Abstract
An alloplasmic wheat line, TA5536, with the “zebra” chromosome z5A was isolated from an Elymus trachycaulus/Triticum aestivum backcross derivative. This chromosome was named “zebra” because of its striped genomic in situ hybridization pattern. Its origin was traced to nonhomologous chromosome 5A of wheat and 1Ht of Elymus; four chromatin segments were derived from chromosome 1Ht and five chromatin segments including the centromere from 5A. In this study, our objective was to determine the mechanism of origin of chromosome z5A, whether by nonhomologous recombination or by multiple translocation events. Different crossing schemes were used to recover recombinants containing various Elymus chromatin segments of the z5A chromosome. In addition, one z5AL telocentric chromosome and three z5AL isochromosomes were recovered. The dissection of the Elymus segments into different stocks allowed us to determine the chromosomal origin of the different chromosome fragments on the basis of the order of the RFLP markers employed and suggested that the zebra chromosome originated from nonhomologous recombination. We present a model of possible mechanism(s) of chromosome evolution and step changes in chromosome number applicable to a wide range of organisms.
CLASSICAL phenomena of changes in multiple sets of basic chromosome number by polyploidy and step changes in basic chromosome number through aneuploidy have been documented for over a century. Stebbins (1950) provided the first synthesis of the enormous literature on chromosome variation and evolution in plants. However, such studies were limited to groups of species that could be hybridized and their meiotic behavior analyzed. With the dawn of the genomics era, comparative genomics and fluorescence in situ hybridization (FISH) are providing new insights into mechanisms of chromosome evolution, including changes in size, morphology, and number at the levels of tribes, families, or higher hierarchies sharing and spanning tens or even hundreds of million years of co-evolutionary history. Typically, one or more rounds of whole-genome duplications, followed by diploidization and step changes in basic chromosome numbers, have been hypothesized. In grasses, Salse et al. (2008) have provided a synthesis of the whole-genome duplications and step changes in basic chromosome numbers, including chromosome fissions, fusions, and translocations that have spawned basic chromosome changes of 1x = 5, 7, 10, or 12 in maize (1x = 10), wheat (1x = 7), rice (1x = 12), and sorghum (1x = 10), sharing 50 million years of evolutionary history. Apart from chromosome fission and fusion, unequal translocations followed by the elimination of a diminutive chromosome have been hypothesized in Arabidopsis species, leading to step changes of decrease in chromosome number from 1x = 8 to 1x = 5 (Lysak et al. 2006; Schubert 2007).
However, there have been only a few experimental demonstrations of step changes in basic chromosome number (reviewed in Schubert 2007). In wheat, there is a long history of interspecific hybridization and experimental introgression aimed at transferring useful genes from alien species into wheat. There is also a large body of literature on chromosome behavior and structural aberrations in interspecific hybrids. One particularly useful aspect of the wheat genetic system is buffering of the genome due to polyploidy that allows study of chromosome structure and behavior in isolation and over a large number of generations. Some key aspects of these studies were summarized by Gill (1991): (1) Interspecific hybridization is mutagenic; (2) many chromosomes from either one of the parents exhibit meiotic drive and show preferential transmission; and (3) cytoplasmic sterility resulting from nucleo-cytoplasmic interactions is common and leads to unilateral introgression of fertility restoration (Rf) and/or genes specific to the cytoplasmic donor in backcross derivatives.
