Abstract
Two-component systems, sensor kinase-response regulator pairs, dominate bacterial signal transduction. Regulation is exerted by phosphorylation of an Asp in receiver domains of response regulators. Lability of the acyl phosphate linkage has limited structure determination for the active, phosphorylated forms of receiver domains. As assessed by both functional and structural criteria, beryllofluoride yields an excellent analogue of aspartyl phosphate in response regulator NtrC, a bacterial enhancer-binding protein. Beryllofluoride also appears to activate the chemotaxis, sporulation, osmosensing, and nitrate/nitrite response regulators CheY, Spo0F, OmpR, and NarL, respectively. NMR spectroscopic studies indicate that beryllofluoride will facilitate both biochemical and structural characterization of the active forms of receiver domains.
Together with their cognate autokinases, response regulators dominate signal transduction in bacteria (1–5) and are also found upstream of protein kinase cascades in eukarya (6–9). Autokinase/response regulator pairs, which are referred to as “two-component” systems, control bacterial cell division (10), development (11), chemotaxis (12), virulence (13–16), and the responses to many changes in nutrient availability (17, 18). Phosphorylation of an Asp in receiver domains of response regulators is used to modulate the function of their corresponding output domains (19), many of which activate or repress transcription (2, 3). The structures of six nonphosphorylated receiver domains have been determined (20–25), and it has been shown that a substantial conformational change occurs after phosphorylation in several cases (26–28). However, the lability of the acyl phosphate linkages in these domains (half-lives of seconds to hours) (2, 3) has hindered structural studies of their phosphorylated active forms.
The response regulator nitrogen regulatory protein C (NtrC), an enhancer-binding protein, functions as a molecular machine to activate transcription by the σ54-holoenzyme form of RNA polymerase (29–31). The NtrC protein of Salmonella typhimurium is composed of three functional domains (32–34): an (amino) N-terminal receiver or regulatory domain that is phosphorylated on Asp-54 (D54), a central output domain that hydrolyzes ATP and activates transcription, and a C-terminal DNA-binding domain. The central domain of NtrC apparently adopts a mononucleotide-binding fold characteristic of a large group of purine nucleotide-binding proteins (35, 36). Phosphorylation of D54 allows NtrC to form large oligomers that are essential for ATP hydrolysis and hence, transcriptional activation (30, 31, 37, 38).
The receiver domain of NtrC (NtrCr) is the only such domain for which the structures of both the phosphorylated and nonphosphorylated forms have been determined (D. Kern, B. F. Volkman, P. Luginbuhl, M. J. Nohaile, S.K., and D.E.W., unpublished material; ref. 21). Although it was possible to maintain phosphorylated NtrCr (P-NtrCr) for long enough to allow determination of its structure by NMR spectroscopy, methods for doing so and for collecting and analyzing the necessary data were nontrivial. We report here that a complex formed between NtrC and beryllofluoride (BeFx) is a fully active analog of P-NtrC, and we present evidence that the same is probably true of BeFx complexes with other response regulators.
Materials and Methods
Phosphate Analogs.
BeFx and aluminofluoride were generated in situ with NaF in the millimolar range and BeCl2 or AlCl3 in the micromolar range (39, 40). Orthovanadate (VO43−) was prepared as described and used in the millimolar range (41). To demonstrate that phosphate analogues had been formed correctly, we showed that all of the above compounds inhibited the ATPase activity of myosin subfragment 1 (Sigma) in the presence of ADP (D.Y. and S.K., unpublished material).
NtrC Proteins.
NtrCD86N, S160F combines two constitutive amino acid substitutions and is our most active constitutive NtrC protein; i.e., it has the highest activity in the absence of phosphorylation (D.Y. and S.K., unpublished material; ref. 42). Phosphorylation further increases its activity. All full-length NtrC proteins (dimer concentration) were maltose-binding protein fusions to NtrC lacking its two natural cysteines (MBP-NtrCC30V, C364A) or its derivatives. MBP-NtrCC30V, C364A has been shown to be as active as native NtrC protein (43, 44).
Assays.
Assays were performed as described: ATPase activity of NtrC (42); transcriptional activation of NtrC (42); Fe-mediated cleavage of NtrC (J. Lee, J. Owens, I. Hwang, C. Meares, and S.K., unpublished material; ref. 45); DNA-binding of OmpR (46) and NarL (47, 48); and FliM peptide binding of CheY (49).
NMR Chemical Shift.
NtrCr, CheY, and Spo0F were expressed from plasmid vectors in Escherichia coli strain BL21 (DE3) and purified by a combination of ion-exchange chromatography and HPLC. Mass spectroscopy verified that each had the correct molecular mass and was >95% pure. For isotope labeling, cells were grown in minimal medium (M9) supplemented with trace metals and biotin and containing labeled [13C]glucose and/or [15N]ammonium chloride (2 g/liter). NMR assignments were obtained by using heteronuclear single-quantum correlation spectroscopy (HSQC), amide proton and nitrogen to Cα carbon correlation experiment (HNCA), and 15N-resolved nuclear overhauser effect spectroscopy (NOESY) for NtrCr, and HSQC, amide proton and nitrogen to Cα and Cβ carbon correlation experiment (HNCACB), HNCA, Cb and Cβ carbon to amide nitrogen and proton via carbonyl carbon correlation experiment (CBCACONH), and [15N]NOESY for CheY.
