Abstract
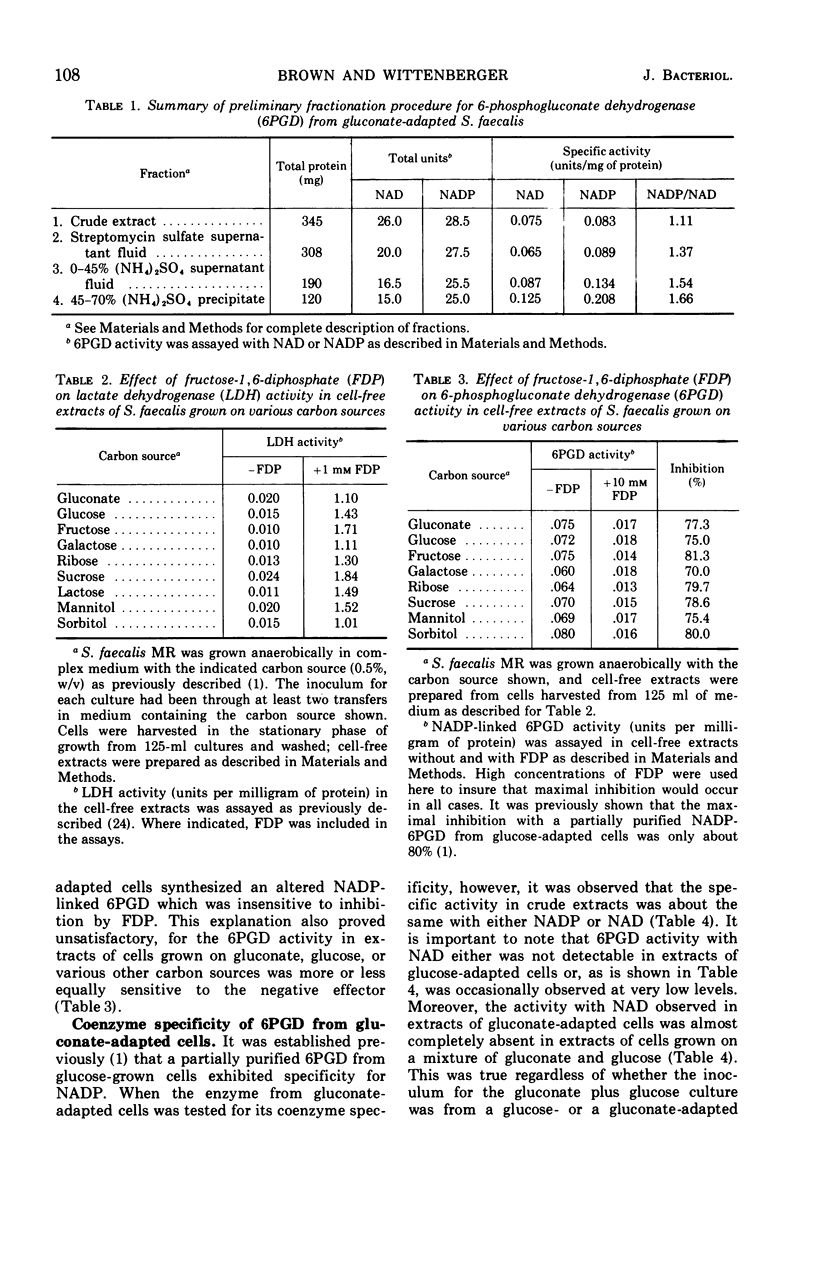
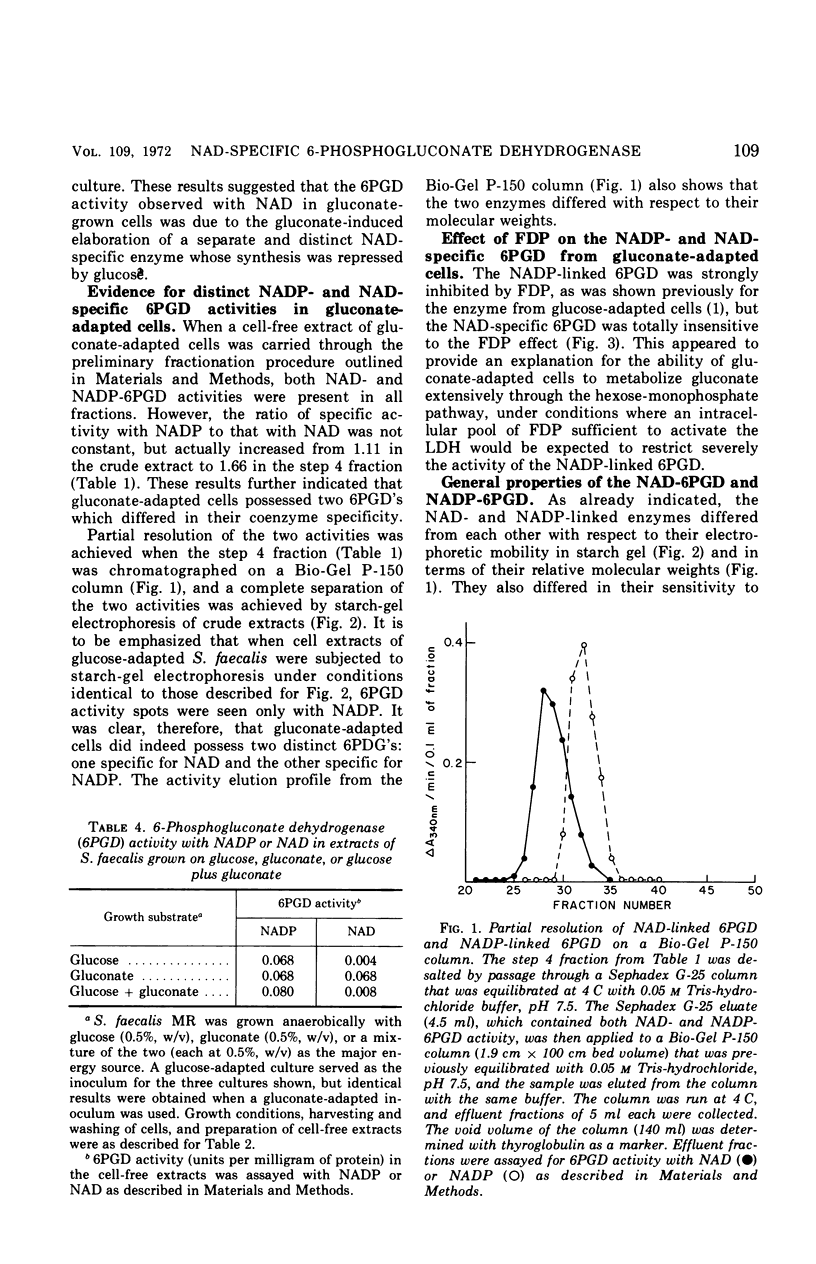
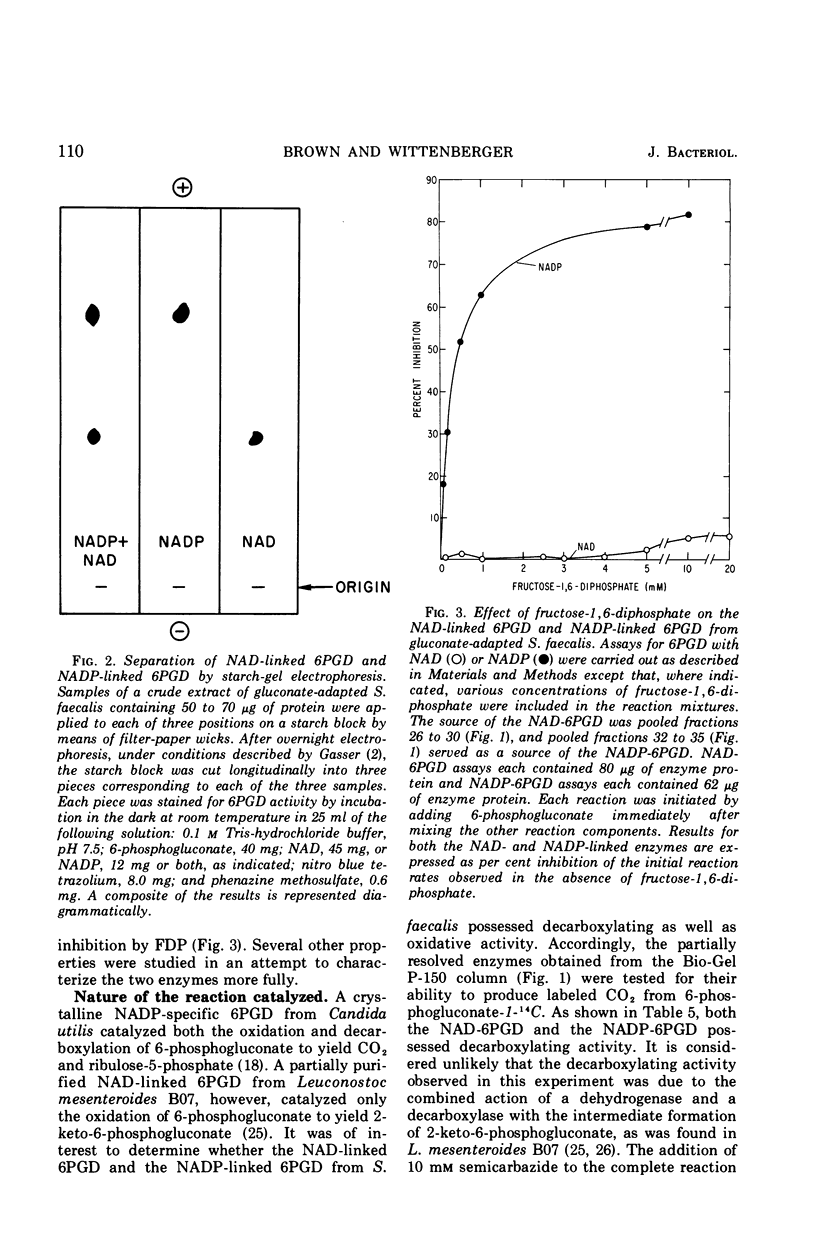
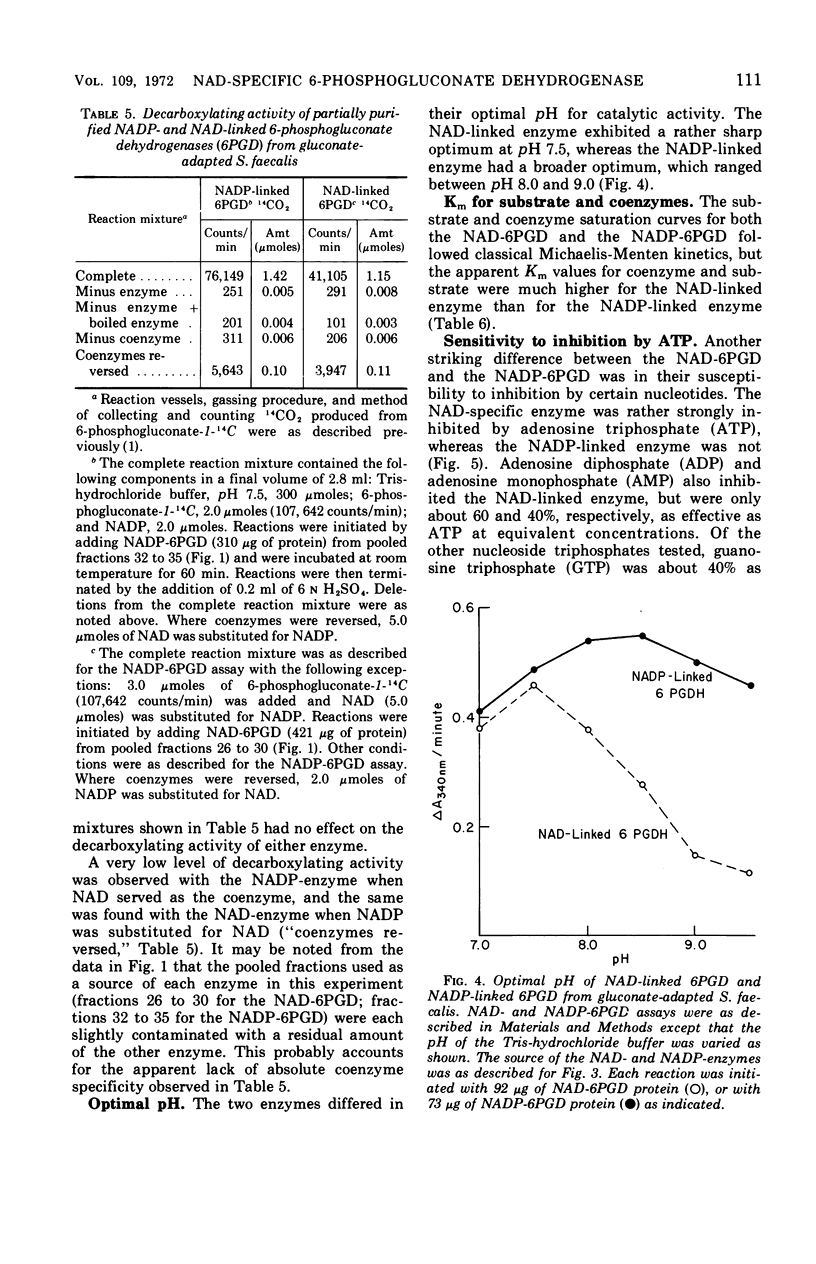
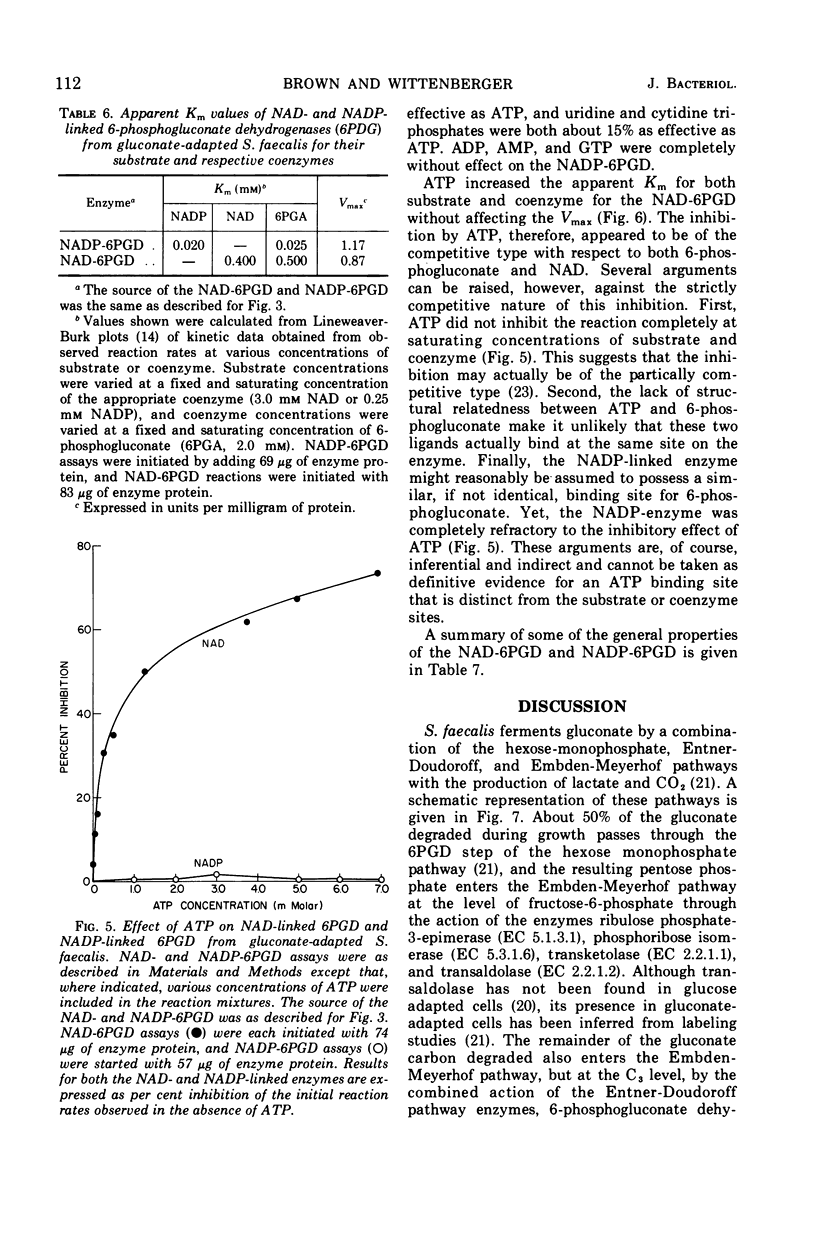
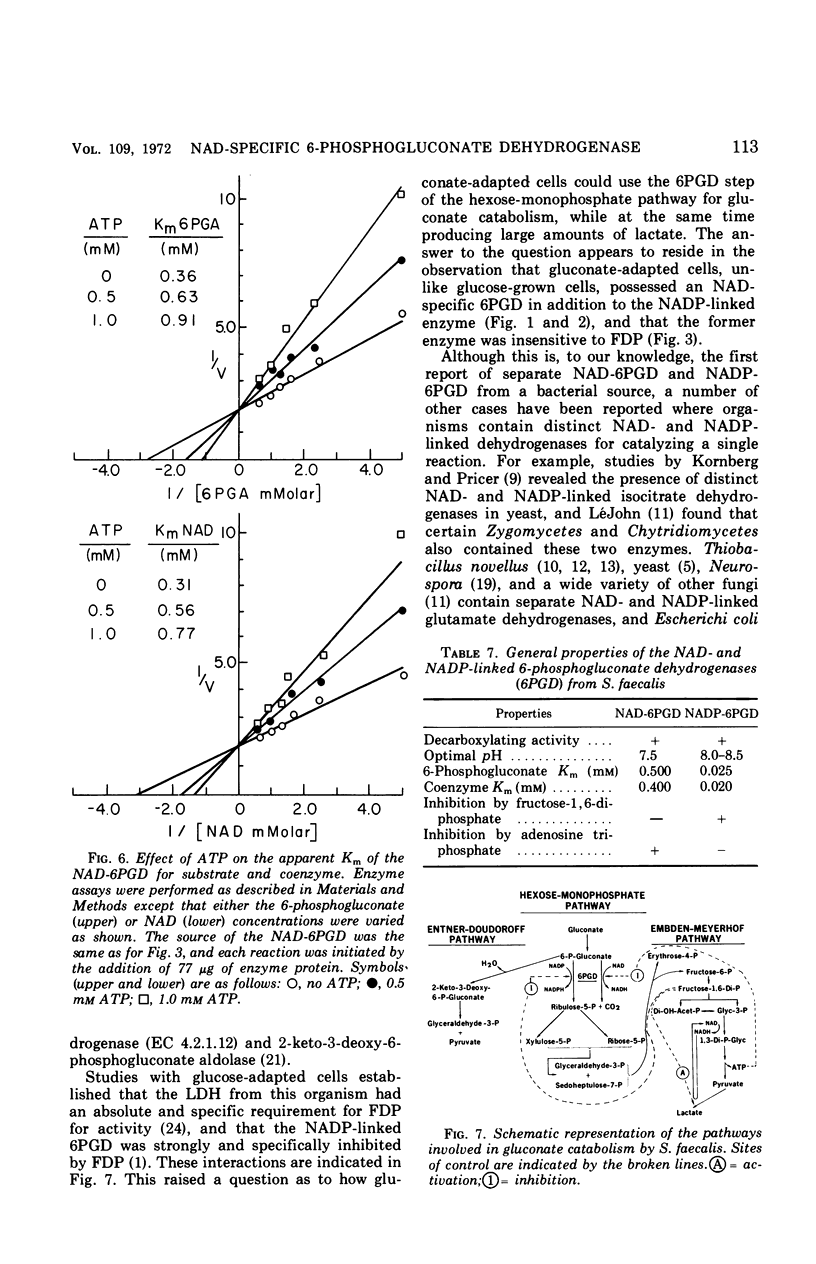
Streptococcus faecalis grown with glucose as the primary energy source contains a single, nicotinamide adenine dinucleotide phosphate (NADP)-specific 6-phosphogluconate dehydrogenase. Extracts of gluconate-adapted cells, however, exhibited 6-phosphogluconate dehydrogenase activity with either NADP or nicotinamide adenine dinucleotide (NAD). This was shown to be due to the presence of separate enzymes in gluconate-adapted cells. Although both enzymes catalyzed the oxidative decarboxylation of 6-phosphogluconate, they differed from one another with respect to their coenzyme specificity, molecular weight, pH optimum, Km values for substrate and coenzyme, and electrophoretic mobility in starch