We have studied one particular wide hybrid between Elymus trachycaulus (Link) Gould ex Shinners (2n = 4x = 28, StStHtHt) and wheat for >25 years. The original hybrid could only be made using E. trachycaulus as female and common wheat as male (Sharma and Gill 1981). Upon backcrossing with wheat, “unfavorable nucleocytoplasmic interactions were evident that led to seed shriveling, embryo abortion, failure of seed or cultured embryos to germinate, seedling death, plant weakness and sterility in the backcross derivatives” (Sharma and Gill 1983a,b). All viable and partially fertile alloplasmic derivatives carried chromosomes 1Ht or 1St or their short arms as telocentrics. Twelve simple or complex translocations involving 1St or 1StS with other E. trachycaulus or wheat chromosomes were recovered (Morris et al. 1990; Jiang et al. 1994). Among them, an alloplasmic wheat line carrying a zebra chromosome designated as z5A—a complex translocation involving 1HtS and wheat chromosome 5A that replaced wheat chromosome 5A—was recovered (Jiang and Gill 1993). Chromosome z5A gave a striped appearance upon genomic in situ hybridization (GISH) and was made up of four segments from E. trachycaulus and five segments including the centromere from wheat. This line was fully fertile and flowered 10 days earlier than the wheat recurrent parent.
In this article, our objective was to determine the mechanism of origin of chromosome z5A, whether by nonhomologous recombination or by multiple translocation events. By separating and genetically isolating the four E. trachycaulus segments of z5A using centric breakage and homologous recombination, we obtained evidence that chromosome z5A likely arose by nonhomologous recombination, resulting in a mosaic chromosome comprised of alternate blocks of Triticum aestivum and E. trachycaulus chromatin segments. A model is presented and the significance of this phenomenon is discussed in relation to chromosome evolution and repatterning forming new chromosomes and also leading to step changes in basic chromosome number in diverse organisms.
MATERIALS AND METHODS
Plant material:
An alloplasmic wheat line, TA5536, carrying the zebra chromosome z5A pair substituting for chromosome 5A of wheat in addition to 20 pairs of wheat chromosomes, was isolated from a derivative of an E. trachycaulus/T. aestivum cv. “Chinese Spring” (CS) hybrid (Sharma and Gill 1983a,b; Morris et al. 1990; Jiang 1993; Jiang and Gill 1993; Jiang et al. 1994). Chromosome z5A was recovered in progeny that had a complete 1Ht chromosome, a 1HtS telosome or isochromosome, or a T1HtS·5AL translocation chromosome in addition to a monosomic wheat chromosome 5A. These observations suggested that the E. trachycaulus chromatin in z5A was derived from chromosome 1Ht (Jiang and Gill 1993). Alloplasmic euploid wheat plants in E. trachycaulus cytoplasm are male sterile with reduced vigor. The short arm of chromosome 1Ht has the fertility restoration gene, Rf-Ht1. The alloplasmic stock TA5536 is vigorous and fully fertile, indicating that the 1Ht segments present in chromosome z5A have the Rf-Ht1 gene. The long arm of chromosome 5A has the major domestication gene Q, which controls the free-threshing character and square spike morphology (Huskins 1946; Unrau et al. 1950; Sears 1952, 1954; Mackey 1954; Simons et al. 2006). TA5536 has a square head and is free threshing, indicating that the chromosome 5A segment containing the Q gene is present in z5A. Meiotic metaphase I pairing revealed that the short arm of z5A paired with the short arm of chromosome 1Ht, whereas the distal region of the long arm of z5A paired with the distal region of 5AL. These data confirmed that chromosome z5A was derived from chromosomes 1Ht of E. trachycaulus and 5A of wheat (Jiang and Gill 1993).
Different crossing schemes were used to separate the four Elymus chromatin bands in chromosome z5A. For obtaining recombinants with different Elymus chromatin bands, the CS double ditelosomic dDt5A stock (20″ + 5AS″ + 5AL″) was crossed as female with TA5536. The F1 was either backcrossed with CS or selfed. To obtain z5A telosomes, the CS monosomic 5A stock was crossed as a female with TA5536. Because chromosome 5A is expected to be absent in ∼75% of the female gametes, the F1 plants were expected to segregate 25% with the chromosome constitution 20″ + z5A′ + 5A′ (42 chromosomes) with normal square spike morphology (two doses of Q) and 75% with 20″ + z5A′ (41 chromosomes) and speltoid (spelled-like) spikes (one dose of Q). Plants with speltoid spikes were selfed and screened for z5A telosomes using GISH. In addition, ditelosomic addition (DtA) stocks DtA1HtS and DtA1HtL were included in Southern hybridization analysis to enable the assignment of DNA markers to either the 1Ht arms or Elymus chromatin bands. All materials are maintained at the Wheat Genetic and Genomic Resources Center, Kansas State University, Manhattan, Kansas.