Results
Functional Evidence That BeFx Activates NtrC.
In the process of determining whether well-studied ATP analogs (41, 50, 51) could inhibit the ATPase activity of S. typhimurium NtrC, we tested their effects on a constitutive mutant form of NtrC, a form that has the capacity to hydrolyze ATP and activate transcription without being phosphorylated (29–31, 42). In contrast to the case for many other purine nucleotide-binding proteins with a mononucleotide fold (39–41, 50, 51), neither ADP⋅BeFx, ADP⋅aluminofluoride, nor ADP⋅VO43− appeared to inhibit the ATPase activity of NtrCD86N, S160F. Strikingly, BeFx, but not aluminofluoride or VO43−, substantially stimulated the ATPase activity of nonphosphorylated NtrCD86N, S160F (D.Y. and S.K., unpublished material). This led us to postulate that BeFx might be forming an acyl phosphate analogue in the receiver or regulatory domain of NtrCD86N, S160F, which would stimulate its constitutive ATPase activity.
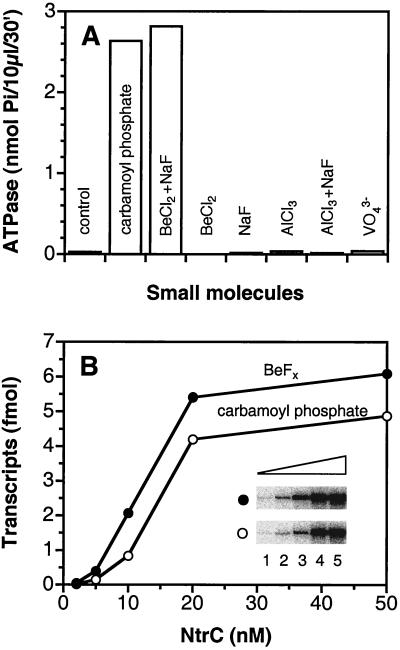
To test the capacity of BeFx to substitute for phosphorylation of NtrC, we compared the effects of BeFx on wild-type NtrC (NtrCWT) to those of the low molecular weight phosphodonor carbamoyl phosphate (52, 53). Receiver domains autophosphorylate from low molecular weight phosphodonors such as carbamoyl phosphate, acetyl phosphate, or phosphoramidate (52, 53) and also catalyze their own dephosphorylation (2, 3). BeFx, which is formed in situ from BeCl2 and NaF (54, 55), was as effective as carbamoyl phosphate in stimulating the ATPase activity of NtrCWT (Fig. 1A). Both BeCl2 and NaF were required and neither aluminofluoride nor VO43− could substitute. Furthermore, BeFx allowed NtrCWT to activate transcription by σ54-holoenzyme, its ultimate biological output (Fig. 1B). Impressively, the activity of NtrCWT in the presence of ≤100 μM BeFx was at least as great as that of the phosphorylated protein (P-NtrCWT) in the presence of 10 mM carbamoyl phosphate. It has been shown that carbamoyl phosphate yields in vitro activities of NtrCWT comparable to those observed when the protein is phosphorylated by its cognate protein phosphodonor, the histidine autokinase NtrB (53).
Figure 1.
Activation of NtrCWT by BeFx. (A) The ATPase activity of NtrCWT (1 μM) was assayed in Hepes buffer, pH 7.3 by monitoring the release of Pi from [γ-32P]ATP at 25°C for 30 min as described (42). Before addition of ATP, other components were incubated for 30 min at 25°C together with the indicated small molecule or combination of small molecules (10 mM carbamoyl phosphate/200 μM BeCl2/200 μM AlCl3/5 mM NaF/1 mM VO43−). (B) Formation of open complexes was assessed in a single-cycle transcription assay on a supercoiled plasmid template pJES534 (1 nM) (37, 42). NtrC was incubated with 100 μM BeCl2 plus 10 mM NaF (●) or 10 mM carbamoyl phosphate (○) at 37°C for 10 min before initiation of the reaction with ATP. (Inset) glnA transcripts. Lanes 1–5, NtrCWT (2, 5, 10, 20, and 50 nM, respectively).
Characterization of BeFx and Its Interaction with NtrC.
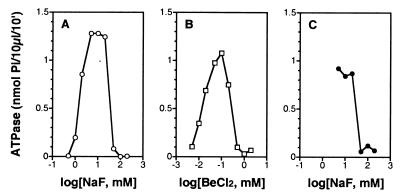
To determine a probable number for x in the activating species BeFx, we tested the effects of different concentrations of NaF on the ATPase activity of NtrCWT in the presence of a fixed concentration of BeCl2 (50 μM) (54, 55). Depending on the excess of F−, different species of BeFx usually predominate in aqueous solution. No ATPase activity was observed for NtrCWT until the concentration of NaF reached 1 mM (Fig. 2A), and maximum activity was reached between 5 and 20 mM NaF. Based on previous evidence (54, 55), (H2O)BeF3− is likely to be the species that yields the active form of NtrC. However, for both theoretical and technical reasons (see below), we cannot exclude BeF42− as an activating species. Whatever the activating species of BeFx, we postulate that it is likely to form an acyl phosphate analog of NtrC, with a bond to an oxygen of the β-carboxyl group of D54. That regulatory effects of BeFx are exerted through D54, as are those of phosphorylation, is indicated by the failure of BeFx to stimulate the ATPase activity of mutant forms of NtrC in which D54 has been replaced by A or N (data not shown).
