Abstract
HIF (hypoxia-inducible factor) is the main transcription factor activated by low oxygen tensions. HIF-1α (and other α subunits) is tightly controlled mostly at the protein level, through the concerted action of a class of enzymes called PHDs (prolyl hydroxylases) 1, 2 and 3. Most of the knowledge of HIF derives from studies following hypoxic stress; however, HIF-1α stabilization is also found in non-hypoxic conditions through an unknown mechanism. In the present study, we demonstrate that NF-κB (nuclear factor κB) is a direct modulator of HIF-1α expression. The HIF-1α promoter is responsive to selective NF-κB subunits. siRNA (small interfering RNA) studies for individual NF-κB members revealed differential effects on HIF-1α mRNA levels, indicating that NF-κB can regulate basal HIF-1α expression. Finally, when endogenous NF-κB is induced by TNFα (tumour necrosis factor α) treatment, HIF-1α levels also change in an NF-κB-dependent manner. In conclusion, we find that NF-κB can regulate basal TNFα and, in certain circumstances, the hypoxia-induced HIF-1α.
Keywords: hypoxia, hypoxia-inducible factor 1 (HIF-1), nuclear factor κB (NF-κB), tumour necrosis factor (TNF)
Abbreviations: ChIP, chromatin immunoprecipitation; EMSA, electrophoretic mobility-shift assay; GLUT, glucose transporter; HEK, human embryonic kidney; HIF, hypoxia-inducible factor; IκB, inhibitor of nuclear factor κB; IKK, IκB kinase; NF-κB, nuclear factor κB; PCNA, proliferating-cell nuclear antigen; PHD, prolyl hydroxylase; qRT-PCR, quantitative reverse transcriptase PCR; ROS, reactive oxygen species; RT, reverse transcriptase; siRNA, small interfering RNA; TNFα, tumour necrosis factor α; VEGF, vascular-endothelial growth factor
INTRODUCTION
HIF (hypoxia-inducible factor) plays key roles in development, physiological processes and pathological conditions as its presence affects survival, cell-cycle progression and metabolism [1,2]. Although first identified as the main transcription factor activated under low oxygen tensions, HIF is a key transcription factor activated by cytokines, oncogenes and ROS (reactive oxygen species) under normoxic conditions [3]. HIF is a heterodimeric factor composed of α and β subunits [also called ARNT (aryl hydrocarbon nuclear translocator)] [1]. Although HIF-1β is constitutively expressed and not regulated by oxygen levels, HIF-α subunits are tightly controlled at the protein level. This occurs, predominantly, through the concerted action of a class of enzymes called PHDs (prolyl hydroxylases) that catalyse prolyl hydroxylation of HIF-α subunits, which incessantly targets them for VHL (von Hippel–Lindau protein)-dependent 26S proteasomal degradation [4]. PHD enzymes require molecular O2, 2-oxoglutarate, iron ions (Fe2+) and ascorbic acid to be fully active, and are inhibited in hypoxic conditions [4].
Four PHDs have been identified so far, but only three, PHDs 1, 2 and 3, have been functionally characterized in terms of HIF modulation [4]. PHD2 is thought to be the dominant PHD in controlling HIF-1α following hypoxia [5]. Furthermore, all three PHDs can be regulated by HIF and by oxygen levels [6–8].
In addition to the HIF-α stabilization mechanism controlled by prolyl hydroxylation, hydroxylation of Asn803 in HIF-1α or Asn851 in HIF-2α, prevents interaction with the CH1 domains of histone acetyltransferases {p300/CBP [CREB (cAMP-response-element-binding protein)-binding protein]} that act as transcriptional co-activators. This modification alters the ability of HIF-1/HIF-2 to transactivate their target genes [9]. Although most knowledge regarding HIF-1 has been derived from studies following hypoxic stress, HIF-1 stabilization has also been found in non-hypoxic settings, such as relatively well-oxygenated regions of tumours, and in diseases such as rheumatoid arthritis and diabetes [10,11]. However, the mechanisms for HIF stabilization under those conditions have not been elucidated. Many of the stimuli that induce HIF-1 in normoxia are known to activate a number of other transcription factors such as NF-κB (nuclear factor κB). It is therefore plausible that cross-talk between these two transcription factors can occur. In addition, the HIF-1α and PHD1 promoters contain NF-κB-binding sites, which have not been functionally characterized. Importantly, HIF-1α stabilization and induction through H2O2 was recently shown to be NF-κB dependent under normoxic conditions [12].
NF-κB is the collective name for a transcription factor that exists as either a hetero- or homo-dimer and is formed by a family of subunits called RelA (p65), RelB, cRel, p50 and its precursor p105 (NF-κB1), and p52 and its precursor p100 (NF-κB2). Some dimers are more prevalent than others and they shuttle between the cytoplasm and nucleus but are predominantly sequestered in the inactive state in the cytoplasm, inhibited by members of the IκB (inhibitor of NF-κB) [13]. Upon stimulation, by compounds such as TNFα (tumour necrosis factor α), oncogenes or UV light, a kinase signalling cascade results in phosphorylation of IκB, signalling ubiquitination-mediated proteasomal degradation, resulting in NF-κB release and translocation into the nucleus [13,14]. The activated NF-κB dimer then binds to target DNA sequences in the nucleus where it regulates transcription of NF-κB target genes. Although NF-κB activation is extremely fast, the cyclic nature of NF-κB activity, in combination with post-translational modifications and hyper- and hypo-acetylated histones at target genes are indicative of further levels of regulatory complexity [15].