Mitotic and meiotic analyses:
Chromosome measurements were performed on 20 z5A and 1Ht chromosomes using 3B of wheat as a standard. Anthers from F1 plants of dDt5A/TA5536 undergoing meiotic metaphase I were fixed in Carnoy's solution I (100% ethanol:glacial acetic acid = 3:1), stained in 1% acetocarmine, and squashed in one drop of 45% acetic acid. Meiotic metaphase I pairing was analyzed in pollen mother cells (PMCs) after GISH.
FISH and GISH analyses:
Seeds were germinated in distilled water on filter paper in a petri dish at room temperature for 2–3 days until roots were ∼2-cm long. Roots were cut, pretreated in ice water for 24 hr, and fixed in Carnoy's solution I. FISH and GISH were according to Zhang et al. (2001).
The Aegilops tauschii Coss. plasmid clone pAet6-J9, containing a 750-bp sequence with high sequence similarity to the gag-pol polyprotein of the Ty3/gypsy retrotransposon cereba, was cloned into the plasmid pCR4Blunt-TOPO (Invitrogen Life Technologies, Carlsbad, CA) (Zhang et al. 2004). This clone contains a centromere-specific repetitive sequence that hybridizes to the centromeric regions of wheat, rye, barley, and maize.
Plasmid DNA was isolated using a QIAprep Spin miniprep kit (QIAGEN, Valencia, CA). Genomic DNA from E. trachycaulus was isolated using the standard CTAB method. One microgram of plasmid DNA was labeled with either rhodamine-6-dUTP using rhodamine-nick translation mix or fluorescein-12-dUTP (Roche Applied Science, Indianapolis) using nick translation according to the manufacturer's protocol. One microgram of total genomic DNA from E. trachycaulus was labeled with fluorescein-12-dUTP. Probes were purified with the QIAquick nucleotide removal kit (QIAGEN).
The composition of the FISH hybridization solution was as described in Zhang et al. (2004). The GISH hybridization solution (∼77% hybridization stringency) contained 50% deionized formamide (Fisher Scientific, Pittsburgh), 1.5× SSC, 10% dextran sulfate (Sigma, St. Louis), 0.17 mg/ml of sheared salmon testes DNA (Sigma), 1.3–2 μg/ml of labeled probe, and unlabeled total genomic DNA of wheat as a blocker. The probe:blocker ratio was ∼1:40–50. The hybridization procedures and post-hybridization washes were according to Zhang et al. (2004). Slides were counterstained with 4′,6-diamidino-2-phenylindole (Molecular Probes, Eugene, OR) or propidium iodide (PI) (Molecular Probes) for FISH when the probe was labeled with rhodamine-6-dUTP or fluorescein-12-dUTP, respectively, and mounted in Vectashield (Vector Laboratories, Burlingame, CA). GISH slides were counterstained with PI. Slides were analyzed with a Zeiss Axioplan 2 epifluorescence microscope and images were captured with a SPOT CCD (charge-coupled device) camera operated with Spot 2.1 software (Diagnostic Instruments, Sterling Heights, MI) and processed with Photoshop v5.5 software (Adobe Systems, San Jose, CA).
Southern hybridization:
Total genomic DNA isolation, digestion, gel electrophoresis, and Southern blot hybridization followed Faris et al. (2000). Probes used were selected from the consensus map of wheat homeologous group 1 chromosomes (Van Deynze et al. 1995).