Figure 2.
Response of the ATPase activity of NtrC to the concentration of NaF or BeCl2. ATPase activity was assessed essentially as described in Fig. 1A, except the reaction time was 10 min. (A) Dependence of the ATPase activity of NtrCWT (0.4 μM) on the concentration of NaF (0.5–200 mM) in the presence of 50 μM BeCl2. (B) Dependence of the ATPase activity of NtrCWT (0.4 μM) on the concentration of BeCl2 (5–2,000 μM) in the presence of 5 mM NaF. (C) Response of the ATPase activity of NtrCD86N, S160F (0.2 μM) to the concentration of NaF (5–200 mM) in the absence of BeCl2.
To assess the affinity of NtrCWT for BeFx, we determined the ATPase activity of the protein at a fixed concentration of NaF (5 mM), at which the most abundant species of BeFx is probably (H2O)BeF3− (54, 55), and different concentrations of BeCl2 (Fig. 2B). ATPase activity was detected even at 5 μM BeCl2, the lowest concentration tested, which was only about sixfold higher than the concentration of NtrC (0.8 μM monomer). The highest ATPase activity was achieved at 100 μM BeCl2. In both the dose-response curves to BeCl2 (Fig. 2B) and NaF (Fig. 2A), inhibition of the ATPase activity of NtrC occurred at high concentrations (≥200 μM BeCl2 or ≥50 mM NaF). Additional experiments indicated that either trace amounts of BeCl2 alone (D.Y. and S.K., unpublished material) or high concentrations of NaF (Fig. 2C) inhibited the ATPase activity of the constitutive form NtrCD86N, S160F. Inhibitory effects of high concentrations of NaF on the ATPase activity of NtrC precluded our determining whether BeF42− might be an activating species (see above).
Structural Evidence That BeFx Activates NtrC.
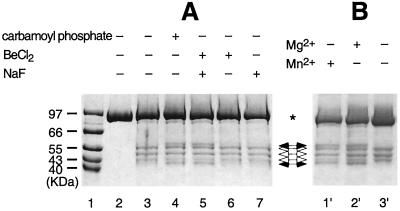
We have two direct lines of evidence that BeFx mimics effects of phosphorylation of NtrC structurally as well as functionally. First, BeFx yields an otherwise phosphorylation-dependent cleavage of NtrCD86C that has been derivatized with an iron chelate (NtrCD86C, Fe) (Fig. 3) (J. Lee, J. Owens, I. Hwang, C. Meares, and S.K., unpublished material; ref. 45). This cleavage, which occurs outside the N-terminal receiver domain of NtrC, is indicative of a conformational change in the protein after activation; this allows the N-terminal domain of one monomer to contact the central or output domain of the opposite monomer in a dimer. Rearrangement at the dimer interface of P-NtrC is postulated to drive the formation of active oligomers. Unlike the ATPase activity of NtrC or its capacity to activate transcription, Fe-mediated protein cleavage does not depend on a divalent cation (J. Lee, J. Owens, I. Hwang, C. Meares, and S.K., unpublished material; ref. 45), and the two phosphorylation- and BeFx-independent cleavages that occur within the receiver domain of NtrC did not require one. By contrast, the BeFx-dependent cleavage did require a divalent cation, and either Mg2+ or Mn2+ could serve (Fig. 3B); thus, like phosphorylation (2, 3, 52, 53), the interaction between BeFx and D54 of NtrC requires a divalent cation. The second line of evidence that the complex between BeFx and NtrC (BeFx⋅NtrC) mimics P-NtrC structurally is provided by NMR spectroscopic studies of BeFx complexed with NtrCr (see below).
Figure 3.
Fe-mediated cleavage of NtrCD86C, Fe. Dependence on BeFx or carbamoyl phosphate (A) and requirement of divalent cation for the BeFx-dependent cleavage (B). NtrC (5–10 μM) was incubated for 30 min at 25°C with the indicated small molecule or combination of small molecules (10 mM carbamoyl phosphate/200 μM BeCl2/10 mM NaF) in cleavage buffer (10 mM Mops, pH 8.0/50 mM KCl/8 mM MgCl2/5% glycerol/1 mM EDTA). Cleavage was then initiated by adding H2O2 and sodium ascorbate to final concentrations of 50 mM, and samples were subjected to analysis by SDS/PAGE as described (J. Lee, J. Owens, I. Hwang, C. Meares, and S.K., unpublished material; ref. 45). Lane 1, molecular weight markers; lane 2, NtrCD86C; lanes 3–7 and 1′-3′, NtrCD86C derivatized with the cysteine-specific Fe chelate (1-[p-(bromoacetamido)benzyl]-EDTA-Fe) (J. Lee, J. Owens, I. Hwang, C. Meares, and S.K., unpublished material; ref. 45). The sample in lane 2′ contained the same components as that in lane 5. MgCl2 was eliminated from the sample in lane 3′ and was replaced with 8 mM MnCl2 in the sample of lane 1′. *, full-length NtrC; (➞), phosphorylation-dependent cleavage bands (two); (➩), phosphorylation-independent cleavage bands. (Although there are four bands total, produced from two independent cleavages, only three bands are visible on this gel (J. Lee, J. Owens, I. Hwang, C. Meares, and S.K., unpublished material).
