Abstract
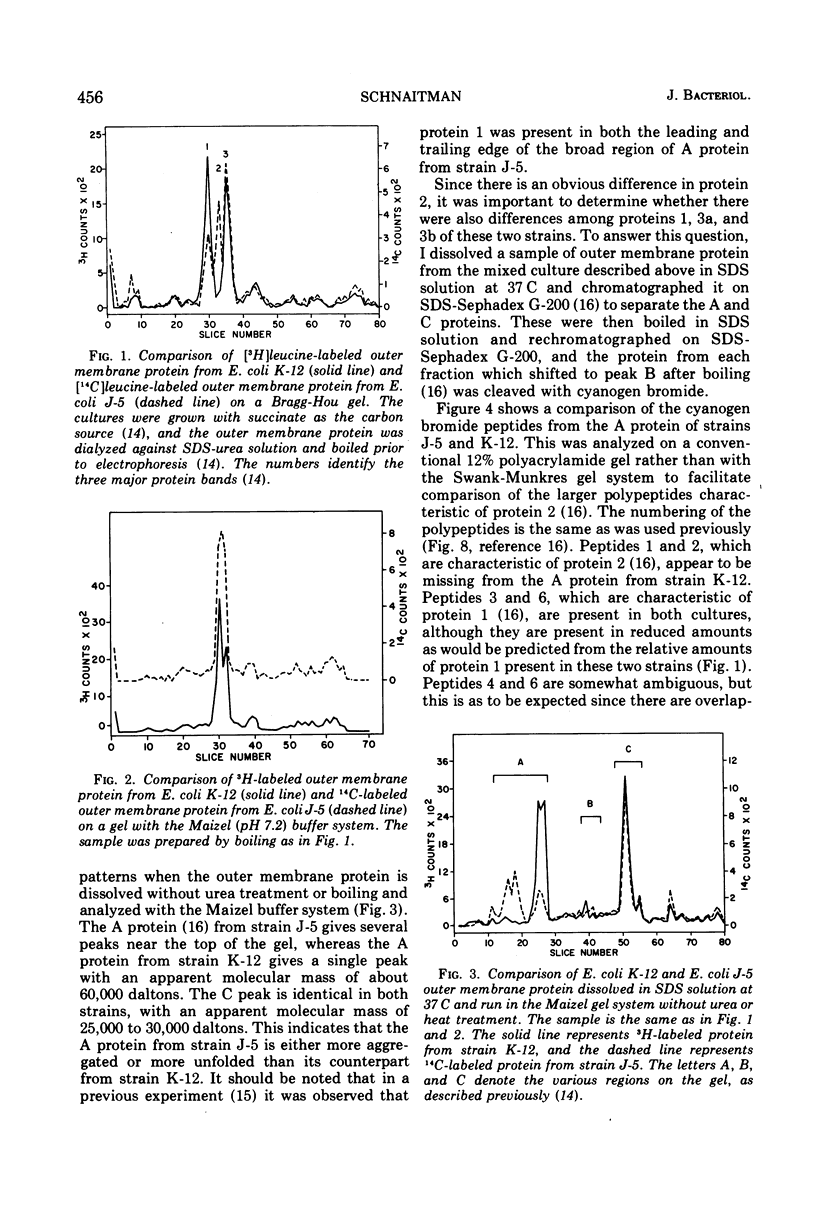
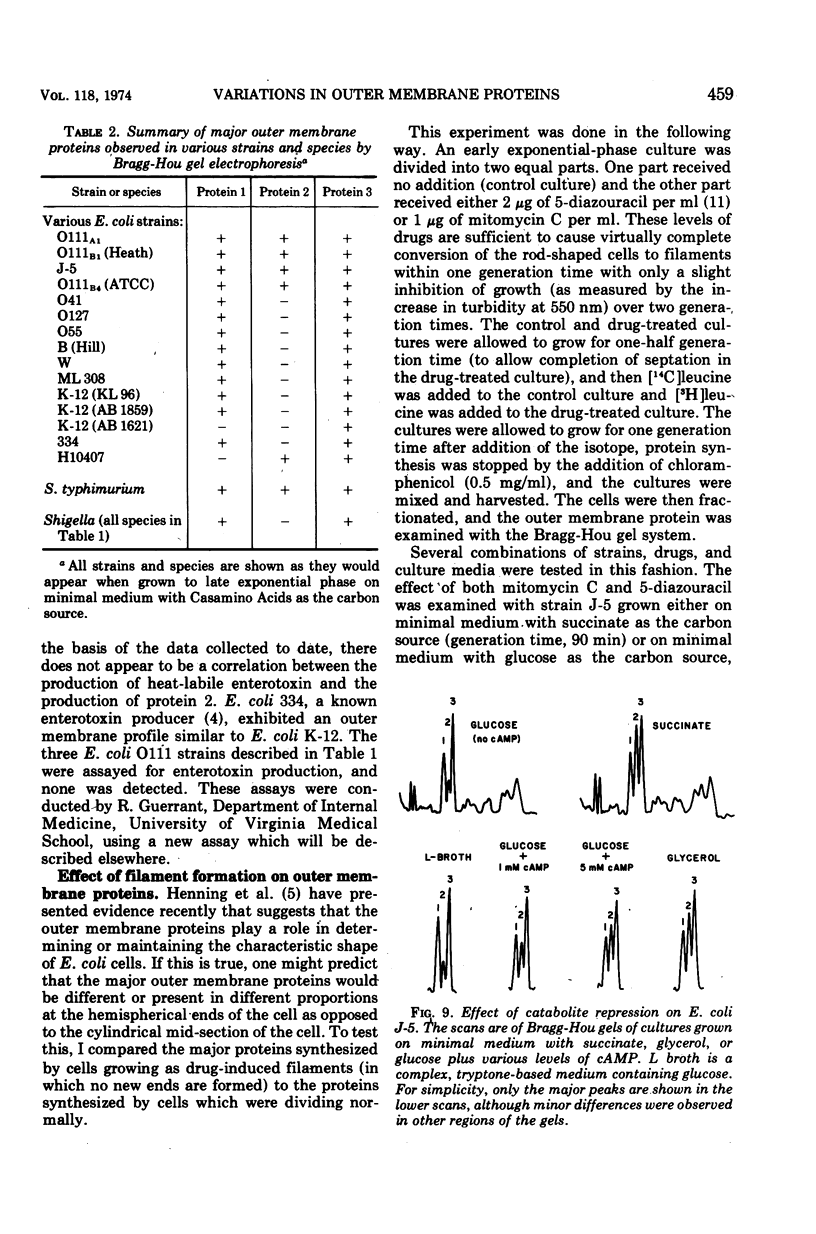
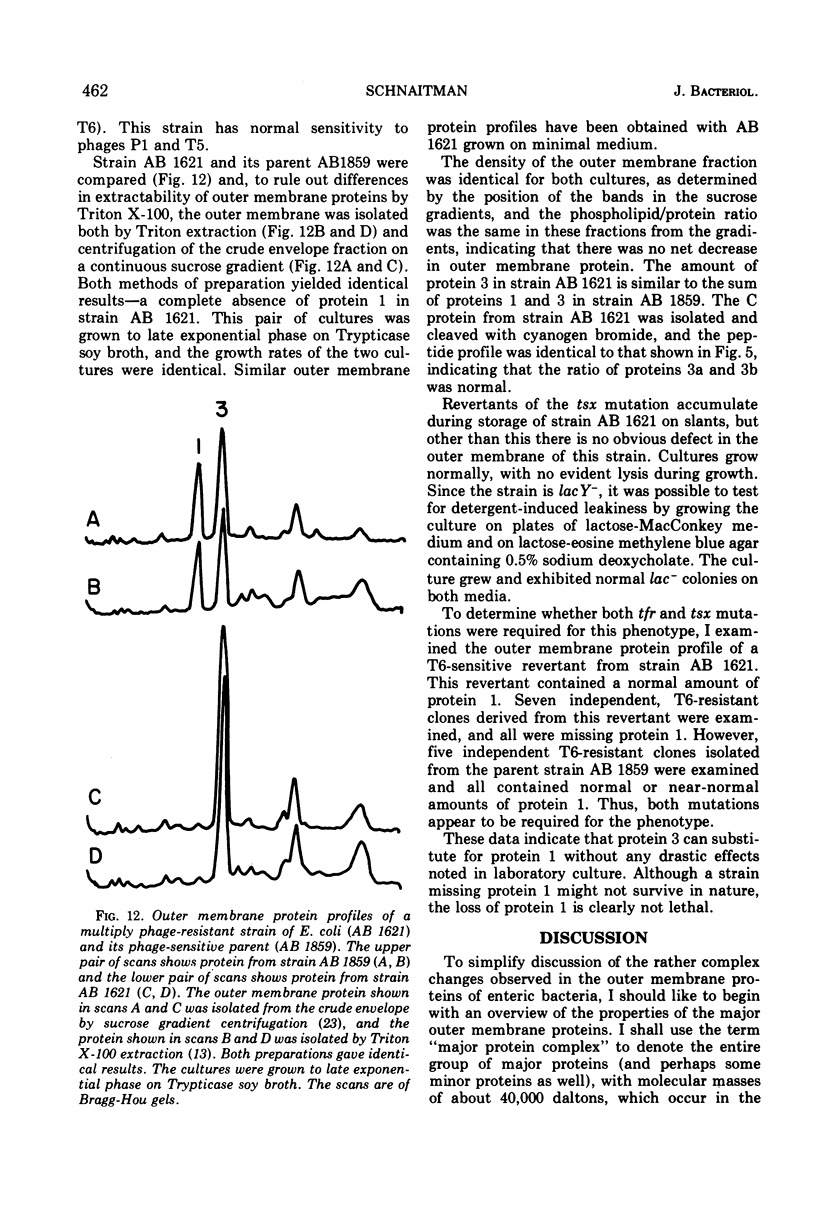
When the 42,000-dalton major outer membrane protein of Escherichia coli O111 is examined on alkaline polyacrylamide gels containing sodium dodecyl sulfate, it is resolved into three distinct bands designated as proteins 1, 2, and 3. Band 3 consists of two distinct polypeptides, proteins 3a and 3b. E. coli K-12 does not make any protein 2, but makes proteins similar to 1, 3a, and 3b as indicated by comparison of cyanogen bromide peptide patterns. Several Shigella species and most other strains of E. coli resemble E. coli K-12 in that they lack protein 2, whereas Salmonella typhimurium is more similar to E. coli O111. In addition to these species and strain differences, cultural differences resulted in differences in the outer membrane protein profiles. Under conditions of catabolite repression, the level of protein 2 in E. coli O111 decreased while the level of protein 1 increased. An enterotoxin-producing strain similar to E. coli O111 produced no protein 1 and an elevated level of protein 2 under conditions of low catabolite repression. The levels of proteins 1 and 3 are also different in different phases of the growth curve, with protein 1 being the major species in the exponential-phase cells and protein 3 being the major species in stationary-phase cells. A multiply phage-resistant mutant of E. coli K-12 with no obvious cell wall defects produced no protein 1 or 2, but made increased amounts of protein 3. Thus, the major outer membrane proteins of E. coli and related species may vary considerably without affecting outer membrane integrity.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ames G. F., Spudich E. N., Nikaido H. Protein composition of the outer membrane of Salmonella typhimurium: effect of lipopolysaccharide mutations. J Bacteriol. 