Abstract
Previous studies have identified an ATP-dependent DNA helicase activity intrinsic to the human minichromosome maintenance (MCM) complex, composed of MCM subunits 4, 6, and 7 [Ishimi, Y. (1997) J. Biol. Chem. 272, 24508–24513]. In contrast to the presence of multiple MCM genes (at least six) in eukaryotes, the archaeon Methanobacterium thermoautotrophicum ΔH (mth) genome contains a single open reading frame coding for an MCM protein. In this study we report the isolation of the mthMCM protein overexpressed in Escherichia coli. The purified recombinant protein was found to exist in both multimeric (≈103 kDa) and monomeric (76 kDa) forms. Both forms of the protein bind to single-stranded DNA, hydrolyze ATP in the presence of DNA, and possess 3′-to-5′ ATP-dependent DNA helicase activities. Thus, a single mthMCM protein contains biochemical properties identical to those associated with the eukaryotic MCM4, -6, and -7 complex. These results suggest that the characterization of the mthMCM protein and its multiple forms may contribute to our understanding of the role of MCM helicase activity in eukaryotic chromosomal DNA replication.
Keywords: replication, ORC1 protein, Cdc6 protein
The minichromosome maintenance (MCM) genes encode a family of proteins first identified by their essential role in the maintenance of ARS-containing minichromosomes in Saccharomyces cerevisiae (1). MCM homologues have been identified in all eukaryotes from yeast to mammals and have highly conserved amino acid sequences (2, 3). The MCM family consists of at least six distinct polypeptides (MCM2–7). In addition to forming a heterohexameric structure, the MCM proteins are capable of forming various subcomplexes in vitro (4–8). Although the MCM proteins were shown to play an important role in the initiation and elongation phases of DNA replication, the precise functions of the MCM complexes are not yet fully understood (reviewed in refs. 3, 9, and 10). In vitro studies have revealed several biochemical properties of the MCM proteins. A heterotrimeric complex of human (h) MCMs (MCM4, -6, and -7) that forms a dimeric structure contains ATP-dependent DNA helicase activity, binds to single-stranded DNA, and possesses DNA-dependent ATPase activity (11). It was also demonstrated that the hMCM2 protein interacts with the hMCM4, -6, and -7 complex to form the heterotetrameric complex, hMCM2, -4, -6, and -7, which is devoid of the activities associated with the hMCM4, -6, and -7 complex (11, 12). Identical observations have been made with MCM proteins isolated from Schizosaccharomyces pombe (spMCM; J.-K.L. and J.H., unpublished observations).
In addition to genetic studies that demonstrated the essential role of the MCM proteins in DNA replication, biochemical studies showed that these proteins are assembled onto origins of replication in S. cerevisiae (13, 14). Furthermore, the proteins appear to interact genetically and physically with the origin recognition complex (ORC) and other proteins that participate in the initiation of DNA replication such as Cdc6p and Cdc45p (14–17). These genetic data obtained from yeast, together with the in vitro biochemical properties, have led to the suggestion that the MCM proteins may function as the replicative helicase in eukaryotes, similar to role of the bacterial DnaB protein in Escherichia coli replication and the large tumor antigen (T Ag) in the replication of simian virus 40 (SV40) DNA (18–20).
Archaea, the third domain of life (21), are believed to replicate DNA in a eukaryotic-like fashion. This conclusion is based in large part on the complete genome sequences of several members of this domain (22–26). Homologues of proteins involved in eukaryotic DNA replication have been identified within these genomes, whereas only limited similarities have been observed for bacterial DNA replication proteins. To date, at least one MCM homologue has been identified in all sequenced archaeal genomes. Each species, however, contains a different number of MCM homologues [up to four polypeptides have been detected in Methanococcus jannaschii (22)].
In the archaeon Methanobacterium thermoautotrophicum ΔH (mth), one MCM homologue has been identified (24). M. thermoautotrophicum is an obligatory anaerobic thermophilic microorganism with an optimal growth temperature of 65–70°C and a generation time of about 5 hr (27). In this study, the isolation and the biochemical characterization of the single mthMCM protein are described. The recombinant protein, expressed and purified from E. coli cells, was found to possess DNA helicase activity similar to that of the eukaryotic MCM4, -6, and -7 complex.
Materials and Methods
Materials.
Labeled and unlabeled dNTPs and rNTPs were obtained from Amersham and Pharmacia-LKB, respectively. Single-stranded (ss) and double-stranded (ds) phage φX174 DNA and plasmid pUC19 were from New England Biolabs. The pET-16b vector was from Novagen and oligonucleotides were synthesized by Gene Link (Hawthorne, NY) and by Integrated DNA Technologies (Coralville, IA). E. coli ssDNA-binding protein (SSB) was from Pharmacia-LKB, and M. thermoautotrophicum replication protein A (RPA) was purified as previously described (28). Rabbit polyclonal antibodies were generated by Cocalico Biologicals (Reamstown, PA). The buffers used and their compositions were as follows: buffer A, which contained 20 mM Tris⋅HCl (pH 7.5), 2 mM DTT, 0.5 mM EDTA, and 10% (vol/vol) glycerol; buffer B, which contained 20 mM Tris⋅HCl (pH 7.5), 0.5 mM EDTA, and 10% glycerol; and buffer L, which contained 50 mM Tris⋅HCl (pH 8.0), 500 mM NaCl, 6 M urea, and 10% glycerol.