There have been several studies demonstrating cross-talk between the NF-κB and HIF signalling pathways, including shared target genes, but a direct link has yet to be elucidated. In the present study, we show that several NF-κB subunits are bound to the HIF-1α promoter. NF-κB depletion results in reduced basal levels of HIF-1α mRNA. In addition, we demonstrate that TNFα-induced NF-κB can increase HIF-1α mRNA, protein and activity levels, leading to transactivation of target genes in normoxia. Changes in HIF-1α levels following hypoxia are only seen when the NF-κB pathway is completely blocked. These results demonstrate that NF-κB can directly modulate the HIF-1α pathway, and that this modulation is sufficient to alter HIF target gene expression in vivo.
MATERIALS AND METHODS
Tissue culture, medium and hypoxia treatments
U2OS osteosarcoma and HEK (human embryonic kidney)-293 cell lines were obtained from the European Collection of Cell Cultures. IKK (IκB kinase) wild-type, IKKα−/−, IKKβ−/− and IKKα/β−/− mouse embryonic fibroblasts were a gift from Professor Inder Verma (Laboratory of Genetics, The Salk Institute for Biological Studies, La Jolla, CA, U.S.A.). All cells were grown in DMEM (Dulbecco's modified Eagle medium; Lonza) supplemented with 10% fetal calf serum (Gibco), 50 units/ml penicillin (BioWhittaker) and 50 μg/ml streptomycin (BioWhittaker) for no more than 30 passages. For hypoxia treatments, cells were placed in 1% O2 using an InVivo 300 workstation (Ruskin).
siRNA (small interfering RNA) transfection and sequences
siRNA duplex oligonucleotides were synthesized by MWG and transfected using Interferin (Polyplus) following the manufacturer's protocol. The oligonucleotides used were: Control, 5′-AACAGUCGCGUUUGCGACUGG-3′ [16]; RelA, 5′-GCUGAUGUGCACCGACAAG-3′ [16]; RelB, 5′-AAUUGGAGAUCAUCGACGAGU-3′; p50/p105, 5′-AAGGGGCUAUAAUCCUGGACU-3′; p52/p100, 5′-AAGAUGAAGAUUGAGCGGCCU-3′ [17]; HIF-1α, 5′-CUGAUGACCAGCAACUUGA-3′; IKKα, 5′-GCAGGCUCUUUCAGGGACA-3′; and IKKβ, 5′-CAGGUGAGCAGAUUGCCAU-3′. The cRel siRNA sequence was as described previously [17].
DNA constructs
Individual NF-κB subunit expression vectors were a gift from Professor Neil Perkins (Division of Gene Regulation and Expression, College of Life Sciences, University of Dundee, Dundee, Scotland, U.K.). HIF-1α promoter luciferase constructs were a gift from Dr Carine Michiels (Department of Biology, University of Namur, Namur, Belgium).
Antibodies
Antibodies against HIF-1α, (R&D Systems), RelA (p65), RelB, cRel, p50 (NF-κB1), Chk1 (checkpoint kinase 1) and p52 (NF-κB2) were from Santa Cruz Biotechnology. Antibodies against GLUT (glucose transporter) 1/3 were from Neomarkers, against HIF-2α were from Novus Biologicals, against HIF-1β, IKKα and IKKβ were from Cell Signalling, against β-actin and PCNA (proliferating-cell nuclear antigen) were from Sigma, against PHD2 were from Abcam, and against Bcl-XL were from Merck Biosciences. Enhanced chemiluminescence (Pierce) was used for detection.
qRT-PCR (quantitative reverse transcriptase PCR)
qRT-PCR was carried out in a 25 μl total reaction mixture with 1 μl of extracted RNA sample (10 pg–100 ng), 12.5 μl of One-Step RT (reverse transcriptase) qPCR master mix Plus for SYBR Green I (Eurogentec), 0.2 μM forward primer, 0.2 μM reverse primer, 0.125 units/ml EuroScript RT and 10.875 μl of RNase-free distilled water. Amplification and detection were performed using a Rotor-Gene 3000 (Corbett Research) and IQ5 Icycler (Bio-Rad) detection system under the following conditions: an initial reverse transcription at 48 °C for 30 min, followed by PCR activation at 95 °C for 10 min and 45 cycles of amplification (15 s at 95 °C and 1 min at 58 °C). During amplification, a detector monitored real-time PCR amplification by quantitative analysis of the fluorescence emissions. Sample values obtained with specific primer sets were normalized to β-actin primer set values.
qRT-PCR primer sequences
For qRT-PCR, the following primer sequences were used: β-actin forward, 5′-GTGGGAGTGGGTGGAGGC-3′ and reverse, 5′-TCAACTGGTCTCAAGTCAGTG-3′; 18S forward, 5′-AAACGGCTACCACATCCAAG-3′ and reverse, 5′-CGCTCCCAAGATCCAACTAC-3′; GLUT1 forward, 5′-GATTGGCTCCTTCTCTGTGG-3′ and reverse, 5′-TCAAAGGACTTGCCCAGTTT-3′; GLUT3 forward, 5′-CAATGCTCCTGAGAAGATCATAA-3′ and reverse, 5′-AAAGCGGTTGACGAAGAGT-3′; HIF-1α forward, 5′-CATAAAGTCTGCAACATGGAAGGT-3′ and reverse, 5′-ATTTGATGGGTGAGGAATGGGTT-3′; and HIF-2α forward, 5′-GCGCTAGACTCCGAGAACAT-3′ and reverse, 5′-TGGCCACTTACTACCTGACCCTT-3′.