1974 Feb;117(2):406–416. doi: 10.1128/jb.117.2.406-416.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragg P. D., Hou C. Organization of proteins in the native and reformed outer membrane of Escherichia coli. Biochim Biophys Acta. 1972 Aug 9;274(2):478–488. doi: 10.1016/0005-2736(72)90193-9. [DOI] [PubMed] [Google Scholar]
- Guerrant R. L., Ganguly U., Casper A. G., Moore E. J., Pierce N. F., Carpenter C. C. Effect of Escherichia coli on fluid transport across canine small bowel. Mechanism and time-course with enterotoxin and whole bacterial cells. J Clin Invest. 1973 Jul;52(7):1707–1714. doi: 10.1172/JCI107352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henning U., Rehn K., Hoehn B. Cell envelope and shape of Escherichia coli K12. Proc Natl Acad Sci U S A. 1973 Jul;70(7):2033–2036. doi: 10.1073/pnas.70.7.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye M., Guthrie J. P. A mutation which changes a membrane protein of E. coli. Proc Natl Acad Sci U S A. 1969 Nov;64(3):957–961. doi: 10.1073/pnas.64.3.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye M., Pardee A. B. Changes of membrane proteins and their relation to deoxyribonucleic acid synthesis and cell division of Escherichia coli. J Biol Chem. 1970 Nov 10;245(21):5813–5819. [PubMed] [Google Scholar]
- Koplow J., Goldfine H. Alterations in the outer membrane of the cell envelope of heptose-deficient mutants of Escherichia coli. J Bacteriol. 1974 Feb;117(2):527–543. doi: 10.1128/jb.117.2.527-543.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizel J. V., Jr Acrylamide-gel electrophorograms by mechanical fractionation: radioactive adenovirus proteins. Science. 1966 Feb 25;151(3713):988–990. doi: 10.1126/science.151.3713.988. [DOI] [PubMed] [Google Scholar]