RESULTS
Characterization of chromosome z5A by GISH, FISH, and meiotic pairing analyses:
Chromosome z5A has a length of 9.47 ± 0.79 μm (67% of 3B length) with an arm ratio (long/short) of 4.6. Chromosome 1Ht has a length of 8.23 ± 0.68 μm (60% of 3B length) with an arm ratio of 1.8. Chromosome 3B is the largest wheat chromosome with a size of 13.8 μm (Gill et al. 1991). GISH revealed that 33% of chromosome z5A (3.13 μm) was derived from E. trachycaulus and 67% (6.34 μm) was derived from wheat. The calculated size of 1HtS (2.99 μm) is similar to that of the 1HtS segment present in chromosome z5A as determined after GISH (3.13 μm). Similarly, the 5AL segment in z5A is 6.34 μm, which is just a little shorter than that of the 5AL reported earlier (7.7 μm, Gill et al. 1991). The measurement data suggested that almost the complete 5AL and 1HtS arms were retained in chromosome z5A.
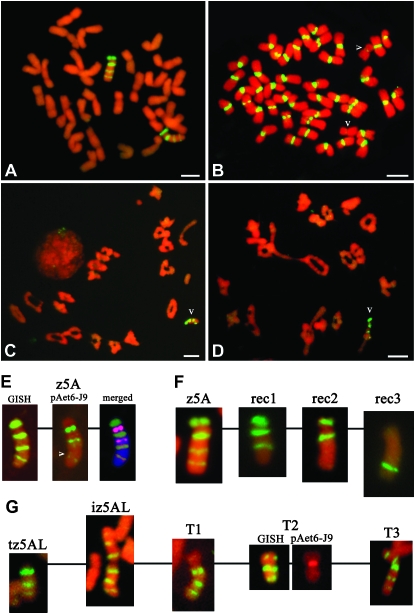
GISH analysis confirmed that chromosome z5A consists of four E. trachycaulus segments designated as E1, E2, E3, and E4, and five wheat segments, including the centromere, designated as W1, W2, W3, W4, and W5 (Figure 1A, Figure 2). The W1 segment contains the centromere derived from chromosome 5A. FISH with the centromeric clone pAet6-J9 on the disomic addition line DA1Ht revealed strong centromeric FISH sites on all wheat chromosomes, whereas chromosome 1Ht showed a very weak centromeric FISH signal (Figure 1B). Clone pAet6-J9 on TA5536 revealed three FISH sites on chromosome z5A (Figure 1E). Sequential GISH, performed on the same slide after stripping off the FISH probe, revealed that the major pAet6-J9 FISH site mapped to the centromeric W1 segment, whereas the second pAet6-J9 FISH site mapped between the W2 and W3 segments and the third minor pAet6-J9 FISH site mapped to the distal region of the E4 segment (Figure 1E). Colocalization of the minor pAet6-J9 FISH site to the E4 segment suggested that this site was derived from E. trachycaulus, in agreement with the much smaller centromeric pAet6-J9 FISH site in chromosome 1Ht compared to those of wheat chromosomes.
