Abstract
The signaling pathways that allow plants to mount defenses against chewing insects are known to be complex. To investigate the role of jasmonate in wound signaling in Arabidopsis and to test whether parallel or redundant pathways exist for insect defense, we have studied a mutant (fad3–2 fad7–2 fad8) that is deficient in the jasmonate precursor linolenic acid. Mutant plants contained negligible levels of jasmonate and showed extremely high mortality (≈80%) from attack by larvae of a common saprophagous fungal gnat, Bradysia impatiens (Diptera: Sciaridae), even though neighboring wild-type plants were largely unaffected. Application of exogenous methyl jasmonate substantially protected the mutant plants and reduced mortality to ≈12%. These experiments precisely define the role of jasmonate as being essential for the induction of biologically effective defense in this plant–insect interaction. The transcripts of three wound-responsive genes were shown not to be induced by wounding of mutant plants but the same transcripts could be induced by application of methyl jasmonate. By contrast, measurements of transcript levels for a gene encoding glutathione S-transferase demonstrated that wound induction of this gene is independent of jasmonate synthesis. These results indicate that the mutant will be a good genetic model for testing the practical effectiveness of candidate defense genes.
Keywords: jasmonic acid, wounding, wound signaling
When chewing insects begin feeding on leaves or other tissues, plant signaling pathways are activated that lead, ultimately, to the synthesis of many different secondary metabolites and specialized chemicals (1–3). It is assumed that these compounds act in plant defense, and increased protection against insect attack has been demonstrated for several classes of chemicals under specialized experimental conditions (4, 5). The best characterized system involves the production of proteinase inhibitors in tomato and other solanaceous plants (6). In tomato, wounding of a single leaf can result in the induction of proteinase inhibitors throughout the aerial portion of the plant. Biochemical and molecular genetic approaches indicate that a peptide hormone, systemin, is a mediator of the systemic signaling required for this induction (7). However, abscisic acid (8) and electrochemical potentials (9) have also been proposed as long-distance signals.
Experiments in which an antisense prosystemin gene was used to block wound signaling (10) and prevent accumulation of proteinase inhibitors resulted in reduced resistance of the plants toward tobacco hornworm larvae (Manduca sexta). Such results indicate that the proteinase inhibitors—which disrupt digestion in the insect’s gut (11)—and the pathways for their induction are major components of plant defense. In tomato, ample evidence indicates that systemin induces proteinase inhibitors and other genes through the synthesis and action of jasmonic acid. The tissue concentrations of jasmonic acid and its lipid precursor linolenic acid increase in tomato leaves after wounding (12) and jasmonate concentrations are also increased by other elicitors of the defense response (13, 14). Application of exogenous jasmonate induces synthesis of proteinase inhibitors (15) and chemical inhibitors of jasmonate synthesis block defense signaling. Finally, a newly described mutant deficient in jasmonate synthesis, def1, fails to accumulate proteinase inhibitors in response to wounding and is considerably more susceptible than wild type to attack by tobacco hornworm larvae (16).
In other plant species, wound-signaling pathways are much less completely defined. Some plants contain systemically inducible proteinase inhibitors (17, 18) but broader ecophysiological studies indicate that many other compounds are produced in plants to deter insect attacks (2, 3). It is not clear how many different signaling systems may have evolved in higher plants or how complex each pathway might be in its organization. For example, studies in Arabidopsis have not identified a systemin homolog and, although the projects for producing expressed sequence tags have identified cDNAs encoding putative proteinase inhibitors, specific assays have failed to demonstrate significant constitutive or wound-induced proteinase inhibitor activity (C. A. Ryan, personal communication). Jasmonate signaling pathways have been shown to control several plant responses in Arabidopsis including pollen development and the accumulation of a vegetative storage protein (19, 20). Jasmonate accumulation in Arabidopsis is induced in wounded tissue as it is in other plants (19). Three classes of Arabidopsis mutants defective in jasmonate synthesis or signaling have been described (19, 21, 22), but these mutants have not been reported to be more susceptible to insect attack. Because so little is known about insect defense in Arabidopsis, these results leave open the possibility that redundant pathways bypass jasmonate signaling or that other mechanisms exist in Arabidopsis that lead to significant protection against insects even in the absence of jasmonate-mediated processes. For example, in common with other members of the Brassicaceae, Arabidopsis contains the constitutive glucosinolate–myrosinase system that is postulated to act as a defense against chewing insects (23, 24).