- Moldow C., Robertson J., Rothfield L. Purification of bacterial membrane proteins. The use of guanidinium thiocyanate and urea. J Membr Biol. 1972;10(2):137–152. doi: 10.1007/BF01867850. [DOI] [PubMed] [Google Scholar]
- Previc E., Richardson S. Growth-physiological changes in Escherichia coli and other bacteria during division inhibition by 5-diazouracil. J Bacteriol. 1969 Jan;97(1):416–425. doi: 10.1128/jb.97.1.416-425.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaitman C. A. Comparison of the envelope protein compositions of several gram-negative bacteria. J Bacteriol. 1970 Dec;104(3):1404–1405. doi: 10.1128/jb.104.3.1404-1405.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaitman C. A. Outer membrane proteins of Escherichia coli. 3. Evidence that the major protein of Escherichia coli O111 outer membrane consists of four distinct polypeptide species. J Bacteriol. 1974 May;118(2):442–453. doi: 10.1128/jb.118.2.442-453.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaitman C. A. Outer membrane proteins of Escherichia coli. I. Effect of preparative conditions on the migration of protein in polyacrylamide gels. Arch Biochem Biophys. 1973 Aug;157(2):541–552. doi: 10.1016/0003-9861(73)90673-5. [DOI] [PubMed] [Google Scholar]
- Schnaitman C. A. Outer membrane proteins of Escherichia coli. II. Heterogeneity of major outer membrane polypeptides. Arch Biochem Biophys. 1973 Aug;157(2):553–560. doi: 10.1016/0003-9861(73)90674-7. [DOI] [PubMed] [Google Scholar]
- Schnaitman C. A. Solubilization of the cytoplasmic membrane of Escherichia coli by Triton X-100. J Bacteriol. 1971 Oct;108(1):545–552. doi: 10.1128/jb.108.1.545-552.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaitman C., Greenawalt J. W. Intracytoplasmic membranes in Escherichia coli. J Bacteriol. 1966 Sep;92(3):780–783. doi: 10.1128/jb.92.3.780-783.