Figure 1.—
GISH and FISH pattern on mitotic (A, B, E, F, and G) and meiotic metaphase chromosomes (C and D) of TA5536 (A, C, D, and E), disomic addition line DA1Ht (B), z5A recombinant chromosomes (rec1–rec3) (F), telosomic chromosome tz5AL, isochromosome iz5AL, and translocation chromosomes (T1–T3) (G). Total genomic DNA from E. trachycaulus and the centromeric clone pAet6-J9 were labeled with fluorescein-12-dUTP (except for the pAet6-J9 FISH pattern on T2 in G) and visualized by yellow-green fluorescence. Chromosomes were counterstained with propidium iodide and fluoresced red. (A) The total genomic DNA from E. trachycaulus generated a striped GISH pattern on a pair of z5A chromosomes in TA5536. The four yellow-green chromosome segments originated from chromosome 1Ht of E. trachycaulus and the four red segments, including the centromere, originated from wheat chromosome 5A. (B) Clone pAet6-J9 hybridized strongly to all 42 wheat centromeres in line DA1Ht, but very weakly to the centromeres of chromosomes 1Ht (arrowheads). (C and D) GISH on the meiotic metaphase I cells in pollen mother cells from anthers of F1 plants of dDt5A/TA5536 with the chromosome constitution 20″ + z5A′ + 5AS′ + 5AL′. Chromosome z5A was univalent (arrowhead) in C and was paired in the distal region of the long arm with the 5AL telosome (arrowhead) in the form of a rod bivalent in D. (E) (Middle) Three FISH sites on chromosome z5A probed with clone pAet6-J9; (left) The sequential GISH result on the same chromosome; and (right) a merged image from the first left and middle images. The chromosome, the GISH signals, and the FISH signals were pseudocolored blue, green, and magenta, respectively. The merged image shows that the top FISH site mapped to the centromeric wheat segment (W1), the second site mapped to the second wheat segment (between W2 and W3), and the third site mapped to the distal region of the last Elymus segment (E4) (arrowhead). (F) GISH pattern on z5A chromosomes and three recombinant chromosomes derived from either selfed or backcrossed derivatives of dDt5A/TA5536. The chromosomes rec1, rec2, and rec3 had the first three (E1, E2, E3), first two (E1, E2), and the last Elymus segment(s) (E4), respectively. Rec3 is a telosomic chromosome. (G) tz5AL and iz5AL show a telosomic chromosome and an isochromosome, respectively; T1 shows a translocation chromosome with the short arm from an unknown wheat chromosome and the long arm from z5AL; T2 (GISH) shows a translocation chromosome with three Elymus bands (one in the short arm and two in the long arm). T2 (pAet6-J9) shows a sequential FISH pattern using clone pAet6-J9 labeled with rhodamine-6-dUTP and visualized by red fluorescence, identifying T2 as a small metacentric chromosome, with the GISH band in the short arm derived from the last Elymus segment of z5A and the two in the long arm corresponding to the middle two Elymus segments of z5A. T3 shows a translocation chromosome with five Elymus bands, two in the short arm corresponding to the last two Elymus segments of z5A, and the long arm of T3 is identical to the long arm of z5A. Bars, 10 μm.
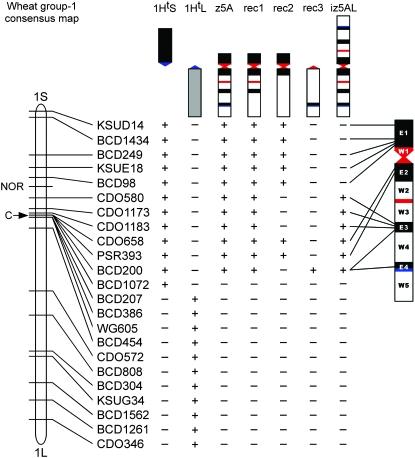
Figure 2.—
Schematic summary of Southern hybridization results and the molecular marker positions on the z5A chromosome. (Top) Schematics for telosomic chromosomes 1HtS and 1HtL, z5A, three recombinant chromosomes (rec1–rec3), and isochromosome iz5AL. A scaled-up z5A chromosome is shown on the right. The Elymus short-arm chromatin is in black, the Elymus long-arm chromatin is in gray, and wheat chromatin is in white. The Elymus centromere is shown in blue and the wheat centromere in red. A schematic of the consensus map of wheat group 1 chromosome is on the left. Only tested markers are listed. The “+” and “−” indicate “presence” and “absence” of markers, respectively. The marker order in Elymus segments E1 and E4 is conserved relative to wheat, whereas the order is reversed in segments E2 and E3. None of the group 1 long-arm markers hybridized to z5A.