ChIP (chromatin immunoprecipitation)
Proteins were cross-linked with formaldehyde for 10 min. After adding 0.125 mol/l glycine, cell lysis buffer [1% SDS, 10 mM EDTA, 50 mM Tris/HCl (pH 8.1), 1 mM PMSF, 1 mg/ml leupeptin and 1 mg/ml aprotonin] was added, followed by sonication and centrifugation (13000 g for 10 min at 4 °C). The supernatant was precleared with sheared salmon sperm DNA and Protein A–Sepharose beads (Sigma). The supernatant was incubated with specific antibodies overnight, and then with Protein A–Sepharose beads for 1 h. After an extensive wash step, the complexes were eluted with buffer (100 mmol/l sodium bicarbonate and 1% SDS) and incubated with proteinase K. DNA was purified using the QIAquick PCR purification kit (Qiagen). PCR was performed with primers for the HIF-1α promoter (forward, 5′-GAACAGAGAGCCCAGCAGAG-3′ and reverse, 5′-CCTGAGGTGGAGGCGGGTTC-3′) flanking the NF-κB-binding site (−197/188 bp) at 64 °C annealing and 72 °C extension for 32 cycles. This primer set spans from −536 to −137 bp from the transcription start site. The HIF-1α control primer set was 5′-TGCTCATCAGTTGCCACTTC- 3′ (forward) and 5′-AAAACATTGCGACCACCTTC-3′ (reverse). This primer set is located in the gene itself, 24937 bp away from the transcription start site. The primers used for HIF-1α target genes promoters were: VEGF (vascular-endothelial growth factor) forward, 5′-ACGTTCCTTAGTGCTGGCGGGTAGGTTTGA-3′ and reverse, 5′-GCACCAAGTTTGTGGAGCTGAGAACGGG-3′; CA9 forward, 5′-GACAAACCTGTGAGACTTTGGCTCC-3′ and reverse, 5′-AGTGACAGCAGCAGTTGCACAGTG-3′; and DEC1 forward, 5′-CACGTGAGACTCATGTGATGAAGCC-3′ and reverse, 5′-AAGCCGAGGAGTAATGGAGAGGCT-3′.
Other experimental procedures
Luciferase assays, whole cell protein, nuclear extracts and EMSA (electrophoretic mobility-shift assay) analysis were performed as described previously ([17a] and references therein). RNA was extracted using a nucleospin RNA II kit (Machery-Nagel). The EMSA probe used for HIF-1α κB site was 5′-GCGTGGGCTGGGGTGGGGACTTGCCGCCTGCGTCGC-3′.
RESULTS
Differential control of the HIF-1α promoter by NF-κB subunits
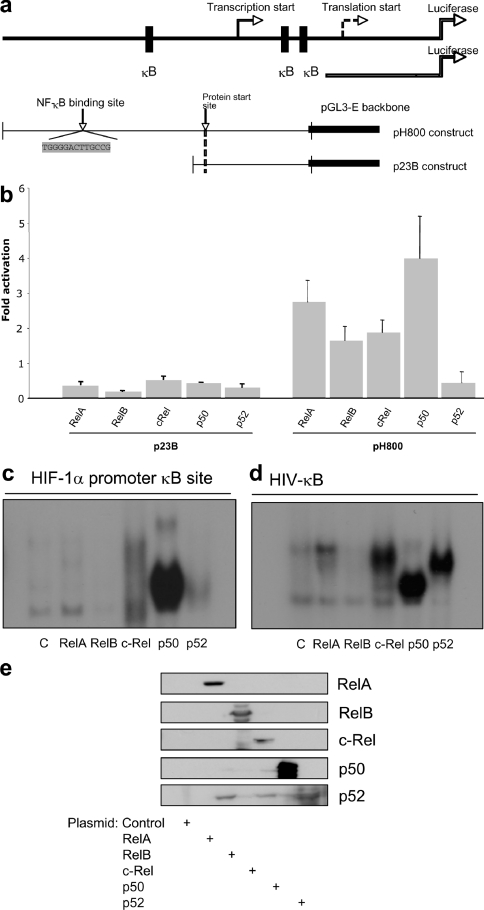
The HIF-1α promoter was originally cloned in 1996 [18] and a study by Michiels and co-workers [19], identified several binding sites for transcription factors such as AP-1 (activator protein 1) and NF-κB [19]. Given the advance in our knowledge of NF-κB consensus sites [14], we reanalysed the HIF-1α promoter (http://www.genomatix.de/products/MatInspector). This analysis identified a putative NF-κB-binding site located −197/188 bp from the initiation site (Figure 1a). Using HIF-1α promoter reporter constructs either with or without the putative NF-κB-binding site [19], we investigated the contribution of individual NF-κB subunits in normoxia (Figures 1a and 1b). All NF-κB subunits could activate the HIF-1α promoter, with the highest effect being observed with p50 and the lowest effect with p52. In contrast, none of the NF-κB subunits activated a truncated version of the HIF-1α promoter construct which lacks the NF-κB site (Figure 1b). To investigate whether NF-κB could bind the putative site on the HIF-1α promoter, we performed EMSAs using nuclear extracts derived from HEK-293 cells which had been transfected with the individual NF-κB subunits (Figure 1c). We could detect intense binding by p50, and weaker binding by p65, c-Rel and p52. No visible binding could be observed with RelB, although the expression levels were comparable (Figure 1e). Binding to the canonical site in the HIV promoter was used to assess the NF-κB binding ability (Figure 1d). These results suggest that NF-κB is able to regulate HIF-1α promoter activity.