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro B. M., Siccardi A. G., Hirota Y., Jacob F. On the process of cellular division in Escherichia coli. Membrane prtein alterations associated with mutations affecting the initiation of DNA synthesis. J Mol Biol. 1970 Aug 28;52(1):75–89. doi: 10.1016/0022-2836(70)90178-6. [DOI] [PubMed] [Google Scholar]
- Siccardi A. G., Shapiro B. M. On the process of cellular division in Escherichia coli. IV. Altered protein composition and turnover of the membranes of thermosensitive mutants defective in chromosomal replication. J Mol Biol. 1971 Mar 28;56(3):475–490. doi: 10.1016/0022-2836(71)90395-0. [DOI] [PubMed] [Google Scholar]
- Skerman F. J., Formal S. B., Falkow S. Plasmid-associated enterotoxin production in a strain of Escherichia coli isolated from humans. Infect Immun. 1972 Apr;5(4):622–624. doi: 10.1128/iai.5.4.622-624.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swank R. T., Munkres K. D. Molecular weight analysis of oligopeptides by electrophoresis in polyacrylamide gel with sodium dodecyl sulfate. Anal Biochem. 1971 Feb;39(2):462–477. doi: 10.1016/0003-2697(71)90436-2. [DOI] [PubMed] [Google Scholar]
- Taylor A. L., Trotter C. D. Linkage map of Escherichia coli strain K-12. Bacteriol Rev. 1972 Dec;36(4):504–524. doi: 10.1128/br.36.4.504-524.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White D. A., Lennarz W. J., Schnaitman C. A. Distribution of lipids in the wall and cytoplasmic membrane subfractions of the cell envelope of Escherichia coli. J Bacteriol. 1972 Feb;109(2):686–690. doi: 10.1128/jb.109.2.686-690.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. C. Isolation and characterization of an Escherichia coli mutant with alteration in the outer membrane porteins of the cell envelope. Biochim Biophys Acta. 1972 Dec 1;290(1):274–289. doi: 10.1016/0005-2736(72)90070-3. [DOI] [PubMed] [Google Scholar]