Herein we report that a mutant of Arabidopsis deficient in linolenic acid is unable to synthesize jasmonates and is strikingly susceptible to devastation by a common saprophagous insect, the fungal gnat larva. Controlled experiments with the mutant and wild-type plants establish that jasmonate is both necessary and sufficient to protect Arabidopsis against insect attack.
METHODS
Plant Material.
The lines of Arabidopsis thaliana used were descended from the Columbia wild type in which mutations were produced by treatment with ethylmethanesulfonate. The fad3–2 fad7–2 fad8 triple mutant line and its allelic relatives are described elsewhere (20). Plants were grown in a commercial potting mixture in controlled environment chambers or in a greenhouse.
Measurements of Jasmonic Acid.
Plants were grown for 3 weeks at 22°C under continuous illumination of 140 μmol of quanta per m2 per sec. At the start of the experiment, half the plants were wounded by crushing each leaf three times with a hemostat. Immediately after wounding and 1, 2, and 4 hours later, samples of wounded and unwounded plants were harvested by cutting the tap root, weighed, and immediately frozen in liquid N2. Plants were stored at −80°C and jasmonate levels were measured by gas chromatography/mass spectrometry as described (25). Roots of wild-type and mutant Arabidopsis were grown in liquid culture as described (26). Roots were harvested and then wounded by cutting them repeatedly with a razor blade. Subsamples of the wounded material and unwounded controls were weighed, wrapped in plastic wrap, and held at room temperature. Immediately after wounding and 3 hours later, samples were frozen in liquid N2. The tissue was stored at −80°C and jasmonate levels were measured as described (25).
Challenging Plants with Bradysia Larva.
Seeds of wild-type and fad3–2 fad7–2 fad8 Arabidopsis were mixed in a 1:1 ratio, then planted at random into 15 pots (average of 9 plants per pot), and grown to the rosette stage under 10 h daylength. After the genotype of each plant had been determined by gas chromatography of a small leaf sample (20), a net enclosure was placed over the 15 pots and populated with 20–25 adult Bradysia flies. On each subsequent day, 8 pots were sprayed with 0.8 ml of H2O and 7 pots were sprayed with 0.8 ml of a dilute aqueous solution of methyl jasmonate. In different experiments, the methyl jasmonate concentration used was either 0.001% or 0.01%. In other experiments, additional chemical treatments were included as described in the text. To estimate the number of larvae present in treated pots, the soil from a pot was stirred into 500 ml of a 60% sucrose solution and allowed to settle. Under these conditions, the gnat larvae floated to the surface and could be counted easily.
Measurement of Transcript Levels in Wild-Type and Mutant Plants.
Total RNA was extracted from the wounded and unwounded plants used for the measurement of jasmonic acid levels and from unwounded plants sprayed with H2O or 0.001% methyl jasmonate. Four micrograms of total RNA from each sample was separated by gel electrophoresis, transferred to nylon membrane, and hybridized to cDNA probes as described (27). Radioactivity from each probe bound to the membrane was visualized by autoradiography and quantified by densitometry of the x-ray film. The probes used were AtVSP encoding a vacuole-localized acid phosphatase (28), DHS1 encoding 3-deoxy-d-arabinoheptulosonate-7-phosphate synthase (29), PAL1 encoding phenylalanine ammonia lyase (30), and GST encoding glutathione S-transferase (31).
RESULTS
Mutant Plants Contain Extremely Low Levels of Jasmonate.