Figure 1. NF-κB subunits can activate the HIF-1α promoter.
(a) Schematic diagram of the HIF-1α promoter–luciferase constructs. (b) HEK-293 cells were co-transfected with 1 μg of HIF-1α promoter–luciferase constructs and 1 μg of each of the NF-κB subunits. Luciferase activity was measured 48 h post-transfection. Values are means+S.D. for a minimum of three independent experiments performed in duplicate. The y-axis shows the fold activation above control plasmid. (c) NF-κB subunits bind to the κB site in the HIF-1α promoter. HEK-293 cells were transfected with 2 μg of each of the NF-κB subunits, nuclear extracts were prepared, and the DNA-binding activity was measured by EMSA using a specific probe for the HIF-1α promoter. (d) Nuclear extracts were prepared as in (c), and the DNA-binding activity was measured using a canonical NF-κB target promoter, HIV. (e) Nuclear extracts were analysed by Western blot for the individual NF-κB subunits.
NF-κB modulates basal HIF-1α mRNA levels
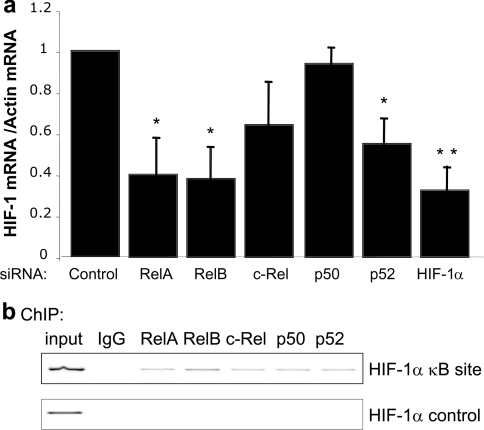
To investigate whether endogenous NF-κB could modulate the HIF-1α promoter, siRNA oligonucleotides directed towards the different NF-κB subunits were used. The siRNA sequences have been previously validated [16,17]. qRT-PCR analysis demonstrated that endogenous NF-κB regulated basal HIF-1α mRNA levels (Figure 2a). Interestingly, reduction of p50 levels had no effect on HIF-1α mRNA levels, possibly indicating a compensatory action by p52 or other NF-κB subunits. The co-operation between p50 and p52 is best depicted in the genetic knockout mice, where p50 and p52 single deletions showed no developmental defects, whereas double deletion of these genes resulted in severe impairment in bone development [20,21]. Endogenous p65, RelB and p52 have the highest effect on HIF-1α mRNA levels (Figure 2a). Despite inducing a slight reduction in HIF-1α mRNA, c-Rel depletion did not induce statistically significant effects (Figure 2a). To assess whether NF-κB is directly regulating HIF-1α mRNA levels, ChIP analysis was performed. Several of the NF-κB subunits could be found at the HIF-1α promoter; however, we could not detect any NF-κB binding in a control region of the gene (Figure 2b). This demonstrates that NF-κB modulates the basal level of HIF-1α mRNA directly.
Figure 2. Endogenous NF-κB subunits control basal HIF-1α mRNA levels.
(a) HEK-293 cells were transfected with the indicated siRNA oligonucleotides, and qRT-PCR was performed. The histogram depicts the relative levels of HIF-1α mRNA normalized to actin mRNA levels. The mean+S.D. were calculated from a minimum of three independent experiments. A Student's t test was performed and *P<0.050 and **P<0.010 when compared with the control. Actual P values are: RelA, P=0.012; RelB, P=0.012; c-Rel, P=0.076; p50, P=0.348; p52, P=0.035; HIF-1α, P=0.007. (b) ChIP analysis using the indicated antibodies and PCR analysis using specific primers for the HIF-1α promoter and HIF-1α control region was performed.
NF-κB modulates HIF-1α protein levels in normoxia
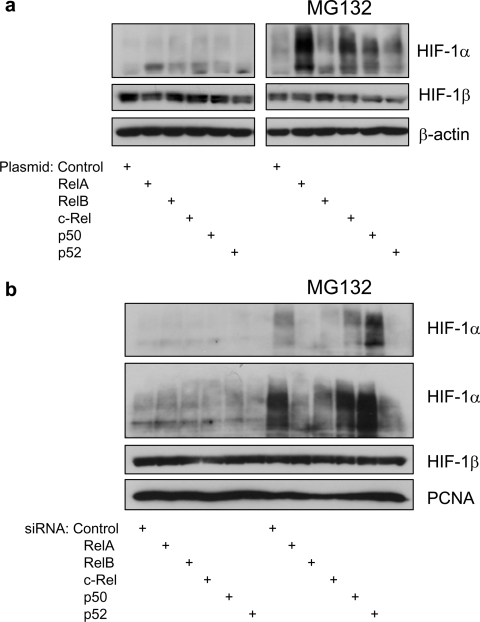
HIF-1α is predominantly regulated at the protein level following hypoxia [22]. Since the studies in the present paper were conducted at normal oxygen levels, PHD activity is not readily inhibited. To investigate whether NF-κB could also modulate HIF-1α protein levels NF-κB overexpression and siRNA-mediated knockdowns were used. Given that HIF-1α protein is rapidly degraded at normal oxygen levels, only small changes could be expected. It was possible to observe increases in protein levels when all of the NF-κB subunits were overexpressed (Figure 3a). However, RelA and c-Rel induced the highest increase followed by p50, p52 and RelB. Changes in HIF-1α protein were best visualized when the proteasome inhibitor MG132 was used (Figure 3a). Conversely, a small reduction of HIF-1α protein levels could be observed when RelA, RelB and p52 were depleted. A small effect was also evident when c-Rel was depleted but no effects could be detected with p50 depletion (Figure 3b). In either overexpression or siRNA-mediated knockdown of NF-κB subunits, HIF-1β levels remained stable (Figure 3).