Evidence That BeFx Activates Other Response Regulators/Receiver Domains.
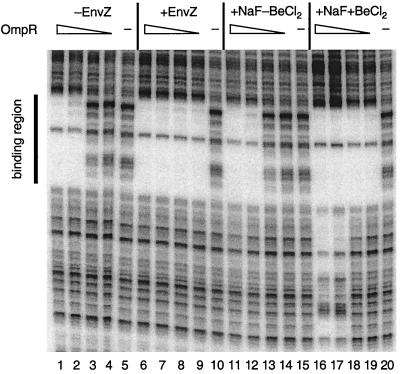
To determine whether effects of BeFx on NtrC could be generalized to other response regulators/receiver domains, we studied four additional proteins: E. coli OmpR and NarL, which activate and repress transcription by σ70-holoenzyme; E. coli CheY, which is a central regulator of the chemotactic response; and Bacillus subtilis Spo0F, which is part of the phosphorelay-controlling initiation of sporulation. OmpR and NarL are two-domain response regulators. OmpR controls expression of outer membrane porin proteins in response to osmolarity and a variety of other signals that are poorly defined (refs. 56 and 57; M.M.I., unpublished material), whereas NarL controls gene expression in response to availability of the respiratory oxidants nitrate and nitrite (58). Although phosphorylation of OmpR affects its DNA-binding activity (59), it is not clear whether the N-terminal domain of OmpR acts positively or negatively. However, unlike the case for NtrC, the N-terminal receiver domain of NarL apparently acts negatively: structural studies provide evidence that the nonphosphorylated receiver domain of NarL blocks DNA-binding by its C-terminal domain (22, 60). Protection from cleavage by DNase I indicated that BeFx⋅OmpR bound to the regulatory region immediately upstream of the ompF promoter better than P-OmpR, whether phosphorylation was achieved with EnvZ (Fig. 4) or with acetyl phosphate (data not shown). Protection from DNase I cleavage and enhancements of cleavage indicated that BeFx⋅NarL and P-NarL bound similarly to the control region for the fdnG operon (data not shown), but under conditions optimal for the activation of NtrC (200 μM BeCl2/10 mM NaF/5 mM MgCl2), BeFx⋅NarL was less active.
Figure 4.
BeFx stimulation of the binding of OmpR to the ompF promoter-regulatory region. Binding of BeFx⋅OmpR to the ompF promoter-regulatory region was assessed by DNase I footprinting as described (46) and was compared with that of P-OmpR. The concentrations of OmpR were: 6.4 μM (lanes 1, 6, 11, and 16); 1.6 μM (lanes 2, 7, 12, and 17); 0.4 μM (lanes 3, 8, 13, and 18); 0.1 μM (lanes, 4, 9, 14, and 19); and 0 μM (lanes 5, 10, 15, and 20). For samples in lanes 6–10, OmpR was phosphorylated by MBP-EnvZ (0.3 μM) as described (46). For samples in lanes 11–20, 10 mM NaF was present, and for those in lanes 16–20, 100 μM BeCl2 was also present. Note: the nonspecific DNA-binding observed in the presence of BeCl2 and NaF in lanes 16 and 17 was not observed when OmpR was phosphorylated by EnvZ (lanes 6 and 7) unless much higher concentrations of OmpR were used (M.M.I., unpublished material).
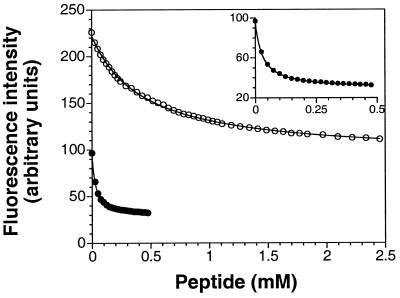
There is no direct in vitro assay for the activity of P-CheY in reversing the direction of flagellar rotation; hence, we monitored the effects of BeFx on CheY by measuring its binding to a peptide from FliM (49), a component of the switch that controls the direction of flagellar rotation. The strength of this binding, which was assessed by quenching of the intrinsic tryptophan fluorescence of CheY, was recently reported to be phosphorylation dependent (49). The dissociation constants for binding of BeFx⋅CheY and inactive CheY to the FliM peptide, 26 and 500 μM, respectively (Fig. 5), were the same as those reported for P-CheY and CheY. By this criterion, BeFx⋅CheY is as active as P-CheY. Like BeFx⋅CheY, a phosphonate derivative of CheYD57C appears to be a biochemical analogue of P-CheY, but it is difficult to prepare (61). Such analogues are extremely useful because the acyl phosphate linkage of P-CheY is exceptionally labile (half-life of seconds) (2, 3).