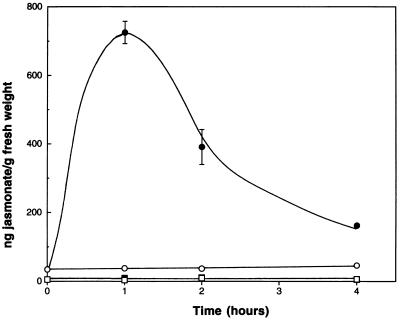
The fad3–2 fad7–2 fad8 mutant of Arabidopsis contains negligible levels of linolenic acid, the lipid precursor of jasmonate (20). Our initial characterization of the fad3–2 fad7–2 fad8 triple mutant was hampered by high mortality of older well-established triple mutant plants. Frequently, 30–50% of the mutant plants died before they could be used in experiments. Death of the plants appeared to be directly attributable to mechanical damage by larvae of Bradysia impatiens (Johannsen), the common fungal gnat. Fungal gnats inhabit many greenhouses and growth chambers and lay their eggs in the potting soil. The larvae feed mainly on decaying organic matter in the top 2 cm of soil before pupating and completing their life cycle to the adult. In normal culture of Arabidopsis, the fungal gnats are, at worst, a minor pest. Heavy infestations can reduce seedling establishment but the larvae do not cause any significant damage to mature plants. We reasoned that killing of the mutants would be consistent with the plants being divested of their regular defenses by a lack of jasmonate signaling. For this reason, we first measured the levels of jasmonic acid in wild-type and mutant plants before and after wounding. Unwounded leaves of wild-type Arabidopsis contained less than 35 ng of jasmonate per g (fresh weight) and the level increased more than 20-fold to 725 ng/g (fresh weight) at 1 hour after wounding, before declining to intermediate values (Fig. 1). By contrast, both unwounded and wounded leaves from fad3–2 fad7–2 fad8 contained levels of jasmonate that were close to the detection limit of the mass spectroscopic assay used, averaging less than 7 ng/g (fresh weight). Very comparable data were obtained when wounding experiments were carried out on root tissue. Wild-type roots contained 34 ng of jasmonate per g (fresh weight) in the absence of wounding and 326 ng/g (fresh weight) at 3 hours after wounding. Wounded and unwounded mutant roots averaged only 8 ng/g (fresh weight). These data are thus entirely similar to those found for leaf tissue.
Figure 1.
Kinetics of jasmonate accumulation in unwounded (open symbols) and wounded (solid symbols) leaf tissue of wild-type (○, •) and fad3–2 fad7–2 fad8 mutant (□, ▪) Arabidopsis. Data are the mean ± SEM of three determinations.
Jasmonate Is Both Necessary and Sufficient for Plant Defense.
Pathways for the breakdown of linolenic acid yield several compounds that have been implicated in plant defense (32, 33). While other evidence suggested that jasmonate was a likely key regulator of the defense pathway (13–15, 34), it remained possible that one of these other compounds was responsible or that susceptibility of mutant plants to attack by gnat larvae resulted from the combined loss of several linolenate derivatives. To explore this issue and to quantify the consequences of defective wound signaling in the fad3–2 fad7–2 fad8 mutant, we carried out experiments in which mixed stands of wild-type and mutant plants were challenged with modest populations of fungal gnats. In these experiments, mutant plants in control pots (those sprayed with water) were subject to increasing damage to rosette leaves, and by the end of the experiment, approximately 80% of the plants had been killed as a result of gnat larvae severing the stems, petioles, and/or tap roots of the plants (Fig. 2 A and B). By contrast, wild-type plants in these same pots remained largely undamaged and only one wild-type plant died out of a total of 117 included in the experiments. Because wild-type and mutant plants were randomly interplanted within the pots, these results demonstrate a very high level of protection in the wild-type plants and a correspondingly strong preference of the fungal gnat larvae for mutant plants. Other pots of plants were sprayed with dilute solutions of methyl jasmonate. The fad3–2 fad7–2 fad8 plants in these pots exhibited only 12% mortality (Fig. 2A) and most plants showed little or no damage from gnat larvae grazing (Fig. 2C).
Figure 2.
Death and protection of mutant plants from Bradysia larvae attack. (A) A mixed population of wild-type and fad3–2 fad7–2 fad8 plants were grown in a net enclosure populated with 20–25 adult Bradysia flies. Each day, eight pots were sprayed with 0.8 ml of H2O and seven pots were sprayed with with 0.8 ml of a dilute aqueous solution of methyl jasmonate. Data from two experiments are shown in which the methyl jasmonate concentration used was either 0.001% (open symbols) or 0.01% (solid symbols). the graph shows the percentage survival of 117 wild-type plants (○ and • both treatments), 73 mutant plants treated with H2O (□, ▪) and 73 mutant plants treated with methyl jasmonate (Δ, ▴). (B and C) For clarity, wild-type and mutant seeds were sown in pots in two rows but were otherwise treated as described above. The photographed plants correspond to day 50 in A. (B) Compared with wild-type controls (on the left), mutant plants (on the right) sprayed with water show extensive damage 20 days after the introduction of adult Bradysia flies. Some leaves on mutant plants have been almost completely eaten. Wilting of other leaves was attributed to damage to the petiole or to the roots of the plants. (C) Mutant plants sprayed with 0.01% methyl jasmonate (on the right) remained healthy and vigorous within the same environment.