Figure 3. NF-κB subunits control basal levels of HIF-1α protein.
(a) HEK-293 cells were transfected with the indicated DNA constructs, and whole cell lysates were obtained 48 h post-transfection. At 3 h prior to harvest, half of the samples were incubated with 50 μM MG132. Western blot analysis for the indicated proteins was then performed on these extracts. (b) HEK-293 cells were transfected with the indicated siRNA oligonucleotides, and whole cell lysates were obtained 48 h post-transfection. At 3 h prior to harvest, half of the samples were incubated with 50 μM MG132. Western blot analysis for the indicated proteins was then performed on these extracts.
TNFα-induced NF-κB modulates HIF-1α expression
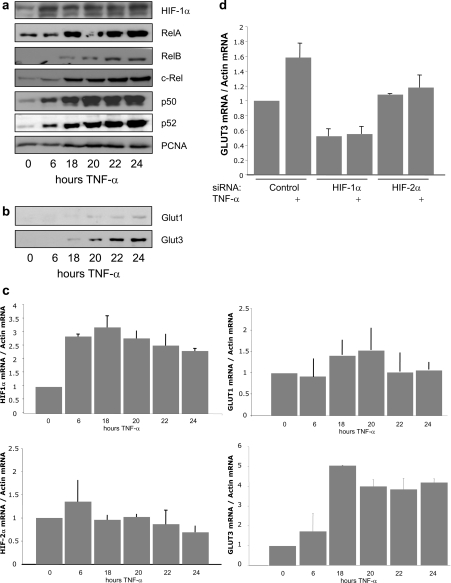
Activation of NF-κB is best associated with inflammatory responses [13]. It is rapidly activated following cytokines such as TNFα or bacterial products such as LPS (lipopolysaccharide). In fact, these stimuli have both been described to activate HIF-1α as well, although the mechanism has not been fully elucidated [23,24]. Given that prolonged TNFα exposure is present in many inflammatory diseases and certain cancers, we investigated whether HIF-1α was induced under such conditions (Figure 4). We exposed cells to TNFα for several hours and analysed nuclear fractions (Figure 4a). It was possible to detect nuclear accumulation of all NF-κB subunits and also a significant increase in HIF-1α (Figure 4a). Furthermore, this prolonged TNFα treatment also induced increases in two HIF target genes: GLUT1 and 3 (Figure 4b). To test whether the observed increases in HIF-1α, GLUT1 and GLUT3 protein resulted from higher mRNA levels, qRT-PCR was performed for the indicated times following TNFα treatment. Interestingly, it was possible to observe an increase in HIF-1α, GLUT3 and to a lesser extent GLUT1 mRNA with no change in HIF-2α mRNA (Figure 4c). These results indicate that the HIF-1α and GLUT3 protein increases resulted from increased gene transcription, whereas the increase in HIF-2α levels was possibly due to protein stability effects. Furthermore, siRNA-mediated depletion of HIF-1α or HIF-2α demonstrated that TNFα-induced GLUT3 is HIF-1α-dependent (Figure 4d and Supplementary Figure S1 at http://www.BiochemJ.org/bj/412/bj4120477add.htm), indicating that TNFα-induced HIF-1α is active transcriptionally.
Figure 4. TNFα treatment induces NF-κB and HIF-1α.
(a) HEK-293 cells were treated with 10 ng/ml TNFα for the indicated times, nuclear extracts were prepared, and these extracts were analysed by Western blot for the indicated proteins. PCNA was used as a loading control. (b) Cells were treated as in (a), and cell extracts were analysed by Western blot for the indicated HIF-1α targets. (c) Cells were treated as in (a), but mRNA was extracted and qRT-PCR was performed for the indicated gene transcripts. The histogram depicts relative levels of specific mRNA transcripts normalized to actin mRNA levels. The mean+S.D. were calculated from a minimum of three independent experiments. (d) Cells were transfected with the indicated siRNA oligonucleotides and treated with 10 ng/ml TNFα, 24 h prior to total mRNA extraction. qRT-PCR was performed as in (c).
TNFα-induced HIF-1α activity is NF-κB dependent
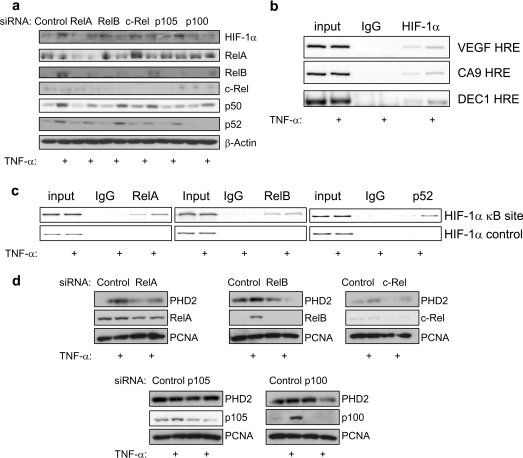
To test whether the observed changes in HIF-1α were NF-κB dependent, siRNA for the different NF-κBs was combined with TNFα treatment. Depletion of all of the subunits, with the exception of p105/p50, resulted in an impairment of TNFα-induced HIF-1α expression (Figure 5a). In addition, depletion of HIF-1α resulted in impaired TNFα induction of PHD2 (Supplementary Figure S2 at http://www.BiochemJ.org/bj/412/bj4120477add.htm), indicating that this is a HIF-1α-dependent event. Furthermore, an inducible recruitment of HIF-1α to the promoters of some of its target genes was also detected (Figure 5b).