Figure 5.
BeFx stimulation of the binding of CheY to a FliM peptide (residues 1–16) (49). Fluorescence intensity of CheY (10 μM) at 340 nm as a function of added FliM peptide in the absence (○) or presence (● and Inset) of 2 mM BeCl2/20 mM NaF/20 mM MgCl2.
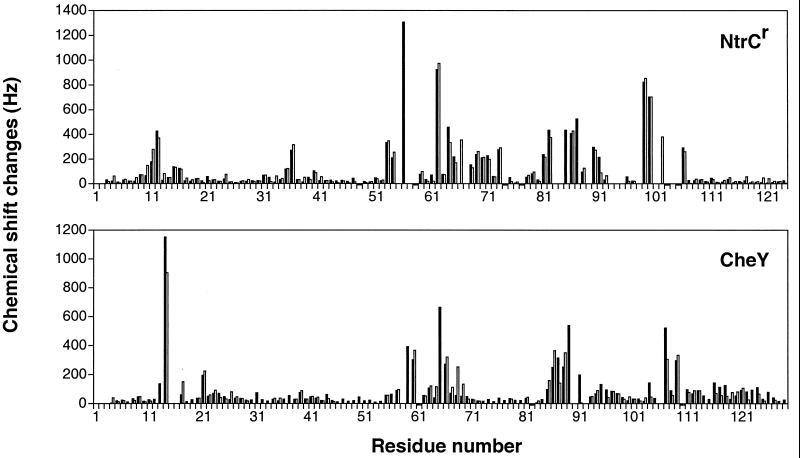
To assess the structural similarity of BeFx-activated receiver domains to the corresponding phosphorylated domains, we used NMR spectroscopy to evaluate the changes in chemical shift that resulted from these two means of activation of NtrCr, CheY, and Spo0F. Chemical shifts for isotope-labeled proteins have been reported for the unmodified inactive state of each protein (21, 23, 62, 63), and they have also been determined for P-NtrCr and P-CheY (refs. 27 and 28; D. Kern, B. F. Volkman, P. Luginbuhl, M. J. Nohaile, S.K., and D.E.W., unpublished material). Titrations of NtrCr and CheY with BeCl2 and NaF produced new resonances in the slow-exchange limit, coexisting with those of the inactive forms. By adjusting the concentrations of Be2+, F−, and Mg2+, it was possible to convert the proteins quantitatively to BeFx complexes. Resonances from the BeFx⋅NtrCr and BeFx⋅CheY complexes were assigned by using triple resonance three-dimensional experiments on 13C/15N-enriched proteins, making possible direct comparisons with the corresponding phosphorylated forms. The chemical shift differences between BeFx⋅NtrCr and NtrCr were remarkably similar to those between P-NtrCr and NtrCr (Fig. 6), particularly for the residues that showed the largest changes. Although chemical shift changes are notoriously difficult to interpret directly, the correlation in the patterns argues strongly that very similar structural changes are likely to occur after activation by either mechanism. In particular, it has been shown recently that there is a substantial reorganization of the face comprised of helices three and four and strands four and five in P-NtrCr (D. Kern, B. F. Volkman, P. Luginbuhl, M. J. Nohaile, S.K., and D.E.W., unpublished material) and based on changes in chemical shift, the same is likely to be true in BeFx⋅NtrCr.
Figure 6.
Chemical shift changes of amide nitrogens and protons, (ΔδN2 + ΔδH2)1/2, with Δδ in Hz, after activation of NtrCr or CheY. Solid bars indicate the difference between the BeFx-activated and inactive states of NtrCr or CheY, whereas open bars represent the phosphorylated and inactive states of NtrCr or CheY. Small negative bars designate Pro which has no NH.
Because of instability of the aspartyl phosphate, assignment of resonances for P-CheY was very difficult and the assignments are less complete than for unmodified CheY (27); nevertheless, after correcting for one very likely misassigned resonance in P-CheY that shifts substantially (27), profiles of the differences in chemical shift between BeFx⋅CheY and CheY are very similar to those between P-CheY and CheY (Fig. 6). Thus, for both NtrCr and CheY, differences in chemical shift for the BeFx complexes recapitulate those for the corresponding phosphorylated (activated) forms.
For Spo0F, it has not yet been possible to achieve quantitative phosphorylation (64) and hence, a comparison of NMR data for BeFx⋅Spo0F and P-Spo0F cannot be made; however, several of the distinctively shifted resonances that show large changes after activation of both NtrCr and CheY, show similar changes for BeFx⋅Spo0F (data not shown). This provides evidence that it, too, is in an active state, although biochemical studies will be required to verify this.