Preparation of Substrates for Helicase and Gel Mobility-Shift Assays.
Four DNA oligomers were synthesized and used for the preparation of the helicase substrates. These included the following: a 98-mer, 5′-GAATACAAGCTTGGGCTGCAGGTCGACTCTAGAGGATCCCCGGGCGAGCTCGAATTCGGGTCTCCCTATAGTGAGTCGTATTAATTTCGATAAGCCAG-3, corresponding to the longer strand of partial duplex substrates (referred to as 98-mer, as described below); a 38-mer, 5′-GAATACACGGAATTCGAGCTCGCCCGGGGATCCTCTAG-3′, an oligonucleotide partially complementary (29 nt) to the 98-mer. The partial duplex formed by annealing this 38-mer and the 98-mer is referred to as HS-M; a 30-mer, 5′-AGAGTCGACCTGCAGCCCAAGCTTGTATTC-3′, an oligomer complementary to the 5′ end of the 98-mer, leaving a single strand 3′-tailed region on the longer DNA, is referred to as HS-3′T; a second 30-mer, 5′-CTGGCTTATCGAAATTAATACGACTCACTA-3′, an oligomer complementary to the 3′ end of the 98-mer, resulting in a single strand 5′-tailed region on the 98-mer, is referred to as HS-5′T. After labeling of the short oligomeric DNAs with [γ-32P]ATP and T4 polynucleotide kinase and annealing to the 98-mer DNA, the annealed products were gel purified as described (29). Gel mobility-shift assays employed the 41-mer 5′-AATCATAGATAGTATCTCCGTGCAAGATAATCACGAGTATC-3′ (41-mer), labeled with [γ-32P]ATP and T4 polynucleotide kinase.
The substrate used to examine the maximum length of duplex displaced by the mthMCM helicase activity was prepared by elongating a singly primed M13 mp19 ssDNA by using Sequenase (United States Biochemical) in the presence of the four dNTPs and ddGTP according to the manufacturer's protocol. The oligonucleotide primer (M13 mp19, positions 6311–6334) was labeled with 32P by using T4 polynucleotide kinase and [γ-32P]ATP. The DNA was then purified by using a gel filtration column to remove nucleotides and excess unannealed labeled primers. The resulting substrate contained duplex regions that varied in length between 34 and 1000 bp.
Cloning of the mthMCM Protein.
The MCM gene (MTH1770) was amplified by PCR from mthDNA (kindly provided by John Reeve, Ohio State University, Columbus) and was cloned, after sequencing, between the NdeI and BamHI sites of the bacterial expression vector pET-16b (Novagen) (called pET16-MCM). The construct contained a His10-tag at the N terminus of the protein. The cloning of the two M. thermoautotrophicum homologues of CDC6/ORC1 [ORFs MTH1412 and MTH1599 (24)] will be described elsewhere.
Expression and Purification of Recombinant Proteins.
The mthMCM protein was overexpressed as follows: E. coli cells BL21(DE3) pLysS (Novagen), harboring the different plasmids, were grown at 37°C in 4 liters of Luria–Bertani (LB) medium in the presence of appropriate antibiotics. When the culture reached an OD600 of 0.5, protein expression was induced by incubation in the presence of 2 mM isopropyl β-d-thiogalactoside (IPTG) for 3 hr, after which time the cells were harvested, yielding 11.2 g (wet weight) of cells. The mthMCM protein was purified from E. coli cells as follows: Bacterial lysates were prepared by sonication in 25 ml of buffer L. After centrifugation for 20 min at 36,000 × g, extracts were mixed with 1 ml of Ni chelate (ProBond resin, Invitrogen) for 2 hr at 4°C with gentle shaking. The mixtures was then poured onto a column, washed with 25 ml of buffer L containing 10 mM imidazole, and eluted with 3 ml of buffer L containing 500 mM imidazole. The latter fraction was diluted 5-fold with buffer B and loaded onto a 5-ml HiTrap-Q column (Pharmacia-LKB), equilibrated with buffer A containing 100 mM NaCl. The column was washed with 25 ml of buffer A containing 200 mM NaCl and developed with a 50-ml linear gradient of NaCl from 100 mM to 600 mM in buffer A. The pooled protein peak (5.8 mg, peaking at 450 mM NaCl) was dialyzed overnight against 2 liters of buffer A containing 500 mM NaCl. A portion of the MCM protein (between 80% and 90% pure) was further purified by glycerol gradient centrifugation or by gel filtration. These purified fractions were used in the experiments described below. Protein concentrations were determined by the Bradford method (Bio-Rad) with bovine serum albumin (BSA) as the standard. Proteins were stored at −70°C. The activities associated with the mthMCM protein were stable to repeated freezing and thawing.
