an underestimation of the frequencies of DSBs if low levels of hardly detectable DSBs occur all along the chromosome with or without preference for the promoter regions; and (iii) finally, the possibility that some crossovers might be initiated by other potentially recombinogenic lesions, such as single-strand DNA nicks or gaps, although nicks have not been detected in the vicinity of several DSB regions (15–17). Further studies will try to discriminate between these possibilities. First, the genetic distances have to be examined in the SK1 strain to accurately correlate DSB and crossover frequencies along the whole chromosome.
Figure 4.
Comparisons between the genetic and physical distances and the amount of meiotic DSBs for 12 intervals of chromosome III. Genetic distances are from Mortimer et al. (42), and physical distances are as in Fig. 2. The percent of DSBs was calculated as the sum of all DSBs in each interval. Domains with and without DSBs are represented on the physical map by ▪ and ░⃞, respectively. Differently shaded areas represent regions of chromosome III where frequencies of crossovers and DSBs are correlated at high (▪) or low (□) levels, or poorly correlated (░⃞).
The second question concerns the frequency of DSBs per chromosome per meiosis because it is assumed that the occurrence of at least one crossover per pair of homologous chromosomes is necessary to ensure their proper segregation at the reductional division of meiosis (2). Our estimate of the cumulative frequency of DSBs for chromosome III as a whole is 0.81 DSB per molecule. As four chromatids are present during prophase I of meiosis, and assuming that half of the DSB repair events are associated with an adjacent crossover, as found for recombinational hot-spots (43), our minimal estimate of 1.62 crossovers (0.81 × 4/2) is sufficient to ensure at least one crossover per meiosis along chromosome III.
It is generally recognized that among organisms, the total amount of recombination does not vary in proportion to the genome size. For example, the yeast genome, which is 13.5 Mb long, has a total of ≈4,500 cM (42) and contains ≈6,000 genes (44), whereas the human and mouse genomes are estimated to be 3,300 Mb long (250-fold the yeast genome size), have 3300 cM but may contain about 70,000–100,000 genes (15-fold the yeast gene number) (45). Altogether, the variations of gene numbers and the existence of two controls, at the gene and at the chromosomal levels, provide modulations that can contribute to the variation of recombination features between different regions of a chromosome, within a genome, sex-specific differences (46) and between organisms. The present data suggests that one factor that might modulate the amount of recombination within a genome is the number of genes (promoters), which presumably varies less among organisms than does genome size.
Acknowledgments
We thank F. Fabre, L. Grivell, M. Jacquet, M. Lichten, P. Slonimski, F. Sor, and D. Voytas for providing chromosome III clones; N. Kleckner for providing the yeast strain; and C. Newlon for communication of unpublished data. We thank C. Esnaut and M. Duguet for their contribution during the earliest development of this project; all members of the A. Nicolas laboratory, F. Fabre, G. Simchen, and P. Slonimski for discussions and comments on the manuscript; K. Smith for English correction. This work was successively performed in the Centre National de la Recherche Scientifique, Unité de Recherche Associée 1354 and 1292, Unité Mixte de Recherche 144, and supported by grants from the Groupement de Recherches et d’Etudes sur les Génomes, the Association de Recherche Contre le Cancer, the Ligue Nationale contre le Cancer and the Actions Concertées Coordonnées SV8 program from the Ministère de l’Education Nationale, de l’Enseignement Supérieur et de la Recherche. F.B. was supported by graduate student fellowships from the Ministère de l’Education Nationale, de l’Enseignement Supérieur et de la Recherche and from the Association pour la Recherche sur le Cancer.