Discussion
BeFx forms an acyl phosphate analogue with all six of the response regulators we have tested: NtrC, OmpR, NarL, CheY, Spo0F, and DctD (D.Y. and B. T. Nixon, unpublished material), an activator of σ54-holoenzyme in which the receiver domain acts negatively (65). As assessed by both functional and structural criteria, BeFx forms an excellent aspartyl phosphate analogue in NtrC and appears to do so for CheY. Preliminary structural studies indicate that BeFx⋅Spo0F may mimic P-Spo0F, and functional studies indicate the same for BeFx⋅OmpR and BeFx⋅NarL. [1H–15N] heteronuclear single-quantum correlation spectroscopy spectra of BeFx complexes with NtrCr, CheY, and Spo0F were readily obtained and those of BeFx⋅NtrCr and BeFx⋅CheY showed changes in chemical shifts that are characteristic of the corresponding phosphorylated proteins (Fig. 6) (refs. 21, 27, 62, and 63; D. Kern, B. F. Volkman, P. Luginbuhl, M. J. Nohaile, S.K., and D.E.W., unpublished material). Hence, determination of the structures of BeFx complexes with a number of receiver domains will be straightforward, and comparison of these structures should allow discrimination between general and specific features of the conformational changes that occur after activation of different receiver domains. Moreover, active BeFx complexes of receiver domains should also facilitate study of their interactions with their corresponding output domains, their cognate autokinases/phosphatases, and other components in their signal transduction pathways (6–9, 66).
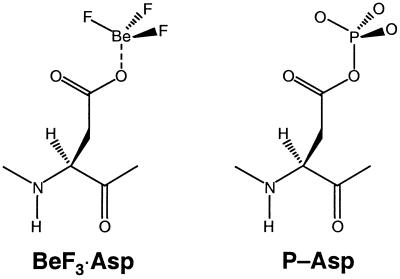
In retrospect, it is perhaps not surprising that BeFx forms an acyl phosphate analogue with the active site Asp of receiver domains (Fig. 7). This is similar to its well-known role in forming the ATP (GTP) analogue ADP⋅BeFx (GDP⋅BeFx) in which it also mimics an acid anhydride linkage (50). The obligatorily tetrahedral geometry of Be with F and O ligands (50, 54, 55) suggests that phosphorylated receiver domains are active in their “ground state” rather than in the transition state for hydrolysis of the phosphate, a state that would occur as a consequence of autophosphatase activity or regulated dephosphorylation. In the latter case, ligands around the terminal phosphorus would be expected to be planar.
Figure 7.
Schematic drawing of the postulated BeF3⋅Asp complex and its biological counterpart, phosphorylated Asp (P-Asp), in bacterial response regulators. A divalent cation (not indicated) is required for the formation of both species.
Acknowledgments
We thank Dorothee Kern, Boris Magasanik, John S. Parkinson, Gregory Petsko, and our colleagues Luhong He, Eric Soupene, Wally van Heeswijk, and Daniel Zimmer for comments on the manuscript, and Ore Carmi and Susan Kato for assistance in the preparation of the manuscript. This work was supported by National Institutes of Health Grant GM38361 to S.K., and by Department of Energy Contract DE-AC03-76SF00098 and Instrumentation Grant DE FG05-86ER75281 and National Science Foundation Grants DMB 86-09305 and BBS 87-20134 to D.E.W.
Abbreviations
- NtrC
nitrogen regulatory protein C
- D54
Asp-54
- NtrCr
receiver domain of NtrC
- P-NtrCr
phosphorylated NtrCr
- BeFx
beryllofluoride
- VO43−
orthovanadate
- NtrCWT
wild-type NtrC
References
- 1.Nixon B T, Ronson C W, Ausubel F M. Proc Natl Acad Sci USA. 1986;83:7850–7854. doi: 10.1073/pnas.83.20.7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parkinson J S, Kofoid E C. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 3.Stock J B, Surette M G, Levit M, Park P. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 25–51. [Google Scholar]
- 4.Mizuno T, Kaneko T, Tabata S. DNA Res. 1996;3:407–414. doi: 10.1093/dnares/3.6.407. [DOI] [PubMed] [Google Scholar]
- 5.Mizuno T. DNA Res. 1997;4:161–168. doi: 10.1093/dnares/4.2.161. [DOI] [PubMed] [Google Scholar]
- 6.Ota I M, Varshavsky A. Science. 1993;262:566–569. doi: 10.1126/science.8211183. [DOI] [PubMed] [Google Scholar]
- 7.Maeda T, Wurgler-Murphy S M, Saito H. Nature (London) 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- 8.Ruis H, Schüller C. BioEssays. 1995;17:959–965. doi: 10.1002/bies.950171109. [DOI] [PubMed] [Google Scholar]