Meiotic metaphase I pairing was analyzed in 197 PMCs of plants with the chromosome constitution 20″ + z5A′ + 5AS′ + 5AL′. Chromosome z5A was univalent in 37 PMCs (19%) (Figure 1C) and paired in the distal region of the long arm with the 5AL telosome as a rod bivalent in 160 PMCs (81%) (Figure 1D). These data confirmed that the distal W5 segment in chromosome z5A was derived from the distal region of 5AL. The chiasmata were almost exclusively in the distal regions of z5A and 5AL, with no chiasma formed in proximal regions.
Identification of z5A recombinants:
The F1 hybrids of the cross dDt5A/TA5536 were either backcrossed with CS or selfed. One hundred and nineteen BC1 plants and 68 F2 plants were screened for recombinants using GISH. Almost all detected recombination events (55 of 56) occurred in the W4 segment, resulting in 28 recombinants having the first three E. trachycaulus segments (E1, E2, and E3) on a complete z5A chromosome, designated as rec1, and 27 recombinants with only the E4 segment on a 5AL telosome, designated as rec3 (Figure 1F). The frequent recovery of the rec1 and rec3 recombinants further suggested that the W4 segment in z5A is homologous and not rearranged relative to the corresponding region of wheat chromosome 5A. Only one recombinant event occurred in the W3 segment and resulted in a z5A recombinant chromosome with only the E1 and E2 segments (rec2, Figure 1F). The recovery of the rec2 recombinant likewise suggested that the W3 segment in z5A is homologous and not rearranged relative to the corresponding region of wheat chromosome 5A.
Separation of the first E. trachycaulus chromatin segment (E1) from the remaining three segments (E2, E3, and E4):
F1 hybrids from the cross of a monosomic 5A plant with TA5536 with speltoid spikes, and thus putatively monosomic for z5A, were selfed. One hundred and ten F2 individuals were screened for telocentric chromosomes using GISH. One plant with a tz5AL telosomic chromosome and a normal z5A chromosome (2n = 41 + t) having a square spike (two doses of the Q gene), one plant with a z5AL isochromosome (iz5AL) (2n = 40 + iz5AL) (square spike, two doses of the Q gene), and two plants with an iz5AL and a normal z5A chromosome (2n = 41 + iz5AL) (compactoid spike, three doses of the Q gene) were recovered (Figure 1G). In addition, plants with three different types of translocation chromosomes were identified (Figure 1G), including (1) T1, with the short arm from an unknown wheat chromosome and the long arm from z5AL; (2) T2, with three Elymus chromatin bands (one band in the short arm and two bands in the long arm); and (3) T3, with five Elymus chromatin bands. The centromere-specific probe pAet6-J9 was used to determine the location of the centromere in T2 by FISH and identified T2 as a small metacentric chromosome (Figure 1G). The GISH pattern of T2 suggested that the GISH band in the short arm was derived from the E4 segment of z5A, whereas the two Elymus segments in the long arm corresponded to the E2 and E3 segments of z5A. The GISH pattern of T3 suggested that the two Elymus segments in the short arm correspond to the E3 and E4 segments of z5A, whereas the long arm of T3 is identical to the long arm of z5A. The T3 translocation stock had speltoid spikes and low fertility, with only 20 seeds produced on 16 tillers. Although z5A long-arm telosome and isochromosomes were recovered, no z5A short-arm telosome was obtained.
Southern hybridization analysis:
Chromosome 1Ht is homeologous to the group 1 chromosomes of wheat. Molecular markers spanning the short and long arms of wheat group 1 chromosomes were used to map the Elymus segments in chromosome z5A by Southern hybridization analysis (Figure 2). The recovered recombinants, derived tz5AL telosome, and iz5AL isochromosomes allowed the assignment of wheat group 1 short-arm markers to specific Elymus segments in chromosome z5A. None of the group 1 long-arm markers hybridized to z5A, indicating that all the Elymus segments in z5A were derived from the short arm of chromosome 1Ht. The presence of centromeric marker BCD1072 in 1HtS and its absence in z5A indicated that not all the short arm of 1Ht is present in chromosome z5A. Jiang and Gill (1993) similarly reported that the short-arm marker PSR161, previously mapped to 1HtS, was absent in chromosome z5A. Later it was shown that marker PSR161 mapped to the functional centromere of wheat group 1 chromosomes (Francki et al. 2002), indicating that the 1HtS chromatin missing in z5A is closely associated with the centromere.