Glycerol Gradient Centrifugation.
A portion of the fraction isolated after the HiTrap-Q step (100 μg) was applied to a 5-ml 15–35% glycerol gradient in buffer A containing 200 mM NaCl. After centrifugation at 45,000 rpm (190,000 × g) for 13 hr in a Beckman SW 50.1 rotor at 4°C, fractions (200 μl) were collected from the bottom of the tube. The distribution of the mthMCM protein was determined after SDS/10% PAGE and staining with Coomassie brilliant blue (R-250) and by the assay of each fraction for activities associated with the mthMCM protein.
Gel-Filtration Separation.
A portion of the purified protein fraction (HiTrap-Q column fraction, 180 μg in 200 μl of buffer A containing 200 mM NaCl) was applied to a Superose-6 gel-filtration column (HR10/30; Pharmacia-LKB) equilibrated with buffer A containing 200 mM NaCl. Fractions (300 μl) were collected and analyzed for the presence of the mthMCM protein and associated enzymatic activities. The distribution of mthMCM protein was detected after SDS/10% PAGE and staining with Coomassie brilliant blue (R-250) as well as by ATPase assays as described below.
ATPase Assay.
ATPase activity was measured in reaction mixtures (15 μl) containing 25 mM Hepes–NaOH (pH 7.5), 50 mM sodium acetate, 5 mM magnesium acetate, 1 mM DTT, 0.1 mg/ml BSA, 1.5 nmol of [γ-32P]ATP (1.5 × 104 cpm/pmol), the indicated amount of polynucleotide, and enzyme fraction. After incubation at 60°C for 60 min, an aliquot (1 μl) was spotted onto a polyethyleneimine-cellulose thin-layer plate, and ATP and Pi were separated by chromatography in 1 M formic acid/0.5 M LiCl. The extent of ATP hydrolysis was quantitated by phosphorimager (Fuji) analysis.
DNA Helicase Assay.
DNA helicase activity was measured in reaction mixtures (15 μl) containing 25 mM Hepes–NaOH (pH 7.5), 25 mM sodium acetate, 7.5 mM magnesium acetate, 5 mM ATP, 1 mM DTT, 0.1 mg/ml BSA, the indicated 32P-labeled partial duplex DNA substrate (10 fmol, 3000 cpm/fmol), and enzyme fraction. After incubation at 55°C for 1 hr, 4 μl of 5× loading buffer (50 mM EDTA/0.5% SDS/0.1% xylene cyanol/0.1% bromphenol blue/50% glycerol) was added, and aliquots were loaded onto a 12% polyacrylamide gel containing 0.1% SDS in 1× TBE (90 mM Tris/90 mM boric acid/1 mM EDTA, pH 8.3) and electrophoresed for 1.5 hr at 150 V.
Gel Mobility-Shift Assay.
Complexes formed between the mthMCM protein and ssDNA were detected by a gel mobility-shift assay of reaction mixtures (15 μl) containing 25 mM Hepes–NaOH (pH 7.5), 50 mM sodium acetate, 7.5 mM magnesium acetate, 1 mM ATP, 1 mM DTT, 0.1 mg/ml BSA, 20 fmol of 5′-32P-labeled 41-mer ssDNA (3000 cpm/fmol), and enzyme. After incubation at 55°C for 20 min, an aliquot of the reaction mixture was electrophoresed for 3 hr at 120 V through a 4% polyacrylamide gel containing 6 mM magnesium acetate and 5% glycerol in 0.5× TBE at 4°C.
Filter Binding Assay.
A nitrocellulose filter binding assay was carried out in reaction mixtures (30 μl) containing binding buffer (20 mM Hepes–NaOH, pH 7.5/5 mM MgCl2/2 mM DTT/100 μg/ml BSA/150 mM NaCl), 50 fmol of 5′-32P-labeled oligo(dT)50 (3000 cpm/fmol), and various levels of the mthMCM protein at 70°C for 10 min. After incubation, the mixture was filtered through an alkaline-washed nitrocellulose filter (Millipore, HA 0.45 mm) (30), which was then washed with binding buffer. The radioactivity adsorbed to the filter was measured by liquid scintillation counting. mthRPA (28) was used as a control to measure DNA binding under the above conditions.
Results
mthMCM Protein Exists in Both Multimeric and Monomeric Forms and Contains an Intrinsic DNA Helicase Activity.
To determine the biochemical properties of the MCM homologue in M. thermoautotrophicum, the protein was purified and its biochemical properties were determined. The gene encoding the mthMCM protein [ORF MTH1770 (24)] was inserted into an E. coli expression vector and expressed as a fusion protein containing an N-terminal His10-tag (see Materials and Methods). When overexpressed, only a small percentage of the MCM protein was soluble. Therefore, the protein was extracted from E. coli cells in the presence of urea and renatured by using the procedure described in Materials and Methods.
