Abstract
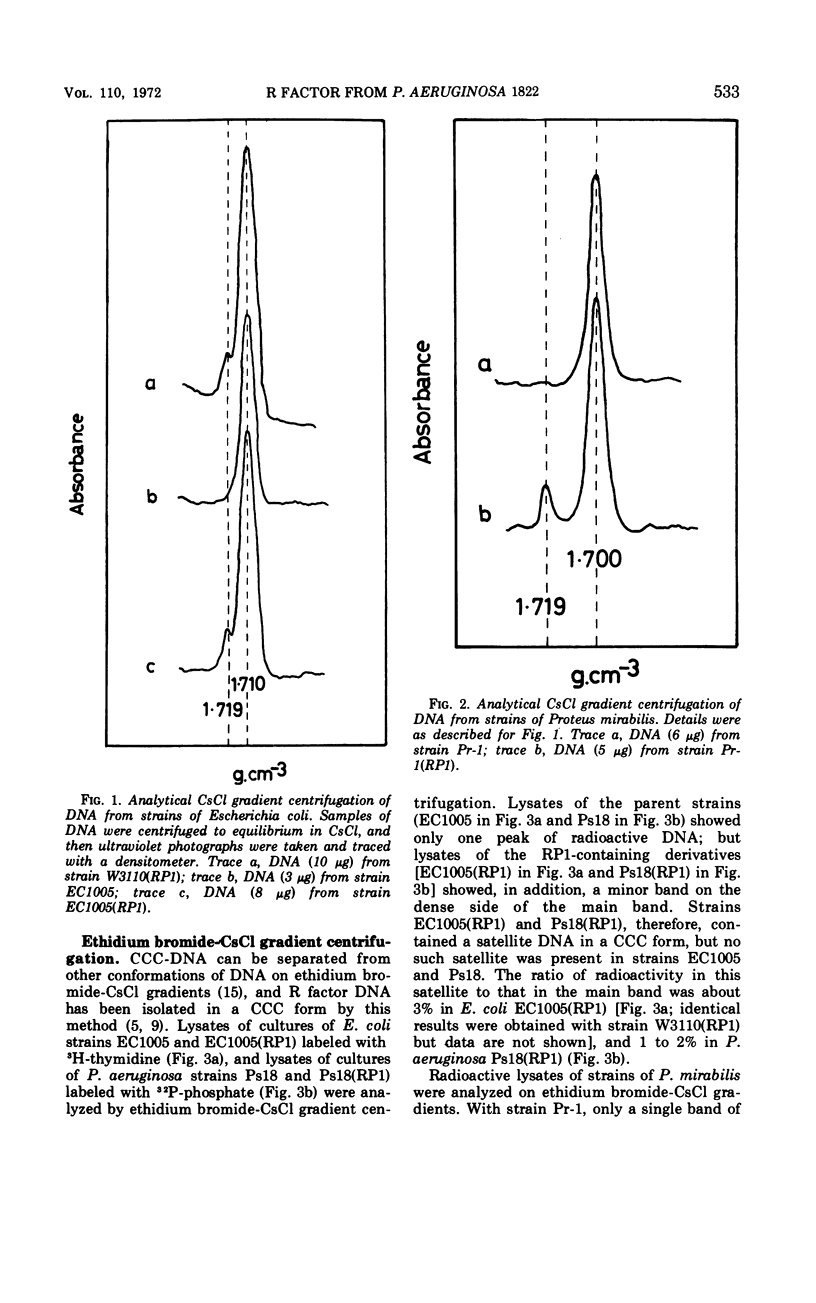
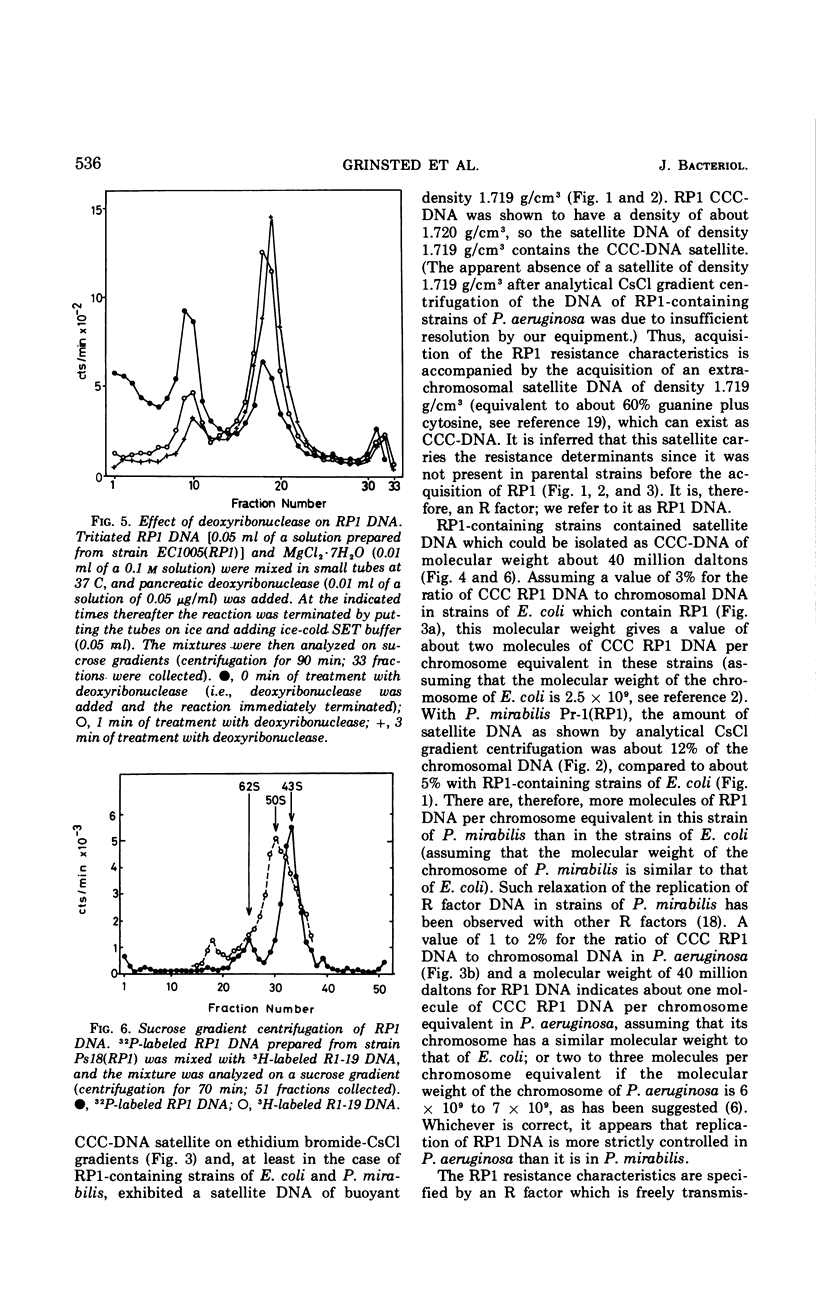
RP1, a group of genes specifying resistance to carbenicillin, neomycin, kanamycin, and tetracycline and originating in a strain of Pseudomonas aeruginosa, was freely transmissible between strains of P. aeruginosa, Escherichia coli, and Proteus mirabilis. Acquisition of the multiple drug resistance specified by RP1 by these strains was accompanied by acquisition of an extrachromosomal satellite of covalently closed circular deoxyribonucleic acid of molecular weight about 40 million daltons and of buoyant density 1.719 g/cm3 (60% guanine plus cytosine).
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bak A. L., Christiansen C., Stenderup A. Bacterial genome sizes determined by DNA renaturation studies. J Gen Microbiol. 1970 Dec;64(3):377–380. doi: 10.1099/00221287-64-3-377. [DOI] [PubMed] [Google Scholar]
- Cooper S., Helmstetter C. E. Chromosome replication and the division cycle of Escherichia coli B/r. J Mol Biol. 1968 Feb 14;31(3):519–540. doi: 10.1016/0022-2836(68)90425-7. [DOI] [PubMed] [Google Scholar]
- Datta N., Hedges R. W., Shaw E. J., Sykes R. B., Richmond M. H. Properties of an R factor from Pseudomonas aeruginosa. J Bacteriol. 1971 Dec;108(3):1244–1249. doi: 10.1128/jb.108.3.1244-1249.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullbrook P. D., Elson S. W., Slocombe B. R-factor mediated beta-lactamase in Pseudomonas aeruginosa. Nature. 1970 Jun 13;226(5250):1054–1056. doi: 10.1038/2261054a0. [DOI] [PubMed] [Google Scholar]
- Kontomichalou P., Mitani M., Clowes R. C. Circular R-factor molecules controlling penicillinase synthesis, replicating in Escherichia coli under either relaxed or stringent control. J Bacteriol. 1970 Oct;104(1):34–44. doi: 10.1128/jb.104.1.34-44.