Abstract
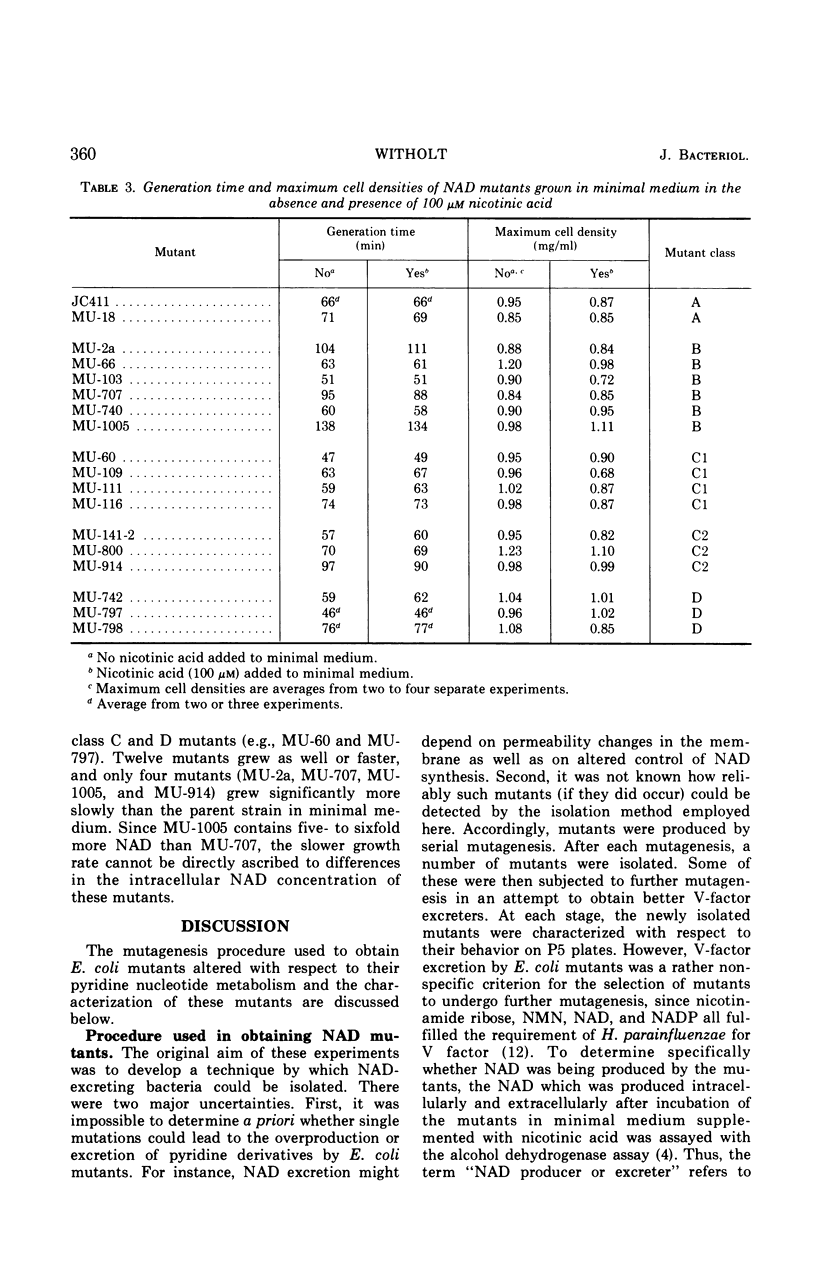
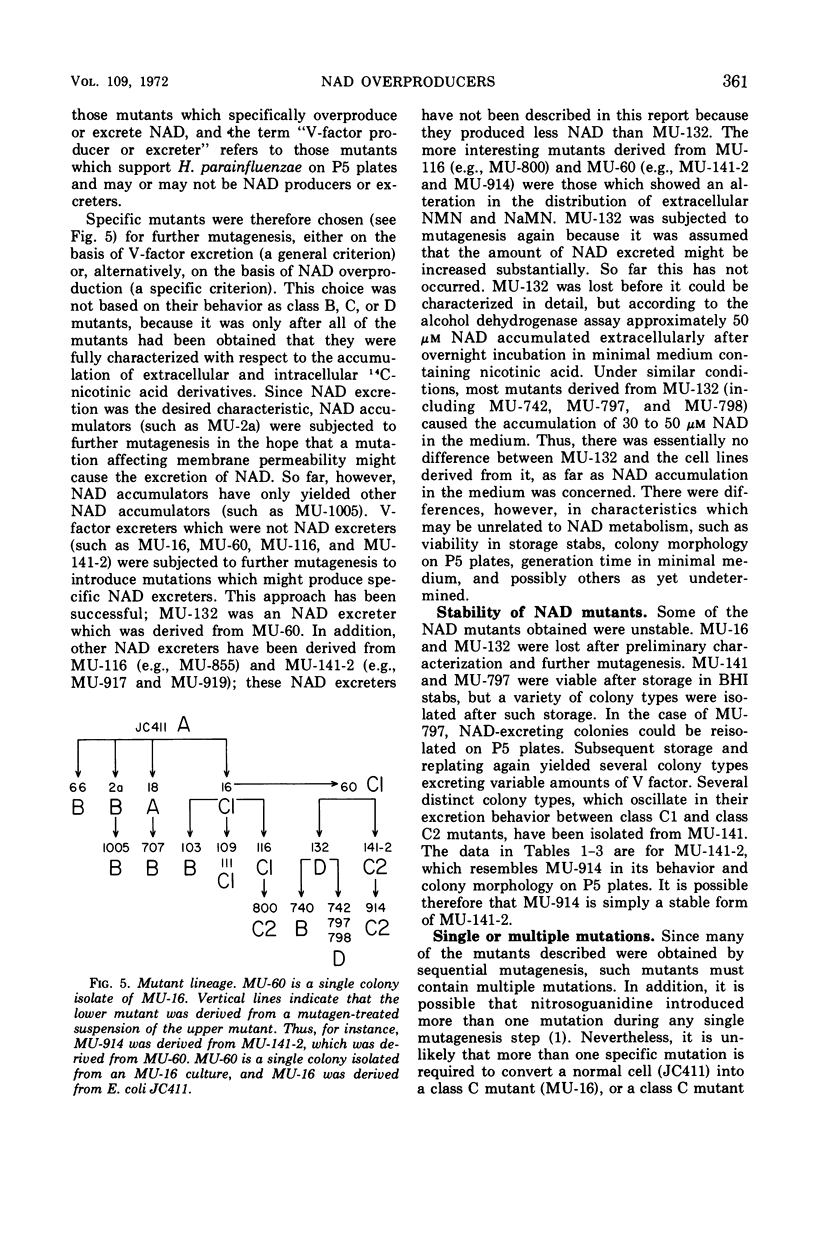
A procedure has been developed for isolating mutants which are defective with respect to nicotinamide adenine dinucleotide (NAD) metabolism. It is based on the well known V-factor requirement of Haemophilus parainfluenzae. This procedure was used to isolate a series of mutants from Escherichia coli. The pyridine metabolism of wild-type and mutant E. coli cells falls in one of four distinct classes. Class A includes wild-type E. coli and represents strains that are normal with respect to pyridine metabolism. Class B mutants have altered internal pools of NAD. The intracellular NAD concentration of different class B mutants varies over a 10-fold range. Class C mutants excrete pyridine mononucleotides, and class D mutants excrete NAD. The production of pyridine nucleotides by class C and D mutants exceeds that of wild-type E. coli by a factor of at least ten. The mutant strains generally have normal generation times and achieve normal cell densities in minimal medium.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ANDREOLI A. J., IKEDA M., NISHIZUKA Y., HAYAISHI O. Quinolinic acid: a precursor to nicotinamide adenine dinucleotide in Escherichia coli. Biochem Biophys Res Commun. 1963 Jul 18;12:92–97. doi: 10.1016/0006-291x(63)90241-9. [DOI] [PubMed] [Google Scholar]
- Bendler J. W., Goodgal S. H. Prophage S2 mutants in Haemophilus influenzae: a technique for their production and isolation. Science. 1968 Oct 25;162(3852):464–465. doi: 10.1126/science.162.3852.464. [DOI] [PubMed] [Google Scholar]
- Dahmen W., Webb B., Preiss J. The deamido-diphosphopyridine nucleotide and diphosphopyridine nucleotide pyrophosphorylases of Escherichia coli and yeast. Arch Biochem Biophys. 1967 May;120(2):440–450. doi: 10.1016/0003-9861(67)90262-7. [DOI] [PubMed] [Google Scholar]