To test the efficacy of other putative defense compounds derived from linolenic acid, we repeated the spraying experiment described above (using H2O and 0.001% jasmonate as negative and positive controls, respectively) but including an additional spraying treatment using an aqueous solution containing 0.01% trans-3-hexenol, 0.01% trans-2-hexenal, 0.01% cis-3-hexenol, and 0.01% traumatic acid. These four compounds are major products derived from linolenic acid by the hydroperoxide lyase pathway (32). The 0.001% jasmonate treatment was effective in protecting the plants, whereas a mixture of the other four compounds, each at a 10-fold higher concentration, did not protect fad3–2 fad7–2 fad8 mutant plants any better than H2O alone (data not shown). Thus, the protective effect appears to be specific for the allene oxide branch of the lipoxygenase pathway of linolenic acid metabolism.
Thus, these data demonstrate that the largely saprophagous fungal gnat larvae were not a significant pest on the wild-type Arabidopsis but that jasmonate-mediated signaling was both necessary and sufficient to provide protection of the fad3–2 fad7–2 fad8 mutant plants against larvae attack.
Jasmonate Has No Direct Effect on Fungal Gnat Larvae.
We investigated the possibility that methyl jasmonate might directly affect the survival of the Bradysia larvae by two different approaches. First, a repeat of the spraying experiment described above was conducted. Fourteen days after the introduction of flies and the start of spraying, two pots sprayed with water and three pots sprayed with 0.001% methyl jasmonate were removed from the experiment and the number of gnat larvae in the soil of each pot was counted. The average number of larvae per pot in the two pots sprayed with water was 342 ± 131 and the three pots sprayed with 0.001% methyl jasmonate was 486 ± 84.
In a second type of experiment, Bradysia larvae were collected from pots of soil, placed on moist filter paper in Petri dishes, and sprayed on two consecutive days with either water or 0.01% methyl jasmonate. After 48 hours, 74% of the larvae treated with water and 80% of the larvae treated with methyl jasmonate remained alive and active. Thus, neither of these experiments indicated any increased mortality or reduced activity in the samples of larvae exposed to methyl jasmonate.
Expression of Wound-Induced Genes in Wild-Type and Mutant Plants.
Because our results indicate that jasmonate is essential for effective defense of Arabidopsis against the fungal gnat larvae, the fad3–2 fad7–2 fad8 mutant provides a new means to investigate genes involved in the biologically effective wound response and to determine what levels of expression of these genes are required to confer meaningful protection, at least within the context of the Arabidopsis–Bradysia interaction and probably more broadly. As a first step in using this system, we measured the transcript levels of a series of genes that have been shown, in Arabidopsis or in other plants, to be induced by wounding.