- 9.Schaller G E. Essays Biochem. 1997;32:101–111. [PubMed] [Google Scholar]
- 10.Quon K C, Marczynski G T, Shapiro L. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 11.Fabret C, Feher V A, Hoch J A. J Bacteriol. 1999;181:1975–1983. doi: 10.1128/jb.181.7.1975-1983.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silversmith R E, Bourret R B. Trends Microbiol. 1999;7:16–22. doi: 10.1016/s0966-842x(98)01409-7. [DOI] [PubMed] [Google Scholar]
- 13.Dziejman M, Mekalanos J J. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 305–317. [Google Scholar]
- 14.Haldimann A, Fisher S L, Daniels L L, Walsh C T, Wanner B L. J Bacteriol. 1997;179:5903–5913. doi: 10.1128/jb.179.18.5903-5913.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Groisman E A. BioEssays. 1998;20:96–101. doi: 10.1002/(SICI)1521-1878(199801)20:1<96::AID-BIES13>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 16.Novak R, Henriques B, Charpentier E, Normark S, Tuomanen E. Nature (London) 1999;399:590–593. doi: 10.1038/21202. [DOI] [PubMed] [Google Scholar]
- 17.Ninfa A J, Atkinson M R, Kamberov E S, Feng J, Ninfa E G. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 67–88. [Google Scholar]
- 18.Wanner B L. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 203–221. [Google Scholar]
- 19.Ninfa A J, Magasanik B. Proc Natl Acad Sci USA. 1986;83:5909–5913. doi: 10.1073/pnas.83.16.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stock A M, Mottonen J M, Stock J B, Schutt C E. Nature (London) 1989;337:745–749. doi: 10.1038/337745a0. [DOI] [PubMed] [Google Scholar]
- 21.Volkman B F, Nohaile M J, Amy N K, Kustu S, Wemmer D E. Biochemistry. 1995;34:1413–1424. doi: 10.1021/bi00004a036. [DOI] [PubMed] [Google Scholar]
- 22.Baikalov I, Schroder I, Kaczor-Grzeskowiak M, Grzeskowiak K, Gunsalus R P, Dickerson R E. Biochemistry. 1996;35:11053–11061. doi: 10.1021/bi960919o. [DOI] [PubMed] [Google Scholar]
- 23.Feher V A, Zapf J W, Hoch J A, Whiteley J M, McIntosh L P, Rance M, Skelton N J, Dahlquist F W, Cavanagh J. Biochemistry. 1997;36:10015–10025. doi: 10.1021/bi970816l. [DOI] [PubMed] [Google Scholar]
- 24.Djordjevic S, Goudreau P N, Xu Q, Stock A M, West A H. Proc Natl Acad Sci USA. 1998;95:1381–1386. doi: 10.1073/pnas.95.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sola M, Gomis-Ruth F X, Serrano L, Gonzalez A, Coll M. J Mol Biol. 1999;285:675–687. doi: 10.1006/jmbi.1998.2326. [DOI] [PubMed] [Google Scholar]
- 26.Drake S K, Bourret R B, Luck L A, Simon M I, Falke J J. J Biol Chem. 1993;268:13081–13088. [PMC free article] [PubMed] [Google Scholar]
- 27.Lowry D F, Roth A F, Rupert P B, Dahlquist F W, Moy F J, Domaille P J, Matsumura P. J Biol Chem. 1994;269:26358–26362. [PubMed] [Google Scholar]
- 28.Nohaile M, Kern D, Wemmer D, Stedman K, Kustu S. J Mol Biol. 1997;273:299–316. doi: 10.1006/jmbi.1997.1296. [DOI] [PubMed] [Google Scholar]
- 29.Magasanik B. In: Regulation of Gene Expression in Escherichia coli. Lin E C C, Lynch A S, editors. Austin, TX: R. G. Landes Co.; 1996. pp. 281–290. [Google Scholar]
- 30.Rombel I, North A, Hwang I, Wyman C, Kustu S. Cold Spring Harbor Symp Quant Biol. 1998;63:157–166. doi: 10.1101/sqb.1998.63.157. [DOI] [PubMed] [Google Scholar]
- 31.Porter S C, North A K, Kustu S. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 147–158. [Google Scholar]
- 32.Drummond M, Whitty P, Wootton J. EMBO J. 1986;5:441–447. doi: 10.1002/j.1460-2075.1986.tb04230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.North A K, Klose K E, Stedman K M, Kustu S. J Bacteriol. 1993;175:4267–4273. doi: 10.1128/jb.175.14.4267-4273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morett E, Segovia L. J Bacteriol. 1993;175:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osuna J, Soberon X, Morrett E. Protein Sci. 1997;6:543–555. doi: 10.1002/pro.5560060304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, Passaglia L, Rombel I, Yan D, Kustu S. J Bacteriol. 1999;181:5443–5454. doi: 10.1128/jb.181.17.5443-5454.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Porter S C, North A K, Wedel A B, Kustu S. Genes Dev. 1993;7:2258–2273. doi: 10.1101/gad.7.11.2258. [DOI] [PubMed] [Google Scholar]