Separation of the four E. trachycaulus segments in chromosome z5A also allowed the determination of the marker order in these segments. Southern hybridization analysis revealed that the marker order in the Elymus segments E1 and E4 was conserved relative to wheat, whereas they were reversed in segments E2 and E3 (Figure 2). The reversed marker order in the latter two segments was probably caused by a paracentric inversion in the short arm of chromosome 1Ht. The short-arm marker BCD200 was mapped to both E3 and E4 E. trachycaulus segments, suggesting that a duplication event occurred during the origin of chromosome z5A.
DISCUSSION
Southern hybridization analysis revealed that the four E. trachycaulus segments present in chromosome z5A were derived from the short arm of 1Ht. However, 1HtS-specific centromeric marker PSR161 and the adjacent marker BCD1072 were missing in z5A, indicating that not all of the 1HtS arm was included in the formation of 5A. The mapping data further indicated the presence of a paracentric inversion in 1HtS.
Although Southern hybridization analysis did not allow the assignment of 5AL markers to the wheat segments in z5A, the structure of these segments can be inferred from GISH and meiotic pairing analyses. This study confirmed previous reports by showing that the distal part of the short arm of chromosome z5A paired with the distal region of the short arm of 1Ht and that the distal region of the long arm of z5A paired with the distal region of wheat arm 5AL (Jiang and Gill 1993). In the presence of the major meiotic pairing locus Ph1 in wheat, homology at chromosome ends triggers the formation of chiasmate metaphase I associations (Curtis et al. 1991; Jones et al. 2002; Qi et al. 2002). Thus, E1 was derived from the distal region of 1HtS, and W5 was derived from the distal region of 5AL.
GISH analysis showed that the functional centromere was derived from wheat and was located within the W1 segment. The homology of wheat segments W3 and W4 was inferred from the recovered recombinants rec2, rec1, and rec3. In the presence of Ph1, recombination occurs normally only between homologous regions in wheat. The recovery of these recombinants indicated that the W3 and W4 segments in chromosome z5A are not structurally rearranged but are homologous to corresponding regions in chromosome 5AL.
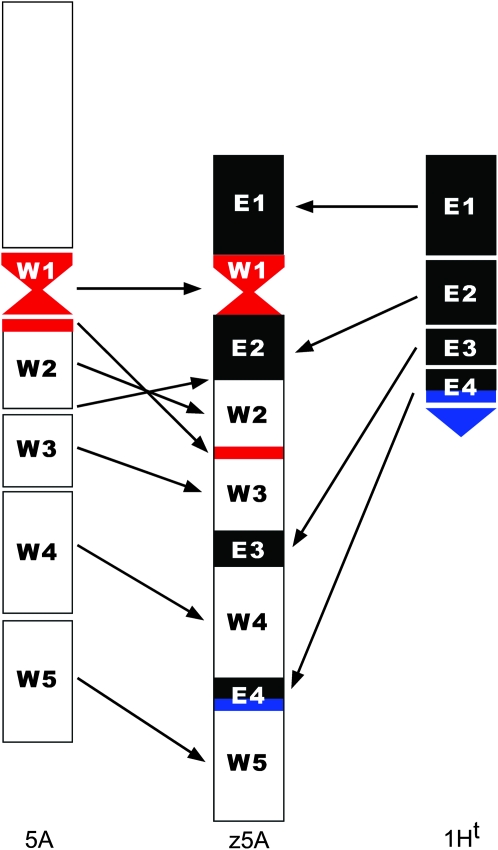
During the formation of the zebra chromosome, many changes occurred not only in chromosome 1Ht of Elymus, but also in chromosome 5A of wheat. As seen from our sequential FISH and GISH results, the original centromere on wheat chromosome 5A was split into two portions in z5A chromosome (Figure 1E, pAet6-J9 on z5A; see also Figure 3). Although two of the group 5 long-arm markers, including Q (this study and Jiang and Gill 1993) and H1 (Jiang and Gill 1993), also are present in the z5A chromosome, some wheat markers in chromosome 5AL were deleted in z5A.