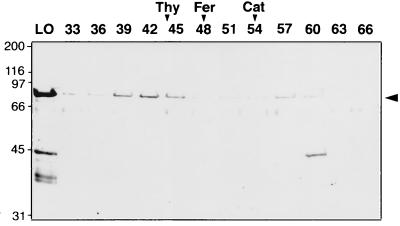
The structure of the purified mthMCM protein was determined by gel filtration. SDS/PAGE analysis of fractions obtained by gel filtration (Fig. 1) detected two protein peaks. The majority of the protein (representing 80% of the applied protein) was detected at a position (fraction 40) indicating that it is larger than thyroglobulin (660 kDa), suggesting that mthMCM protein was oligomeric in structure. The estimated molecular mass of this oligomer was about 950 kDa, suggesting that this protein may form a dodecamer complex. The second protein peak (representing 20% of the protein) was detected at a position (fraction 57) expected for monomeric form of the mthMCM protein. A contaminant of ≈40 kDa was observed in the monomeric region. This material, however, did not elute coincidentally with the mthMCM monomer protein in either sizing procedure used (see below). Because of this observation and the coincidence between the various activities associated with the monomeric mthMCM protein, no further consideration of this contaminant was addressed below
Figure 1.
Gel filtration analysis of the mthMCM protein preparations. The purified HiTrap-Q column fraction of the mthMCM protein (0.2 ml, 140 μg) was loaded onto a Superose-6 gel filtration column and analyzed. Aliquots (30 μl) of the fraction were subjected to SDS/10% PAGE analysis followed by Coomassie blue staining. The peak positions of thyroglobulin (Thy, 669 kDa), ferritin (Fer, 440 kDa), and catalase (Cat, 232 kDa) are indicated at the top. LO, material loaded onto the column.
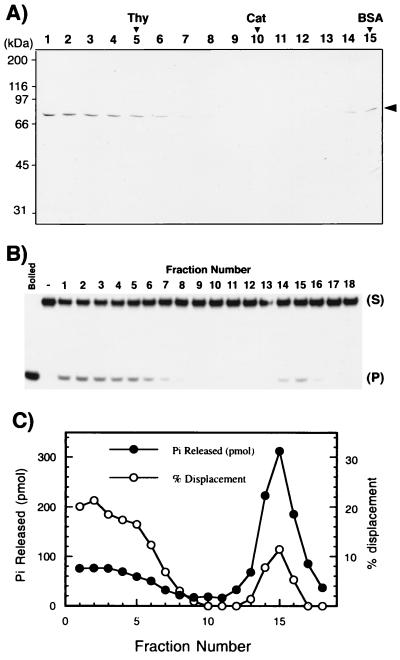
We next examined the distribution of DNA helicase and ATPase activities of the mthMCM protein by glycerol gradient centrifugation. Consistent with gel filtration studies, the mthMCM protein yielded the protein peaks at fractions 2 and 15 (Fig. 2A). Helicase assays were performed with 2 μl of each glycerol gradient fraction in the presence of 10 fmol of 32P-labeled 38-mer oligonucleotide (HS-M) annealed to the 98-mer oligonucleotide as described in Materials and Methods. The displacement of the labeled strand from the duplex substrate was determined by SDS/PAGE analysis. As shown in Fig. 2B, the helicase activity and mthMCM protein sedimented coincidentally and were detected in both peaks (Fig. 2 A and B). Though the amount of helicase activity associated with the first peak (fractions 1–6) was greater than that associated with the MCM protein present in the second peak (fractions 14–16) (Fig. 2 A and B), their specific activities were identical. Similar results were obtained when the gel-filtration fractions were analyzed for helicase activity (data not presented).
Figure 2.
Glycerol gradient sedimentation analysis of the mthMCM preparation. (A) The purified HiTrap-Q column fraction of the mthMCM protein (0.1 ml, 75 μg) was loaded onto a 5-ml 15–35% glycerol gradient and centrifuged for 13 hr at 45,000 rpm in a Beckman SW50.1 rotor at 4°C. Eighteen fractions were collected from the bottom, and aliquots (15 μl) of each fraction were loaded onto an SDS/10% polyacrylamide gel, subjected to electrophoresis, and stained with Coomassie blue. Arrows indicate the positions of marker proteins. (B) Distribution of DNA helicase activity in glycerol gradient fractions. The assay for helicase activity was carried out with 2 μl of each fraction in reaction mixtures containing 10 fmol of the HS-M substrate (S). P, product. (C) Distribution of DNA-dependent ATPase activity. The ATPase activity was assayed in the presence of φX174 ssDNA (30 ng/μl), using 2 μl of each fraction.