- Herbst E. J., Snell E. E. THE NUTRITIONAL REQUIREMENTS OF HEMOPHILUS PARAINFLUENZAE 7901. J Bacteriol. 1949 Sep;58(3):379–386. doi: 10.1128/jb.58.3.379-386.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IMSANDE J. Pathway of diphosphopyridine nucleotide biosynthesis in Escherichia coli. J Biol Chem. 1961 May;236:1494–1497. [PubMed] [Google Scholar]
- Kline B. C., Schoenhard D. E. Biochemical characterization of sulfur assimilation by Salmonella pullorum. J Bacteriol. 1970 Apr;102(1):142–148. doi: 10.1128/jb.102.1.142-148.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAMBORG M., STOLZENBACH F. E., KAPLAN N. O. The nicotinic acid analogue of diphosphopyridine nucleotide. J Biol Chem. 1958 Apr;231(2):685–694. [PubMed] [Google Scholar]
- LEDER I. G., HANDLER P. Synthesis of nicotinamide mononucleotide by human erythrocytes in vitro. J Biol Chem. 1951 Apr;189(2):889–899. [PubMed] [Google Scholar]
- Lundquist R., Olivera B. M. Pyridine nucleotide metabolism in Escherichia coli. I. Exponential growth. J Biol Chem. 1971 Feb 25;246(4):1107–1116. [PubMed] [Google Scholar]
- PREISS J., HANDLER P. Biosynthesis of diphosphopyridine nucleotide. I. Identification of intermediates. J Biol Chem. 1958 Aug;233(2):488–492. [PubMed] [Google Scholar]
- SCHAECHTER M., MAALOE O., KJELDGAARD N. O. Dependency on medium and temperature of cell size and chemical composition during balanced grown of Salmonella typhimurium. J Gen Microbiol. 1958 Dec;19(3):592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- Spencer R. L., Preiss J. Biosynthesis of diphosphopyridine nucleotide. The purification and the properties of diphospyridine nucleotide synthetase from Escherichia coli b. J Biol Chem. 1967 Feb 10;242(3):385–392. [PubMed] [Google Scholar]