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowbury E. J., Lilly H. A., Kidson A., Ayliffe G. A., Jones R. J. Sensitivity of Pseudomonas aeruginosa to antibiotics: emergence of strains highly resistant to carbenicillin. Lancet. 1969 Aug 30;2(7618):448–452. doi: 10.1016/s0140-6736(69)90163-9. [DOI] [PubMed] [Google Scholar]
- NOVICK R. P. ANALYSIS BY TRANSDUCTION OF MUTATIONS AFFECTING PENICILLINASE FORMATION IN STAPHYLOCOCCUS AUREUS. J Gen Microbiol. 1963 Oct;33:121–136. doi: 10.1099/00221287-33-1-121. [DOI] [PubMed] [Google Scholar]
- NOVICK R. P. Micro-iodometric assay for penicillinase. Biochem J. 1962 May;83:236–240. doi: 10.1042/bj0830236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisioka T., Mitani M., Clowes R. Composite circular forms of R factor deoxyribonucleic acid molecules. J Bacteriol. 1969 Jan;97(1):376–385. doi: 10.1128/jb.97.1.376-385.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P. Extrachromosomal inheritance in bacteria. Bacteriol Rev. 1969 Jun;33(2):210–263. doi: 10.1128/br.33.2.210-263.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PERRET C. J. Iodometric assay of penicillinase. Nature. 1954 Nov 27;174(4439):1012–1013. doi: 10.1038/1741012a0. [DOI] [PubMed] [Google Scholar]
- POLLOCK M. R., TORRIANI A. M. Purification et caractéristiques physicochimiques de la pénicillinase de Bacillus cereus. C R Hebd Seances Acad Sci. 1953 Jul 20;237(3):276–278. [PubMed] [Google Scholar]
- Radloff R., Bauer W., Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in HeLa cells. Proc Natl Acad Sci U S A. 1967 May;57(5):1514–1521. doi: 10.1073/pnas.57.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe E., Jones R. J., Lowbury E. J. Transfer of anibioic resistanceetween Pseudomonas aeruginosa, Escherichia coli, and other gram-negative bacilli in rns. Lancet. 1971 Jan 23;1(7691):149–152. doi: 10.1016/s0140-6736(71)91930-1. [DOI] [PubMed] [Google Scholar]