The ATPase activity of the glycerol gradient fractions was analyzed by incubating an aliquot (2 μl) of each glycerol gradient fraction in the presence of φx174 ssDNA. As shown in Fig. 2C, ATPase activity cosedimented with both peaks of mthMCM protein. The multimeric form catalyzed the hydrolysis of 18 pmol of ATP per min per μg of protein, whereas the monomeric form hydrolyzed 110 pmol/min per μg of protein. Identical results were obtained with the two forms isolated by gel filtration (data not presented). Thus, the specific activity of the mthMCM monomer was nearly 6-fold greater than that of the multimeric form. These observations contrast with the specific activity of the DNA helicase activity, which did not differ between these two forms. The significance of these observations is discussed below.
Effect of DNA on the ATPase Activity of mthMCM Protein.
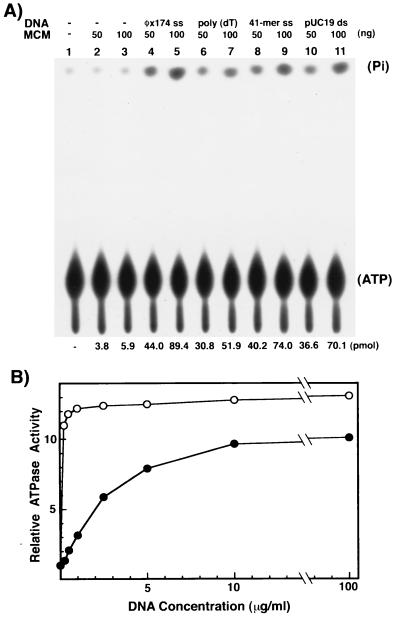
The hydrolysis of the ATP by the eukaryotic MCM4, -6, and -7 complex is markedly stimulated by ssDNA and to a lesser extent by dsDNA (ref. 11; J.-K.L. and J.H., unpublished observation). For this reason, the influence of several DNAs on the ATPase activity associated with the multimeric form the mthMCM protein was examined. As shown in Fig. 3A, the ATPase activity of the mthMCM protein was stimulated 13-fold by the addition of φX174 ssDNA (compare lanes 4 and 5 with lanes 2 and 3, respectively). Short ssDNA oligonucleotides also stimulated the ATPase activity (compare lanes 6 and 8 with lane 2 and lanes 7 and 9 with lane 3) but to a lesser extent than φX174 ssDNA. An 11-fold stimulation of the ATPase activity was also observed with dsDNA (compare lane 10 with lane 2 and lane 11 with lane 3). This observation is in contrast to the observations made with the eukaryotic MCM4, -6, and -7 complex. In the latter case, dsDNA stimulated ATPase activity poorly (less than 2-fold) (11).
Figure 3.
The mthMCM protein contains DNA-dependent ATPase activity. (A) The influence of various DNA preparations on the ATPase activity of the mthMCM protein. The ATPase activity of the multimeric glycerol gradient peak was determined in the absence or presence of the various indicated DNAs (30 ng/μl). The numbers at the bottom of the autoradiogram indicate the amount of 32Pi released (quantitated by phosphorimager analysis). (B) Influence of DNA concentration on ATPase activity. ATPase activity assay was measured in the presence of the indicated amount of φX174 ssDNA (○) or pUC19 dsDNA (●) and the mthMCM multimeric glycerol gradient peak fraction (80 ng). The activity observed with no DNA added (5 pmol) was set at a value of 1; ATP hydrolysis in the presence of DNA at various concentrations was calculated relative to this value.
As shown in Fig. 3B, the stimulation of ATPase activity plateaued at lower concentrations of ssDNA (≈1 μg/ml) than dsDNA (≈10 μg/ml). Under the conditions used (up to 100 μg/ml DNA), the extent of ATP hydrolysis in the presence of dsDNA did not reach the level observed with ssDNA.
The finding that dsDNA stimulated the ATPase activity of the mthMCM protein but marginally affected the ATPase activity of the hMCM4, -6, -7 complex (or the spMCM4, -6, -7 complex) may be due to the reaction conditions used. M. thermoautotrophicum is a thermophile with optimal growth at 60–70°C (27). For this reason, ATPase assays were performed at high temperatures (60°C). At this temperature portions of the duplex DNA are likely to denature, forming ss regions, which may account for the stimulation of ATPase activity observed with dsDNA. The ATPase activity associated with the eukaryotic enzymes was analyzed at a lower temperature (37°C), and thus the amount of ss regions formed in dsDNA is likely to be considerably less, possibly accounting for the limited stimulation of the ATPase activity observed.
mthMCM Protein Binds ssDNA.