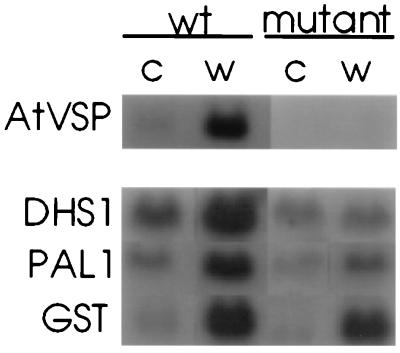
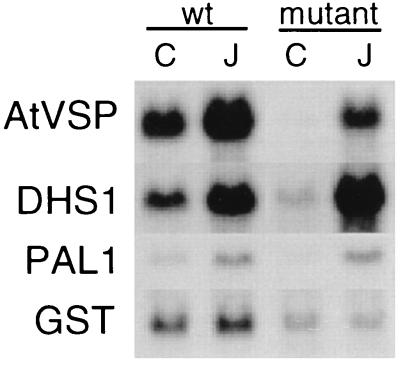
The gene AtVSP encodes a wound-inducible protein homologous to a soybean vacuolar acid phosphatase. This gene is regulated similarly to the gene encoding proteinase inhibitor II, which is involved in insect defense in tomato (28). The gene DHS1 encodes 3-deoxy-d-arabinoheptulosonate-7-phosphate synthase, a key enzyme in the production of lignin (29). The PAL1 gene encodes phenylalanine ammonia lyase, the enzyme that regulates flux into the pathway for phytoalexin synthesis (30). The product of the GST gene glutathione S-transferase is thought to help protect plants from oxidative tissue damage during wounding or pathogen attack (31). Interestingly, these genes showed distinctly different patterns of expression and induction in wild-type and mutant Arabidopsis (Fig. 3). The basal expression of DHS1 and PAL1 in mutant plants was less than in wild type, suggesting that resting levels of linolenic acid derivatives may modulate levels of these enzymes in unwounded plants. After wounding, transcripts of both these genes were induced approximately 4-fold in wild-type tissue and AtVSP was induced 15-fold. No induction of DHS1 and AtVSP occurred in the mutant, suggesting a strong dependence of jasmonate accumulation for expression in wounded tissue. PAL1 expression in the mutant was induced 2-fold, although transcript levels remained considerably lower than in the wounded wild type. To investigate the role of jasmonate in the induction of these genes, we carried out a complementary experiment in which unwounded wild-type and fad3–2 fad7–2 fad8 plants were sprayed with water or with a 0.001% solution of methyl jasmonate and then harvested 1.5 hours later. The results of this experiment (Fig. 4) indicate that transcripts of DHS1, AtVSP, and PAL1 were strongly induced in the mutant by application of exogenous methyl jasmonate. By contrast, the GST gene, which was strongly induced by wounding in both mutant and wild-type plants (Fig. 3), does not respond to jasmonate treatment.
Figure 3.
Wound induction of gene expression in wild-type (wt) and fad3–2 fad7–2 fad8 Arabidopsis. Transcript levels are shown in unwounded (C) and wounded (W) tissue after 2-hour incubation (4 hours for AtVSP). The probes used were AtVSP encoding a vacuole-localized acid phosphatase, DHS1 encoding 3-deoxy-d-arabinoheptulosonate-7-phosphate synthase, Pal1 encoding phenylalanine ammonia lyase, and GST encoding glutathione S-transferase.
Figure 4.
Jasmonate induction of gene expression in wild-type (wt) and fad3–2 fad7–2 fad8 Arabidopsis. Transcript levels are shown in unwounded tissue that had been sprayed with water (C, control) or with 0.001% methyl jasmonate (J) and harvested after a 1.5-hour incubation. The probes used are those described in Fig. 3.
DISCUSSION
Our current understanding of wound-signaling is incomplete. There is known to be at least some overlap with signaling processes involved in plant responses to fungal and bacterial pathogens (1, 2, 35). For this reason, and because a number of chemical and other signals (7, 8) have been postulated to be involved in wound signaling, it is tempting to draw analogies to other signal–response systems in plants, microbes and animals and suggest that wound signaling might involve branched and interconnecting pathways that allow for crosstalk and some redundancy in providing protection against insect attack (36, 37). The different expression patterns for putative defense genes (Fig. 3) appear to support such a model. In particular, the fact that GST expression is induced by wounding completely independent of jasmonate synthesis points to the existence of a separate wound-signaling system. Despite these possible complexities, the experiments reported herein have provided a simple and clear-cut result. The fad3–2 fad7–2 fad8 mutants, but not neighboring wild-type plants, were devastated by larval attack but could be protected by applications of jasmonate (Fig. 2). Although parallel mechanisms may operate at other stages in the pathway, we have shown that, for the Arabidopsis–Bradysia interaction, the synthesis and action of jasmonate is a nonredundant step if the plant is to mount a biologically effective response. Analogous results have recently been obtained for a tomato mutant with reduced jasmonate synthesis that is susceptible to damage from Manduca sexta larvae (16).