- Roth T. F., Hayashi M. Allomorphic forms of bacteriophage phiX-174 replicative DNA. Science. 1966 Nov 4;154(3749):658–660. doi: 10.1126/science.154.3749.658. [DOI] [PubMed] [Google Scholar]
- Rownd R., Nakaya R., Nakamura A. Molecular nature of the drug-resistance factors of the Enterobacteriaceae. J Mol Biol. 1966 Jun;17(2):376–393. doi: 10.1016/s0022-2836(66)80149-3. [DOI] [PubMed] [Google Scholar]
- SCHILDKRAUT C. L., MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its buoyant density in CsCl. J Mol Biol. 1962 Jun;4:430–443. doi: 10.1016/s0022-2836(62)80100-4. [DOI] [PubMed] [Google Scholar]
- STUDIER F. W. SEDIMENTATION STUDIES OF THE SIZE AND SHAPE OF DNA. J Mol Biol. 1965 Feb;11:373–390. doi: 10.1016/s0022-2836(65)80064-x. [DOI] [PubMed] [Google Scholar]
- Silver R. P., Falkow S. Specific labeling and physical characterization of R-factor deoxyribonucleic acid in Escherichia coli. J Bacteriol. 1970 Oct;104(1):331–339. doi: 10.1128/jb.104.1.331-339.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. T. R-factor gene expression gram-negative bacteria. J Gen Microbiol. 1969 Jan;55(1):109–120. doi: 10.1099/00221287-55-1-109. [DOI] [PubMed] [Google Scholar]
- Sykes R. B., Richmond M. H. Intergeneric transfer of a beta-lactamase gene between Ps. aeruginosa and E. coli. Nature. 1970 Jun 6;226(5249):952–954. doi: 10.1038/226952a0. [DOI] [PubMed] [Google Scholar]
- Vinograd J., Lebowitz J., Radloff R., Watson R., Laipis P. The twisted circular form of polyoma viral DNA. Proc Natl Acad Sci U S A. 1965 May;53(5):1104–1111. doi: 10.1073/pnas.53.5.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WATANABE T. Infective heredity of multiple drug resistance in bacteria. Bacteriol Rev. 1963 Mar;27:87–115. doi: 10.1128/br.27.1.87-115.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]