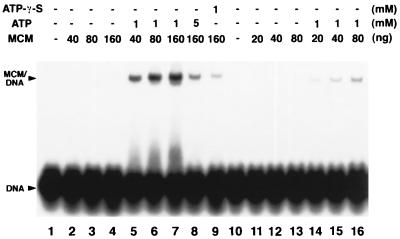
Direct interaction between the mthMCM protein and DNA was demonstrated with the gel mobility-shift and filter binding assays. For these studies, a 5′-32P-labeled ss 41-mer was incubated with increasing levels of both forms of the mthMCM protein in the presence or absence of ATP at 55°C for 20 min, and protein binding to DNA was analyzed as described in Materials and Methods. As shown in Fig. 4, the multimeric form of mthMCM bound to ssDNA and the interaction was stimulated more than 50-fold by the addition of ATP (compare lanes 5–7 with lanes 2–4). When the nonhydrolyzable ATP analogue ATP-γ-S was used, only limited DNA binding was detected (lane 9). Similar results were obtained with the mthMCM monomeric form (lanes 10–16). In the presence of ATP, however, less binding was detected with the monomer compared with the binding observed with the multimeric form (compare lane 5 with 15 and lane 6 with 16). Interestingly, at a higher ATP concentration DNA binding was reduced (lane 8), This reduction in binding activity may be related to the ratio of Mg2+ to ATP used in the reaction.
Figure 4.
ssDNA-binding activity of the different forms of the mthMCM protein. ssDNA-binding activity of the mthMCM protein was determined in the presence or absence of the indicated amounts of ATP or γ-thio-ATP (ATP-γ-S). The multimeric mthMCM glycerol gradient peak fraction (fraction 2), described in Fig. 2A, was used in lanes 1–9, and the monomeric glycerol gradient peak fraction (fraction 15) was used in lanes 10–16.
Biochemical Characterization of the mthMCM Helicase Activity.
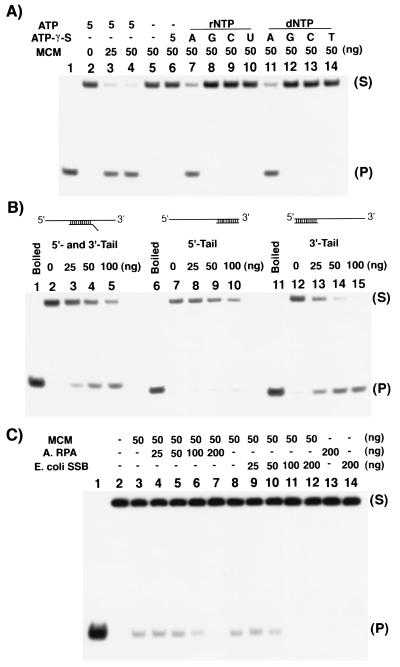
The experiments described above demonstrate that the single-subunit archaeal MCM protein has DNA helicase activity and DNA-dependent ATPase activity and binds ssDNA in the presence of ATP. The following experiments were designed to determine the biochemical properties of the mthMCM helicase activity. First the nucleotide requirement was analyzed. As shown in Fig. 5A, only ATP and dATP (lanes 7 and 11) supported mthMCM helicase activity; none of the other rNTPs (lanes 8–10) or dNTPs (lanes 12–14) supported this activity. Furthermore, ATP hydrolysis was required for helicase activity, since the nonhydrolyzable ATP analogue ATP-γ-S, did not support duplex displacement (lane 6). These results are similar to those observed with the hMCM complex in which helicase activity was detected only in the presence of ATP or dATP (11).
Figure 5.
Properties of the mthMCM DNA helicase activity. (A) NTP requirement for mthMCM helicase activity. DNA helicase activity assays were performed with the indicated amounts of mthMCM protein (multimer, glycerol gradient fraction peak) and 10 fmol of HS-3′T substrate in the absence or presence of 5 mM rNTPs or dNTPs. (B) mthMCM helicase translocates in the 3′-to-5′ direction. DNA helicase activity was assayed with 10 fmol of HS-M, HS-5′T, or HS-3′T and various amounts of the mthMCM multimeric form (glycerol gradient peaks). (C) Effect of SSBs on the mthMCM helicase activity. DNA helicase assays were performed with 5 fmol of HS-3′T substrate. The amounts of E. coli SSB and mthRPA added to reactions were as indicated.
Next, the polarity of translocation of mthMCM helicase activity along DNA was determined, using three different DNA templates (Fig. 5B). One template contained a 32P-labeled oligonucleotide (HS-M) annealed to the central region of the long 98-mer, resulting in the presence of both 5′ and 3′ ssDNA tails (lanes 1–5). The other two 32P-labeled oligonucleotides were annealed to either the 3′ (HS-3′T) or 5′ (HS-5′T) ends of the 98-mer, resulting in a partial duplex containing either a 5′ (lanes 6–10) or a 3′ ssDNA tail (lanes 11–15), respectively. As shown in Fig. 5B, the mthMCM protein displaced the duplex only when a 3′ ssDNA tail was available (lanes 2–5 and 13–15). No displacement was detected with the DNA substrate that contained only the 5′ ssDNA tail. These results indicate that the polarity of translocation of the archaeal MCM protein on DNA is in the 3′-to-5′ direction.
The effects of SSBs on the helicase activity were also examined. These experiments were carried out with either E. coli SSB or with the single-subunit RPA homologue identified in M. thermoautotrophicum (28). As shown in Fig. 5C, the presence of either SSB inhibited the helicase activity. The presence of SSB may block the access of the mthMCM protein to the substrate and thus inhibit its activity.