The fad3–2 fad7–2 fad8 mutant provides additional information about the synthesis and action of oxylipin signals. For example, both linolenic acid and linoleic acid are substrates for lipoxygenase, and it has been suggested that the parallel metabolism of linoleic acid to 9,10-dihydrojasmonic acid might provide an alternative wound signal (38). Since the fad3–2 fad7–2 fad8 mutants contain increased levels of linoleic acid (65% compared with 16% in wild-type Arabidopsis) (9), our results indicate that it is unlikely that 9,10-dihydrojasmonic acid is involved in insect defense. It is known that other input signals, some fungal elicitors for example, can activate the octadecanoid pathway for jasmonate synthesis (1, 2, 35) and that jasmonate is involved in activating genes involved in processes, such as pollen development, that are distinct from protection of the plant against insect attack (20, 28, 38). Our results and these considerations point to a key role for jasmonate in plant responses to a range of stresses.
Our results suggest that the fad3–2 fad7–2 fad8 mutant should be an excellent system to identify wound-induced genes that specifically respond to jasmonic acid and other derivatives of linolenic acid. The various expression patterns shown by four representative genes in Fig. 3 demonstrate the importance of having a clear-cut genetic model in which to compare levels of expression (relative to wild type) in a mutant that is demonstrably susceptible to insect attack. Furthermore, constitutive expression of transgenes in the mutant can be used to test candidate defense genes for their ability to meaningfully reduce damage and mortality from insect attack. Information from such studies will be important for designing novel gene combinations that can improve the insect resistance of plants. Finally, a search for mutations that suppress the insect susceptibility of fad3–2 fad7–2 fad8 Arabidopsis can be used to identify genes whose products act in the signaling cascade downstream of jasmonate.
Acknowledgments
We are grateful to T. Miller, W. Turner (Department of Entomology, Washington State University) and W. Steffan (Museum of Art, Sun City, AZ) for assistance in identification and culture of fungal gnats and to G. A. Howe and C. A. Ryan for helpful discussions and comments on the manuscript. This work was supported by grants from the U.S. Department of Energy (DE-FG06–92ER20077 to J.B.), The United States Department of Agriculture (NRICGP 95–37304-2440 to R.A.C. and E.B.), and The National Science Foundation (MCB95–14034 to J.E.M.), and by the Agricultural Research Center, Washington State University.
References
- 1.Ryan C A. Annu Rev Phytopathol. 1990;28:425–449. [Google Scholar]
- 2.Bowles D J. Annu Rev Biochem. 1990;59:873–908. doi: 10.1146/annurev.bi.59.070190.004301. [DOI] [PubMed] [Google Scholar]
- 3.Baldwin I T. In: Insect–Plant Interactions. Bernays E A, editor. Boca Raton, FL: CRC; 1994. pp. 1–23. [Google Scholar]
- 4.Hilder V A, Gatehouse A M R, Sheerman S E, Barker R F, Boulter D. Nature (London) 1987;330:160–163. [Google Scholar]
- 5.Johnson R, Narváez J, An G, Ryan C. Proc Natl Acad Sci USA. 1989;86:9871–9876. doi: 10.1073/pnas.86.24.9871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green T R, Ryan C A. Science. 1972;175:776–777. doi: 10.1126/science.175.4023.776. [DOI] [PubMed] [Google Scholar]
- 7.Pearce G, Strydom D, Johnson S, Ryan C A. Science. 1991;253:895–989. doi: 10.1126/science.253.5022.895. [DOI] [PubMed] [Google Scholar]
- 8.Peña-Cortés H, Albrecht T, Prat S, Weiler E W, Willmitzer L. Planta. 1993;191:123–128. [Google Scholar]
- 9.Wildon D C, Thain J F, Minchin P E H, Gubb I R, Reilly A J, Skipper Y D, Doherty H M, O’Donnell P J, Bowles D J. Nature (London) 1992;360:62–65. [Google Scholar]
- 10.Orozco-Cardenas M, McGurl B, Ryan C A. Proc Natl Acad Sci USA. 1993;90:8273–8276. doi: 10.1073/pnas.90.17.8273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Broadway R M, Duffey S S. J Insect Physiol. 1986;32:827–833. [Google Scholar]