gels. The two enzymes also differed in their response to certain effector ligands. The NADP-linked enzyme was specifically inhibited by fructose-1,6-diphosphate, but was insensitive to adenosine triphosphate (ATP) and certain other nucleotides. The NAD-specific enzyme, in contrast, was insensitive to fructose-1,6-diphosphate, but was inhibited by ATP. The available data suggest that the NAD enzyme is involved primarily in the catabolism of gluconate, whereas the NADP enzyme appears to function in the production of reducing equivalents (NADPH) for use in various reductive biosynthetic reactions.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown A. T., Wittenberger C. L. Mechanism for regulating the distribution of glucose carbon between the Embden-Meyerhof and hexose-monophosphate pathways in Streptococcus faecalis. J Bacteriol. 1971 May;106(2):456–467. doi: 10.1128/jb.106.2.456-467.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GIBBS M., SOKATCH J. T., GUNSALUS I. C. Product labeling of glucose-1-C14 fermentation by homofermentative and heterofermentative lactic acid bacteria. J Bacteriol. 1955 Nov;70(5):572–576. doi: 10.1128/jb.70.5.572-576.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser F. Electrophoretic characterization of lactic dehydrogenases in the genus Lactobacillus. J Gen Microbiol. 1970 Aug;62(2):223–239. doi: 10.1099/00221287-62-2-223. [DOI] [PubMed] [Google Scholar]
- HOLZER H., SCHNEIDER S. Anreicherung und Trennung einer DPN-spezifischen und einer TPN-spezifischen Glutaminsäure-dehydrogenase aus Hefe. Biochem Z. 1957;329(5):361–369. [PubMed] [Google Scholar]
- KAPLAN N. O. Symposium on multiple forms of enzymes and control mechanisms. I. Multiple forms of enzymes. Bacteriol Rev. 1963 Jun;27:155–169. doi: 10.1128/br.27.2.155-169.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KORNBERG A., PRICER W. E., Jr Di- and triphosphopyridine nucleotide isocitric dehydrogenases in yeast. J Biol Chem. 1951 Mar;189(1):123–136. [PubMed] [Google Scholar]