The processivity of the helicase activity was also examined by using ss M13 mp19 DNA that contained duplex regions between 34 and 1,000 bp in length. In the presence of 150 ng of protein (incubated for 60 min at 55°C), the maximal size of the DNA displaced was 200 nt (data not presented).
Discussion
In S. cerevisiae, ORC is bound to origins of replication throughout the cell cycle. In G1, ORC recruits additional replication proteins, including Cdc6p and the MCM family of proteins, to form the prereplicative complex. Finally, Cdc45p and the replication polymerases are recruited to the origin complex. At the initiation of S phase, Cdc6p is degraded and the MCM proteins and Cdc45p were found to move with the DNA polymerases, presumably as part of the replication fork. These events appear to be conserved in evolution, because similar observations have been made in Xenopus extracts with proteins and complexes homologous to those found in S. cerevisiae. In both yeast and Xenopus, the association of the protein components with chromatin follows the above sequence of events (9, 15).
As described above, we have demonstrated that the single 76-kDa mthMCM protein, isolated after its overexpression in E. coli, exhibits biochemical properties similar to those of the MCM proteins isolated from eukaryotic cells. A complex of hMCM4, -6, and -7 proteins and the spMCM4, -6, and -7 proteins, as well as the mthMCM protein, has ssDNA-binding activity, DNA-dependent ATPase activity, and 3′-to-5′ DNA helicase activity.
These observations are in accord with the proposal that the MCM complex may function as the replicative helicase responsible for unwinding of duplex DNA during chromosomal DNA replication, analogous to that of the E. coli DnaB protein and the simian virus 40 T Ag. Both DnaB and T Ag are single polypeptides that form double-hexameric rings at the origin and encircle DNA (18, 19).
Gel-filtration and glycerol gradient analyses, indicate that the archaeal MCM protein can exist in two major states, either as a monomer or as an oligomer as large as a 12-mer or possibly as a double hexamer. Both forms of the protein contain comparable DNA helicase activity but differ in their level of ssDNA-dependent ATPase activity. Because monomers showed a 6-fold higher specific activity than multimers, it may be that a hexameric form possesses a level of ATPase activity equivalent to the activity of the monomer. The activities associated with the multiple forms of the mthMCM protein are somewhat analogous to those observed with T Ag. Studies with T Ag demonstrated that both monomeric and hexameric forms contain DNA helicase activity. Incubation of the monomeric T Ag with ATP results in the production of multimers and incubation of hexameric T Ag with Mg2+ in the absence of ATP results in the formation of the monomer (31). Similar experiments carried out with the two different forms of the mthMCM protein, however, failed to detect alterations in either structure (data not presented).
In E. coli, the DnaB helicase alone cannot assemble itself around the DNA at the origin. The DnaB hexamer is loaded onto DNA by the DnaC protein in an ATP-dependent reaction. Genetic data in yeast suggest that in eukaryotes the MCM proteins may be loaded onto the DNA and that this reaction requires ORC and Cdc6p (14, 16, 32). In M. thermoautotrophicum, two polypeptides with amino acid sequences related to Cdc6p and/or ORC1p have been identified (ORFs MTH1412 and MTH1599 (24)). Thus, it is possible that one of these proteins is the functional homologue of ORC, whereas the other is the homologue of Cdc6p. When the effects of these two M. thermoautotrophicum proteins on the activities associated with mthMCM were examined, it was shown that both inhibited the helicase activity without affecting the ssDNA-binding and ATPase activities. Weak interaction between MTH1599 and mthMCM was detected by immunoprecipitation. In addition, incubation of the multimeric mthMCM complex with MTH1599 resulted in the dissociation of the multimeric complex and a heterogeneous distribution of the mthMCM protein determined by glycerol gradient centrifugation (data not presented).
The studies described here suggest that the mechanism of replication carried out by archaea may be less complicated than the replication process in eukaryotes. The fact that the mthMCM multimeric complex is composed of one protein rather than six (as in eukaryotes) may help elucidate the function of the eukaryotic MCM proteins. Previous studies with M. thermoautotrophicum replication factor C (RFC) also indicated that the clamp loading complex is composed of only two different protein subunits rather than five distinct subunits found in eukaryotic RFC. These findings suggest that a study of the DNA replication pathway in archaea may contribute importantly to our understanding of eukaryotic replication.
Acknowledgments
We thank Dr. John Reeve for providing us with Mth genomic DNA. This work was supported by National Institutes of Health Grant GM 38559 to J.H., and J.H. is a Professor of the American Cancer Society.