- 38.Wyman C, Rombel I, North A K, Bustamante C, Kustu S. Science. 1997;275:1658–1661. doi: 10.1126/science.275.5306.1658. [DOI] [PubMed] [Google Scholar]
- 39.Lunardi J, Dupuis A, Garin J, Issartel J P, Michel L, Chabre M, Vignais P V. Proc Natl Acad Sci USA. 1988;85:8958–8962. doi: 10.1073/pnas.85.23.8958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Werber M M, Peyser Y M, Muhlrad A. Biochemistry. 1992;31:7190–7197. doi: 10.1021/bi00146a023. [DOI] [PubMed] [Google Scholar]
- 41.Goodno C C. Proc Natl Acad Sci USA. 1979;76:2620–2624. doi: 10.1073/pnas.76.6.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flashner Y, Weiss D S, Keener J, Kustu S. J Mol Biol. 1995;249:700–713. doi: 10.1006/jmbi.1995.0330. [DOI] [PubMed] [Google Scholar]
- 43.Klose K E, North A K, Stedman K M, Kustu S. J Mol Biol. 1994;241:233–245. doi: 10.1006/jmbi.1994.1492. [DOI] [PubMed] [Google Scholar]
- 44.Hwang I, Thorgeirsson T, Lee J, Kustu S, Shin Y K. Proc Natl Acad Sci USA. 1999;96:4880–4885. doi: 10.1073/pnas.96.9.4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greiner D P, Miyake R, Moran J K, Jones A D, Negishi T, Ishihama A, Meares C F. Bioconjugate Chem. 1997;8:44–48. doi: 10.1021/bc9600731. [DOI] [PubMed] [Google Scholar]
- 46.Huang K-J, Igo M M. J Mol Biol. 1996;262:615–628. doi: 10.1006/jmbi.1996.0540. [DOI] [PubMed] [Google Scholar]
- 47.Li J, Kustu S, Stewart V. J Mol Biol. 1994;241:150–165. doi: 10.1006/jmbi.1994.1485. [DOI] [PubMed] [Google Scholar]
- 48.Darwin A J, Tyson K L, Busby S J W, Stewart V. Mol Microbiol. 1997;25:583–595. doi: 10.1046/j.1365-2958.1997.4971855.x. [DOI] [PubMed] [Google Scholar]
- 49.McEvoy M M, Bren A, Eisenbach M, Dahlquist F W. J Mol Biol. 1999;289:1423–1433. doi: 10.1006/jmbi.1999.2830. [DOI] [PubMed] [Google Scholar]
- 50.Chabre M. Trends Biochem Sci. 1990;15:6–10. doi: 10.1016/0968-0004(90)90117-t. [DOI] [PubMed] [Google Scholar]
- 51.Wittinghofer A. Curr Biol. 1997;7:R682–R685. doi: 10.1016/s0960-9822(06)00355-1. [DOI] [PubMed] [Google Scholar]
- 52.Lukat G S, McCleary W R, Stock A M, Stock J B. Proc Natl Acad Sci USA. 1992;89:718–722. doi: 10.1073/pnas.89.2.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feng J, Atkinson M R, McCleary W, Stock J B, Wanner B L, Ninfa A J. J Bacteriol. 1992;174:6061–6070. doi: 10.1128/jb.174.19.6061-6070.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goldstein G. Anal Chem. 1964;36:243–244. [Google Scholar]
- 55.Martin R B. Biochem Biophys Res Commun. 1988;155:1194–1200. doi: 10.1016/s0006-291x(88)81266-x. [DOI] [PubMed] [Google Scholar]
- 56.Pratt L A, Hsing W, Gibson K E, Silhavy T J. Mol Microbiol. 1996;20:911–917. doi: 10.1111/j.1365-2958.1996.tb02532.x. [DOI] [PubMed] [Google Scholar]
- 57.Egger L A, Park H, Inouye M. Genes Cells. 1997;2:167–184. doi: 10.1046/j.1365-2443.1997.d01-311.x. [DOI] [PubMed] [Google Scholar]
- 58.Stewart V, Rabin R S. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 233–252. [Google Scholar]
- 59.Aiba H, Nakasai F, Mizushima S, Mizuno T. J Biochem. 1989;106:5–7. doi: 10.1093/oxfordjournals.jbchem.a122817. [DOI] [PubMed] [Google Scholar]
- 60.Baikalov I, Schroder I, Kaczor-Grzeskowiak M, Cascio D, Gunsalus R P, Dickerson R E. Biochemistry. 1998;37:3665–3676. doi: 10.1021/bi972365a. [DOI] [PubMed] [Google Scholar]
- 61.Halkides C J, Zhu X, Phillion D P, Matsumura P, Dahlquist F W. Biochemistry. 1998;37:13674–13680. doi: 10.1021/bi9806293. [DOI] [PubMed] [Google Scholar]
- 62.Moy F J, Lowry D F, Matsumura P, Dahlquist F W, Krywko J E, Domaille P J. Biochemistry. 1994;33:10731–10742. doi: 10.1021/bi00201a022. [DOI] [PubMed] [Google Scholar]
- 63.Santoro J, Bruix M, Pascual J, Lopez E, Serrano L, Rico M. J Mol Biol. 1995;247:717–725. doi: 10.1006/jmbi.1995.0175. [DOI] [PubMed] [Google Scholar]
- 64.Zapf J W, Hoch J A, Whiteley J M. Biochemistry. 1996;35:2926–2933. doi: 10.1021/bi9519361. [DOI] [PubMed] [Google Scholar]
- 65.Lee J H, Scholl D, Nixon B T, Hoover T R. J Biol Chem. 1994;269:20401–20409. [PubMed] [Google Scholar]
- 66.Perego M, Hoch J A. Trends Genet. 1996;12:97–101. doi: 10.1016/0168-9525(96)81420-x. [DOI] [PubMed] [Google Scholar]