Figure 5. TNFα induces HIF-1α activity in an NF-κB-dependent manner.
(a) HEK-293 cells were transfected with the indicated siRNA oligonucleotides, treated with 10 ng/ml TNFα 24 h prior to harvest, and whole cell lysates were prepared. Extracts were analysed by Western blot using the indicated antibodies. (b) ChIP analysis using the indicated antibodies and PCR of specific regions of the HIF-1α target genes, VEGF, CA9 and DEC1. HRE, hypoxia-response element. (c) As in (b) but the HIF-1α promoter and control regions were analysed. (d) Cells were treated as in (a), and Western blot analysis was performed using the indicated antibodies. PCNA was used as a loading control.
To verify that TNFα was inducing changes at the HIF-1α promoter, ChIPs were performed. It was possible to observe, following 18 h of TNFα treatment, an active recruitment of RelA and p52 to the HIF-1α promoter (Figure 5c). Importantly, TNFα treatment did not induce NF-κB binding to a control region of the HIF-1α gene. Furthermore, depletion of RelA, RelB, c-Rel and p52 reduced TNFα-induced HIF-1α-mediated PHD2 induction (Figure 5d). These results indicate that TNFα-induced changes in HIF-1α levels and activity are NF-κB-dependent.
Complete block of the NF-κB activation pathway prevents hypoxia-induced HIF-1α
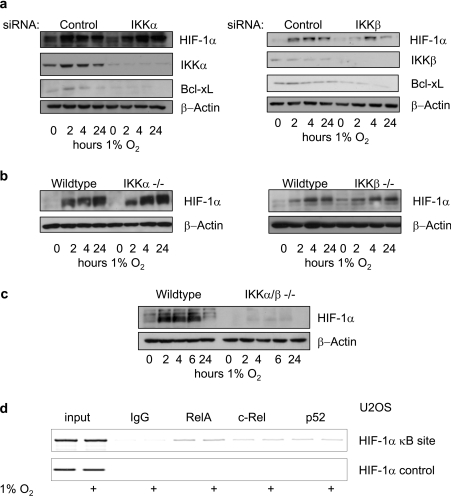
Given that NF-κB can control basal HIF-1α mRNA levels, we investigated whether blocking NF-κB could prevent hypoxia-induced HIF-1. We took advantage of genetic knockout cells and also used siRNA for the upstream kinase complex that regulates NF-κB. We could observe that, when IKKα or IKKβ were depleted, either by genetic knockout or siRNA-mediated silencing, there was very little effect on HIF-1α levels (Figures 6a and 6b), suggesting that protein stabilization can compensate for any impairment in the canonical or non-canonical pathways. However, when both IKKα and IKKβ were depleted, HIF-1 stabilization in response to hypoxia was severely impaired (Figure 6c). This suggests that HIF-1α gene transcription is mediated by several subunits of NF-κB and these derive from both pathways of activation. These results support our mRNA and ChIP results on how NF-κB controls the HIF pathway. Given that hypoxia does not induce increases in HIF-1α mRNA, it was not surprising that no changes in NF-κB recruitment to the HIF-1α promoter were observed following hypoxia treatment (Figure 6d). Once again, we could not detect any NF-κB binding to a control region of the HIF-1α gene (Figure 6d). These results suggest that any impairment seen in HIF-1α stabilization following hypoxia, in the absence of NF-κB, is due to changes in basal mRNA levels and not due to a lack of an active induction of mRNA.
Figure 6. Complete inhibition of NF-κB impairs hypoxia induced HIF-1α levels.
(a) U2OS cells were transfected with the IKKα or IKKβ siRNA oligonucleotides and exposed to 1% O2 for the indicated times prior to harvest. Whole cell lysates were analysed by Western blot. (b) Wild-type, IKKα−/− and IKKβ−/− mouse embryonic fibroblasts were exposed to 1% O2 for the indicated periods of time, and whole cell lysates were prepared. Extracts were analysed by Western blot. (c) Wild-type and IKKα/β−/− mouse embryonic fibroblasts were exposed to 1% O2 for the indicated periods of time, and whole cell lysates were prepared. Extracts were analysed by Western blot. (d) U2OS cells were left untreated or treated for 4 h with 1% O2 prior to harvest, and ChIP analysis using the indicated antibodies was performed. PCR analysis using specific primers for the HIF-1α promoter and HIF-1α control region was performed.
DISCUSSION
Chronic inflammation is self-perpetuating and has been shown to distort the microenvironment as a result of aberrantly active transcription factors. Consequent alterations in growth factor, chemokine, cytokine and ROS balance within the cellular milieu provide the axis of growth and survival needed for de novo development of cancer and metastasis [25]. As such, we hypothesized that novel interactions between two key transcription factors, NF-κB and HIF, existed in this process. In the present study, we demonstrate that HIF-1α basal mRNA levels are controlled by several NF-κB subunits (Figures 1–3, 5 and 6). In addition, we show that TNFα-, and to some extent hypoxia-, induced NF-κB regulates HIF-1α levels and activity, triggering the transcription of target genes such as GLUT3 and PHD2 (Figures 4–6).