Abbreviations
- ORC
origin recognition complex
- sp-
Schizosaccharomyces pombe
- h-
human
- MCM
minichromosome maintenance
- mth-
Methanobacterium thermoautotrophicum ΔH
- ss
single-stranded
- ds
double-stranded
- T Ag
simian virus 40 large tumor antigen
- SSB
ssDNA-binding protein
- RPA
replication protein A
- ATP-γ-S
γ-thio-ATP
References
- 1.Maine G T, Sinha P, Tye B K. Genetics. 1984;106:365–385. doi: 10.1093/genetics/106.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kearsey S E, Labib K. Biochim Biophys Acta. 1998;1398:113–136. doi: 10.1016/s0167-4781(98)00033-5. [DOI] [PubMed] [Google Scholar]
- 3.Chong J P, Thommes P, Blow J J. Trends Biochem Sci. 1996;21:102–106. [PubMed] [Google Scholar]
- 4.Burkhart R, Schulte D, Hu D, Musahl C, Gohring F, Knippers R. Eur J Biochem. 1995;228:431–438. [PubMed] [Google Scholar]
- 5.Ishimi Y, Ichinose S, Omori A, Sato K, Kimura H. J Biol Chem. 1996;271:24115–24122. doi: 10.1074/jbc.271.39.24115. [DOI] [PubMed] [Google Scholar]
- 6.Sherman D A, Pasion S G, Forsburg S L. Mol Biol Cell. 1998;9:1833–1845. doi: 10.1091/mbc.9.7.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adachi Y, Usukura J, Yanagida M. Genes Cells. 1997;2:467–479. doi: 10.1046/j.1365-2443.1997.1350333.x. [DOI] [PubMed] [Google Scholar]
- 8.Chong J P, Mahbubani H M, Khoo C Y, Blow J J. Nature (London) 1995;375:418–421. doi: 10.1038/375418a0. [DOI] [PubMed] [Google Scholar]
- 9.Tye B K. Annu Rev Biochem. 1999;68:649–686. doi: 10.1146/annurev.biochem.68.1.649. [DOI] [PubMed] [Google Scholar]
- 10.Kearsey S E, Maiorano D, Holmes E C, Todorov I T. BioEssays. 1996;18:183–190. doi: 10.1002/bies.950180305. [DOI] [PubMed] [Google Scholar]
- 11.Ishimi Y. J Biol Chem. 1997;272:24508–24513. doi: 10.1074/jbc.272.39.24508. [DOI] [PubMed] [Google Scholar]
- 12.Ishimi Y, Komamura Y, You Z, Kimura H. J Biol Chem. 1998;273:8369–8375. doi: 10.1074/jbc.273.14.8369. [DOI] [PubMed] [Google Scholar]
- 13.Aparicio O M, Weinstein D M, Bell S P. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka T, Knapp D, Nasmyth K. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- 15.Dutta A, Bell S P. Annu Rev Cell Dev Biol. 1997;13:293–332. doi: 10.1146/annurev.cellbio.13.1.293. [DOI] [PubMed] [Google Scholar]
- 16.Donovan S, Harwood J, Drury L S, Diffley J F. Proc Natl Acad Sci USA. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou L, Stillman B. Science. 1998;280:593–596. doi: 10.1126/science.280.5363.593. [DOI] [PubMed] [Google Scholar]
- 18.Baker T A, Bell S P. Cell. 1998;92:295–305. doi: 10.1016/s0092-8674(00)80923-x. [DOI] [PubMed] [Google Scholar]
- 19.Leatherwood J. Curr Opin Cell Biol. 1998;10:742–748. doi: 10.1016/s0955-0674(98)80117-8. [DOI] [PubMed] [Google Scholar]
- 20.Hurwitz J, Dean F B, Kwong A D, Lee S H. J Biol Chem. 1990;265:18043–18046. [PubMed] [Google Scholar]
- 21.Woese C R, Fox G E. Proc Natl Acad Sci USA. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, et al. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 23.Klenk H P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, et al. Nature (London) 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 24.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, et al. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawarabayasi Y, Hino Y, Horikawa H, Yamazaki S, Haikawa Y, Jin-no K, Takahashi M, Sekine M, Baba S, Ankai A, et al. DNA Res. 1999;6:83–101. doi: 10.1093/dnares/6.2.83. , 145–152. [DOI] [PubMed] [Google Scholar]
- 26.Kawarabayasi Y, Sawada M, Horikawa H, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, et al. DNA Res. 1998;5:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- 27.Zeikus J G, Wolfe R S. J Bacteriol. 1972;109:707–715. doi: 10.1128/jb.109.2.707-713.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelman Z, Pietrokovski S, Hurwitz J. J Biol Chem. 1999;274:28751–28761. doi: 10.1074/jbc.274.40.28751. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 30.McEntee K, Weinstock G M, Lehman I R. Proc Natl Acad Sci USA. 1980;77:857–861. doi: 10.1073/pnas.77.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dean F B, Borowiec J A, Eki T, Hurwitz J. J Biol Chem. 1992;267:14129–14137. [PubMed] [Google Scholar]
- 32.Perkins G, Diffley J F. Mol Cell. 1998;2:23–32. doi: 10.1016/s1097-2765(00)80110-0. [DOI] [PubMed] [Google Scholar]