ABBREVIATION
- DSB
double-strand break
References
- 1.Lichten M, Goldman A S H. Annu Rev Genet. 1995;29:423–444. doi: 10.1146/annurev.ge.29.120195.002231. [DOI] [PubMed] [Google Scholar]
- 2.Roeder G S. Proc Natl Acad Sci USA. 1995;92:10450–10456. doi: 10.1073/pnas.92.23.10450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kleckner N. Proc Natl Acad Sci USA. 1996;93:8167–8174. doi: 10.1073/pnas.93.16.8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun H, Treco D, Szostak J. Nature (London) 1989;338:87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- 5.Alani E, Padmore R, Kleckner N. Cell. 1990;61:419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- 6.Zenvirth D, Arbel T, Sherman A, Goldway M, Klein S, Simchen G. EMBO J. 1992;11:3441–3447. doi: 10.1002/j.1460-2075.1992.tb05423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nag D K, Petes T D. Mol Cell Biol. 1993;13:2324–2331. doi: 10.1128/mcb.13.4.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu T-C, Lichten M. Science. 1994;263:515–518. doi: 10.1126/science.8290959. [DOI] [PubMed] [Google Scholar]
- 9.Bullard S A, Kim S, Galbraith A M, Malone R E. Proc Natl Acad Sci USA. 1996;93:13054–13059. doi: 10.1073/pnas.93.23.13054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu T-C, Lichten M. Genetics. 1995;140:55–66. doi: 10.1093/genetics/140.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohta K, Shibata T, Nicolas A. EMBO J. 1994;13:5754–5763. doi: 10.1002/j.1460-2075.1994.tb06913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan Q-Q, Petes T D. Mol Cell Biol. 1996;16:2037–2043. doi: 10.1128/mcb.16.5.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keeney S, Kleckner N. Genes Cells. 1996;1:475–489. doi: 10.1046/j.1365-2443.1996.d01-257.x. [DOI] [PubMed] [Google Scholar]
- 14.de Massy B, Nicolas A. EMBO J. 1993;12:1459–1466. doi: 10.1002/j.1460-2075.1993.tb05789.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Massy B, Rocco V, Nicolas A. EMBO J. 1995;14:4589–4598. doi: 10.1002/j.1460-2075.1995.tb00138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J, Wu T C, Lichten M. EMBO J. 1995;14:4599–4608. doi: 10.1002/j.1460-2075.1995.tb00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu L, Kleckner N. EMBO J. 1995;14:5115–5128. doi: 10.1002/j.1460-2075.1995.tb00194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu F, Petes T D. Genetics. 1996;143:1115–1125. doi: 10.1093/genetics/143.3.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Game J C. Dev Genet. 1992;13:485–497. doi: 10.1002/dvg.1020130610. [DOI] [PubMed] [Google Scholar]
- 20.Klein S, Zenvirth D, Dror V, Barton A B, Kaback D B, Simchen G. Chromosoma. 1996;105:276–284. doi: 10.1007/BF02524645. [DOI] [PubMed] [Google Scholar]
- 21.Rocco V, Nicolas A. Genes Cells. 1996;1:645–661. doi: 10.1046/j.1365-2443.1996.00256.x. [DOI] [PubMed] [Google Scholar]
- 22.Oliver S G, van der Aart Q J, Agostoni-Carbone M L, Aigle M, Alberghina L, et al. Nature (London) 1992;357:38–46. doi: 10.1038/357038a0. [DOI] [PubMed] [Google Scholar]
- 23.Padmore R, Cao L, Kleckner N. Cell. 1991;66:1239–1256. doi: 10.1016/0092-8674(91)90046-2. [DOI] [PubMed] [Google Scholar]