The results of the present study have numerous implications for a number of pathologies where NF-κB and HIF-1 are deregulated, such as rheumatoid arthritis or cancer. However, despite these implications, NF-κB regulation of HIF-1α could be stimulus-specific or even cell-type-specific. The evidence for this comes from the available genetic knockout models. While HIF-1α deletion results in embryonic lethality at day 9–11 [26,27], with defects evident as early as 7 days, combined genetic deletion of IKKα and IKKβ results in lethality at day 12 [28]. Single deletions of IKKα, IKKβ or RelA result in lethality at much later stages [28]. In HIF-1α−/− mice, defects are seen in neural tube formation, cardiovascular malformations and increases in cell death in the cephalic mesemchyme. Of interest, IKKα/β double null mice also have defects in neurulation, but these seem to be associated with increased apoptosis in the neuronal epithelium. It would be interesting to re-analyse these mice embryos for HIF-1α expression in the different tissues.
The results of the present study also demonstrate that TNFα induces HIF-1 activity despite the presence of oxygen. TNFα can produce ROS in cells [29] and these have been shown to inhibit PHD activity [30,31]. Although we have not investigated this possibility, the present results on HIF-1α stabilization and activity would support this. Precise ROS measurements and PHD activity assays would answer these questions.
Online data
Acknowledgments
We thank Professor Neil Perkins (Division of Gene Regulation and Expression, College of Life Sciences, University of Dundee, Dundee, Scotland, U.K.) and Dr Cameron Bracken (School of Molecular and Biomedical Science, University of Adelaide, Adelaide, Australia) for helpful discussion and reagents. We are also grateful to Dr Margaret Ashcroft (Cancer Research UK Centre for Cancer Therapeutics, Institute of Cancer Research, London, U.K.), Dr Carine Michiels (Department of Biology, University of Namur, Namur, Belgium) and Professor Inder Verma (Laboratory of Genetics, The Salk Institute for Biological Studies, La Jolla, CA, U.S.A.) for valuable reagents. We are particularly grateful to Dr Cari Culver and Sharon Mudie for help with the manuscript preparation and all of the people in the Wellcome Trust Centre for Gene Regulation and Expression and the College of Life Sciences at the University of Dundee for support. This research was funded by a BBSRC (Biotechnology and Biological Sciences Research Council) studentship (to P.v.U.), a Royal Society research grant, a Tenovus Scotland small grant, Association for International Cancer Research (to N. K.), Research Councils UK (to S. R.) and a MRC NIRG (Medical Research Council New Investigator Research Grant). The University of Dundee is a Scottish Registered Charity (No. SC015096).
References
- 1.Bardos J. I., Ashcroft M. Negative and positive regulation of HIF-1: a complex network. Biochim. Biophys. Acta. 2005;1755:107–120. doi: 10.1016/j.bbcan.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 2.Garcia J. A. HIFing the brakes: therapeutic opportunities for treatment of human malignancies. Science STKE. 2006 doi: 10.1126/stke.3372006pe25. PE25. [DOI] [PubMed] [Google Scholar]
- 3.Dery M. A., Michaud M. D., Richard D. E. Hypoxia-inducible factor 1: regulation by hypoxic and non-hypoxic activators. Int. J. Biochem. Cell Biol. 2005;37:535–540. doi: 10.1016/j.biocel.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 4.Fandrey J., Gorr T. A., Gassmann M. Regulating cellular oxygen sensing by hydroxylation. Cardiovasc. Res. 2006;71:642–651. doi: 10.1016/j.cardiores.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Berra E., Benizri E., Ginouves A., Volmat V., Roux D., Pouyssegur J. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1α in normoxia. EMBO J. 2003;22:4082–4090. doi: 10.1093/emboj/cdg392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Metzen E., Stiehl D. P., Doege K., Marxsen J. H., Hellwig-Bürgel T., Jelkmann W. Regulation of the prolyl hydroxylase domain protein 2 (phd2/egln-1) gene: identification of a functional hypoxia-responsive element. Biochem. J. 2005;387:711–717. doi: 10.1042/BJ20041736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pescador N., Cuevas Y., Naranjo S., Alcaide M., Villar D., Landazuri M. O., Del Peso L. Identification of a functional hypoxia-responsive element that regulates the expression of the egl nine homologue 3 (egln3/phd3) gene. Biochem. J. 2005;390:189–197. doi: 10.1042/BJ20042121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erez N., Stambolsky P., Shats I., Milyavsky M., Kachko T., Rotter V. Hypoxia-dependent regulation of PHD1: cloning and characterization of the human PHD1/EGLN2 gene promoter. FEBS Lett. 2004;567:311–315. doi: 10.1016/j.febslet.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 9.Lando D., Peet D. J., Gorman J. J., Whelan D. A., Whitelaw M. L., Bruick R. K. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002;16:1466–1471. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Catrina S. B., Okamoto K., Pereira T., Brismar K., Poellinger L. Hyperglycemia regulates hypoxia-inducible factor-1α protein stability and function. Diabetes. 2004;53:3226–3232. doi: 10.2337/diabetes.53.12.3226. [DOI] [PubMed] [Google Scholar]