- 12.Peña-Cortés H, Albrecht T, Prat S, Weiler E W, Willmitzer L. Planta. 1993;191:123–128. [Google Scholar]
- 13.Doares S H, Syrovets T, Weiler E W, Ryan C A. Proc Natl Acad Sci USA. 1995;92:4095–4098. doi: 10.1073/pnas.92.10.4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peña-Cortés H, Fisahn J, Willmitzer L. Proc Natl Acad Sci USA. 1995;92:4106–4113. doi: 10.1073/pnas.92.10.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farmer E E, Ryan C A. Plant Cell. 1992;4:129–134. doi: 10.1105/tpc.4.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howe G A, Lightner J, Browse J, Ryan C A. Plant Cell. 1996;8:2067–2077. doi: 10.1105/tpc.8.11.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bradshaw H D F, Hollick J B, Parsons T J, Clarke H R G, Gordon M P. Plant Mol Biol. 1989;14:51–59. doi: 10.1007/BF00015654. [DOI] [PubMed] [Google Scholar]
- 18.Cordero M J, Ravent-s D, San Segundo B. Plant J. 1994;6:141–150. doi: 10.1046/j.1365-313x.1994.6020141.x. [DOI] [PubMed] [Google Scholar]
- 19.Bell E, Creelman R A, Mullet J E. Proc Natl Acad Sci USA. 1995;92:8675–8679. doi: 10.1073/pnas.92.19.8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McConn M, Browse J. Plant Cell. 1996;8:403–416. doi: 10.1105/tpc.8.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Staswick P E, Su W, Howell S H. Proc Natl Acad Sci USA. 1992;89:6837–6840. doi: 10.1073/pnas.89.15.6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feys B J F, Benedetti C E, Penfold C N, Turner J G. Plant Cell. 1995;6:751–759. doi: 10.1105/tpc.6.5.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thangstad O P, Winge P, Husebye H, Bones A. Plant Mol Biol. 1993;23:511–524. doi: 10.1007/BF00019299. [DOI] [PubMed] [Google Scholar]
- 24.Xue J, Jørgensen M, Pihlgren U, Rask L. Plant Mol Biol. 1995;27:911–922. doi: 10.1007/BF00037019. [DOI] [PubMed] [Google Scholar]
- 25.Creelman R A, Tierney M L, Mullet J E. Proc Natl Acad Sci USA. 1992;89:4938–4941. doi: 10.1073/pnas.89.11.4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miquel M, Browse J. J Biol Chem. 1992;267:1502–1509. [PubMed] [Google Scholar]
- 27.Bell E, Mullet J E. Mol Gen Genet. 1991;230:456–462. doi: 10.1007/BF00280303. [DOI] [PubMed] [Google Scholar]
- 28.Berger S, Bell E, Sadka A, Mullet J E. Plant Mol Biol. 1995;27:933–942. doi: 10.1007/BF00037021. [DOI] [PubMed] [Google Scholar]
- 29.Keith B, Dong X, Ausubel F M, Fink G R. Proc Natl Acad Sci USA. 1991;88:8821–8825. doi: 10.1073/pnas.88.19.8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wanner L A, Li G, Ware D, Sommssich I E, Davis K R. Plant Mol Biol. 1995;27:327–338. doi: 10.1007/BF00020187. [DOI] [PubMed] [Google Scholar]
- 31.Kim C S, Kwak J M, Nam H G, Kim K C, Cho B H. Plant Cell. 1994;13:340–343. doi: 10.1007/BF00232633. [DOI] [PubMed] [Google Scholar]
- 32.Croft K P C, Juttner F, Slusarenko A J. Plant Physiol. 1993;101:13–24. doi: 10.1104/pp.101.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farmer E E. Plant Mol Biol. 1994;26:1423–1437. doi: 10.1007/BF00016483. [DOI] [PubMed] [Google Scholar]
- 34.Sembdner G, Parthier B. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:569–589. [Google Scholar]
- 35.Mueller M J, Brodschelm W, Spannagl E, Zenk M H. Proc Natl Acad Sci USA. 1993;90:7490–7494. doi: 10.1073/pnas.90.16.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doares S H, Narváez-Vásquez J, Conconi A, Ryan C A. Plant Physiol. 1995;108:1741–1746. doi: 10.1104/pp.108.4.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ellard-Ivey M, Douglas C J. Plant Physiol. 1996;112:183–192. doi: 10.1104/pp.112.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blechert S, Brodschelm W, Hölder S, Kammerer L, Kutchan T M, Mueller M J, Xia Z-Q, Zenk M H. Proc Natl Acad Sci USA. 1995;92:4099–4105. doi: 10.1073/pnas.92.10.4099. [DOI] [PMC free article] [PubMed] [Google Scholar]