- 24.Louis E J, Naumova E S, Lee A, Naumov G, Haber J E. Genetics. 1994;136:789–802. doi: 10.1093/genetics/136.3.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collins I, Newlon C S. Mol Cell Biol. 1994;14:3524–3534. doi: 10.1128/mcb.14.5.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newlon C S, Collins I, Dershowitz A, Deshpande A M, Greenfeder S A, Ong L Y, Theis J F. Cold Spring Harbor Symp Quant Biol. 1993;58:415–423. doi: 10.1101/sqb.1993.058.01.048. [DOI] [PubMed] [Google Scholar]
- 27.Schultes N P, Szostak J W. Mol Cell Biol. 1991;11:322–328. doi: 10.1128/mcb.11.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White M A, Dominska M, Petes T D. Proc Natl Acad Sci USA. 1993;90:6621–6625. doi: 10.1073/pnas.90.14.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rocco V, de Massy B, Nicolas A. Proc Natl Acad Sci USA. 1992;89:12068–12072. doi: 10.1073/pnas.89.24.12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donovan S, Diffley J F X. Curr Opin Genet Dev. 1996;6:203–207. doi: 10.1016/s0959-437x(96)80051-7. [DOI] [PubMed] [Google Scholar]
- 31.Yoshikawa A, Isono K. Yeast. 1990;6:383–401. doi: 10.1002/yea.320060504. [DOI] [PubMed] [Google Scholar]
- 32.Amati B B, Gasser S M. Cell. 1988;54:967–978. doi: 10.1016/0092-8674(88)90111-0. [DOI] [PubMed] [Google Scholar]
- 33.Rattray A J, Symington L S. Genetics. 1993;134:175–188. doi: 10.1093/genetics/134.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reynolds A E, McCarroll R M, Newlon C S, Fangman W L. Mol Cell Biol. 1989;9:4488–4494. doi: 10.1128/mcb.9.10.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laurenson P, Rine J. Microbiol Rev. 1992;56:543–560. doi: 10.1128/mr.56.4.543-560.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gotta M, Laroche T, Formenton A, Maillet L, Scherthan H, Gasser S M. J Cell Biol. 1996;134:1349–1363. doi: 10.1083/jcb.134.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maillet L, Boscheron C, Gotta M, Marcand S, Gilson E, Gasser S M. Genes Dev. 1996;10:1796–1811. doi: 10.1101/gad.10.14.1796. [DOI] [PubMed] [Google Scholar]
- 38.Boeke J D, Sandmeyer S B. In: Yeast Transposable Elements. Broach J R, Pringle J R, Jones E W, editors. Vol. 1. Plainview, NY: Cold Spring Harbor Lab. Press; 1991. pp. 193–261. [Google Scholar]
- 39.Wu X, Haber J E. Cell. 1996;87:277–285. doi: 10.1016/s0092-8674(00)81345-8. [DOI] [PubMed] [Google Scholar]
- 40.Dujon B, Alexandri D, Andre B, Ansorge W, Baladron W, et al. Nature (London) 1994;369:371–378. [Google Scholar]
- 41.Sharp P M, Lloyd A T. Nucleic Acids Res. 1993;21:179–183. doi: 10.1093/nar/21.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mortimer R K, Contopoulou R, King J S. Yeast. 1992;8:817–902. doi: 10.1002/yea.320081002. [DOI] [PubMed] [Google Scholar]
- 43.Fogel S, Mortimer R K, Lusnak K. In: The Molecular Biology of the Yeast Saccharomyces cerevisiae. Strathern J N, Jones E W, Broach J R, editors. Vol. 1. Plainview, NY: Cold Spring Harbor Lab. Press; 1981. pp. 289–339. [Google Scholar]
- 44.Goffeau A, Barrell B G, Bussey H, Davis R W, Dujon B, Feldman H, Galibert F, Hoheisel J D, Jacq C, Johnston M, Louis E J, Mewes H W, Murakami Y, Philippsen P, Tettelin H, Oliver S G. Science. 1996;274:546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 45.Miklos G L G, Rubin G M. Cell. 1996;86:521–529. doi: 10.1016/s0092-8674(00)80126-9. [DOI] [PubMed] [Google Scholar]
- 46.Thomas B J, Rothstein R. Cell. 1991;64:1–3. doi: 10.1016/0092-8674(91)90199-9. [DOI] [PubMed] [Google Scholar]