- 11.Taylor P. C., Sivakumar B. Hypoxia and angiogenesis in rheumatoid arthritis. Curr. Opin. Rheumatol. 2005;17:293–298. doi: 10.1097/01.bor.0000155361.83990.5b. [DOI] [PubMed] [Google Scholar]
- 12.Bonello S., Zahringer C., BelAiba R. S., Djordjevic T., Hess J., Michiels C., Kietzmann T., Gorlach A. Reactive oxygen species activate the HIF-1α promoter via a functional NFκB site. Arterioscler. Thromb. Vasc. Biol. 2007;27:755–761. doi: 10.1161/01.ATV.0000258979.92828.bc. [DOI] [PubMed] [Google Scholar]
- 13.Hayden M. S., Ghosh S. Shared principles in NF-κB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 14.Perkins N. D., Gilmore T. D. Good cop, bad cop: the different faces of NF-κB. Cell Death Differ. 2006;13:759–772. doi: 10.1038/sj.cdd.4401838. [DOI] [PubMed] [Google Scholar]
- 15.Perkins N. D. Post-translational modifications regulating the activity and function of the nuclear factor κB pathway. Oncogene. 2006;25:6717–6730. doi: 10.1038/sj.onc.1209937. [DOI] [PubMed] [Google Scholar]
- 16.Anderson L. A., Perkins N. D. Regulation of RelA (p65) function by the large subunit of replication factor C. Mol. Cell. Biol. 2003;23:721–732. doi: 10.1128/MCB.23.2.721-732.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schumm K., Rocha S., Caamano J., Perkins N. D. Regulation of p53 tumour suppressor target gene expression by the p52 NF-κB subunit. EMBO J. 2006;25:4820–4832. doi: 10.1038/sj.emboj.7601343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a.Rocha S., Garrett M. D., Campbell K. J., Schumm K., Perkins N. D. Regulation of NF-κB and p53 through activation of ATR and Chk1 by the ARF tumour suppressor. EMBO J. 2005;24:1157–1169. doi: 10.1038/sj.emboj.7600608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iyer N. V., Leung S. W., Semenza G. L. The human hypoxia-inducible factor 1α gene: HIF1A structure and evolutionary conservation. Genomics. 1998;52:159–165. doi: 10.1006/geno.1998.5416. [DOI] [PubMed] [Google Scholar]
- 19.Minet E., Ernest I., Michel G., Roland I., Remacle J., Raes M., Michiels C. HIF1A gene transcription is dependent on a core promoter sequence encompassing activating and inhibiting sequences located upstream from the transcription initiation site and cis elements located within the 5′UTR. Biochem. Biophys. Res. Commun. 1999;261:534–540. doi: 10.1006/bbrc.1999.0995. [DOI] [PubMed] [Google Scholar]
- 20.Franzoso G., Carlson L., Xing L., Poljak L., Shores E. W., Brown K. D., Leonardi A., Tran T., Boyce B. F., Siebenlist U. Requirement for NF-κB in osteoclast and B-cell development. Genes Dev. 1997;11:3482–3496. doi: 10.1101/gad.11.24.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iotsova V., Caamano J., Loy J., Yang Y., Lewin A., Bravo R. Osteopetrosis in mice lacking NF-κB1 and NF-κB2. Nat. Med. 1997;3:1285–1289. doi: 10.1038/nm1197-1285. [DOI] [PubMed] [Google Scholar]
- 22.Rocha S. Gene regulation under low oxygen: holding your breath for transcription. Trends Biochem. Sci. 2007;32:389–397. doi: 10.1016/j.tibs.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 23.Jung Y., Isaacs J. S., Lee S., Trepel J., Liu Z. G., Neckers L. Hypoxia-inducible factor induction by tumour necrosis factor in normoxic cells requires receptor-interacting protein-dependent nuclear factor κB activation. Biochem. J. 2003;370:1011–1017. doi: 10.1042/BJ20021279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frede S., Stockmann C., Freitag P., Fandrey J. Bacterial lipopolysaccharide induces HIF-1 activation in human monocytes via p44/42 MAPK and NF-κB. Biochem. J. 2006;396:517–527. doi: 10.1042/BJ20051839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perwez Hussain S., Harris C. C. Inflammation and cancer: an ancient link with novel potentials. Int. J. Cancer. 2007;121:2373–2380. doi: 10.1002/ijc.23173. [DOI] [PubMed] [Google Scholar]
- 26.Iyer N. V., Kotch L. E., Agani F., Leung S. W., Laughner E., Wenger R. H., Gassmann M., Gearhart J. D., Lawler A. M., Yu A. Y., Semenza G. L. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1α. Genes Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryan H. E., Lo J., Johnson R. S. HIF-1α is required for solid tumor formation and embryonic vascularization. EMBO J. 1998;17:3005–3015. doi: 10.1093/emboj/17.11.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Q., Estepa G., Memet S., Israel A., Verma I. M. Complete lack of NF-κB activity in IKK1 and IKK2 double-deficient mice: additional defect in neurulation. Genes Dev. 2000;14:1729–1733. [PMC free article] [PubMed] [Google Scholar]
- 29.Babbar N., Casero R. A., Jr Tumor necrosis factor-α increases reactive oxygen species by inducing spermine oxidase in human lung epithelial cells: a potential mechanism for inflammation-induced carcinogenesis. Cancer Res. 2006;66:11125–11130. doi: 10.1158/0008-5472.CAN-06-3174. [DOI] [PubMed] [Google Scholar]
- 30.Bell E. L., Chandel N. S. Mitochondrial oxygen sensing: regulation of hypoxia-inducible factor by mitochondrial generated reactive oxygen species. Essays Biochem. 2007;43:17–27. doi: 10.1042/BSE0430017. [DOI] [PubMed] [Google Scholar]
- 31.Taylor C. T. Mitochondria and cellular oxygen sensing in the HIF pathway. Biochem. J. 2008;409:19–26. doi: 10.1042/BJ20071249. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.