- Katsuki H., Takeo K., Kameda K., Tanaka S. Existence of two malic enzymes in Escherichia coli. Biochem Biophys Res Commun. 1967 May 5;27(3):331–336. doi: 10.1016/s0006-291x(67)80102-5. [DOI] [PubMed] [Google Scholar]
- London J., Meyer E. Y. Malate utilization by a group D Streptococcus: regulation of malic enzyme synthesis by an inducible malate permease. J Bacteriol. 1970 Apr;102(1):130–137. doi: 10.1128/jb.102.1.130-137.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LéJohn H. B. AMP-activation of an allosteric NAD -dependent glutamate dehydrogenase. Biochem Biophys Res Commun. 1967 Jul 10;28(1):96–102. doi: 10.1016/0006-291x(67)90412-3. [DOI] [PubMed] [Google Scholar]
- LéJohn H. B. Enzyme regulation, lysine pathways and cell wall structures as indicators of major lines of evolution in fungi. Nature. 1971 May 21;231(5299):164–168. doi: 10.1038/231164a0. [DOI] [PubMed] [Google Scholar]
- LéJohn H. B., McCrea B. E. Evidence for two species of glutamate dehydrogenases in Thiobacillus novellus. J Bacteriol. 1968 Jan;95(1):87–94. doi: 10.1128/jb.95.1.87-94.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai T., Tokushige M., Nagai J., Katsuki H. Physiological functions of NAD- and NADP-linked malic enzymes in Escherichia coli. Biochem Biophys Res Commun. 1971 May 21;43(4):875–881. doi: 10.1016/0006-291x(71)90698-x. [DOI] [PubMed] [Google Scholar]
- PLATT T. B., FOSTER E. M. Products of glucose metabolism by homofermentative streptococci under anaerobic conditions. J Bacteriol. 1958 Apr;75(4):453–459. doi: 10.1128/jb.75.4.453-459.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PONTREMOLI S., DE FLORA A., GRAZI E., MANGIAROTTIG, BONSIGNORE A., HORECKER B. L. Crystalline D-gluconate 6-phosphate dehydrogenase. J Biol Chem. 1961 Nov;236:2975–2980. [PubMed] [Google Scholar]
- SANWAL B. D., LATA M. The occurrence of two different glutamic dehydrogenases in Neurospora. Can J Microbiol. 1961 Jun;7:319–328. doi: 10.1139/m61-039. [DOI] [PubMed] [Google Scholar]
- SOKATCH J. T., GUNSALUS I. C. Aldonic acid metabolism. I. Pathway of carbon in an inducible gluconate fermentation by Streptococcus faecalis. J Bacteriol. 1957 Apr;73(4):452–460. doi: 10.1128/jb.73.4.452-460.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeo K., Murai T., Nagai J., Katsuki H. Allosteric activation of DPN-linked malic enzyme from Escherichia coli by aspartate. Biochem Biophys Res Commun. 1967 Dec 15;29(5):717–722. doi: 10.1016/0006-291x(67)90276-8. [DOI] [PubMed] [Google Scholar]
- Wittenberger C. L., Angelo N. Purificationa and properties of a fructose-1,6-diphosphate-activated lactate dehydrogenase from Streptococcus faecalis. J Bacteriol. 1970 Mar;101(3):717–724. doi: 10.1128/jb.101.3.717-724.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]