Figure 3.—
Proposed origin of chromosome z5A by chromosome breakage and fusion of the broken chromatin blocks. (Left) Wheat chromosome 5A, (middle) z5A, and (right) telosome 1HtS, from which the four Elymus segments present in z5A were derived. The centromeres of wheat and Elymus are shown in red and blue, respectively. The original centromere of wheat chromosome 5A was split into two portions in z5A. The major part is the functional centromere, located within the W1 segment, and the smaller part is located in W2. The third and smallest centromere was derived from part of the centromere of chromosome 1Ht (E4). Marker analysis indicated that the entire short arm of 1Ht was not present in z5A. GISH and meiotic pairing analyses showed that the E1 segment was derived from the distal region of 1HtS and W4 was derived from the distal region of 5AL. Because the marker order in segments E1 and E4 is conserved relative to wheat, but reversed in segments E2 and E3, either a paracentric inversion in the short arm preexisted in 1Ht or fusion of the broken chromatin blocks occurred, as indicated.
In addition to the recombinants, telosome, and isochromosomes, three different types of translocation chromosomes were recovered. The overall high translocation frequency (3 of 110) involving the z5A chromosome indicated that this chromosome is prone to breakage and translocation.
Jiang and Gill (1993) postulated two possible origins for the zebra chromosome. One was that it arose by illegitimate recombination between nonhomologous chromosome arms 1HtS of E. trachycaulus and 5AL of wheat. In this scenario, the marker order of the four Elymus and five wheat segments is expected to be linear from one telomere to the other on chromosome z5A. The second possibility was that chromosome z5A arose from multiple breakage and fusion translocation events, and, in this case, the marker order is expected to be random. With the exception of Elymus segments E2 and E3, where the marker orders are reversed relative to wheat—most likely resulting from a paracentric inversion in 1HtS—the marker orders or homologies of the remaining Elymus and wheat segments are linear from one telomere to the other. Such an order would be highly unlikely if chromosome z5A originated by multiple translocation events.
On the basis of our data, a possible mode of origin of chromosome z5A is shown in Figure 3. The alloplasmic wheat plant was double monosomic for 5A and 1HtS and the two nonhomologous chromosomes may have prealigned prior to S-phase of meiosis or mitosis in germinal tissues during transition from the vegetative to reproductive phase. Chromosome z5A might have originated from nonhomologous recombination during the DNA double-strand break (DSB) repair process (Gorbunova and Levy 1999; Pacher et al. 2007) during the S-phase. Initially, at least five breaks occurred in chromosome 5A, resulting in six chromatin blocks. Similarly, four breaks in the 1HtS telosome split it into five chromatin blocks. Rejoining of the prealigned 5AL and 1HtS blocks in succession resulted in the formation of chromosome z5A. It was accompanied by the complete loss of 5AS, including the telomere and the loss of a very small distal centromeric end of 1HtS. The formation of chromosome z5A was not the result of a reciprocal segment exchange.
Amazingly, the z5A has the centromere of 5A of wheat embedded in E1 and E2 segments from 1HtS of E. trachycaulus. Its short arm, including the telomere, was derived from E. trachycaulus and its long arm is a chimera of 5AL and 1HtS segments with the distal part and the telomere derived from 5AL. This represents an aneuploid change in chromosome number from two to one and the beginning of the evolutionary history of a new z5A chromosome.
The aneuploid changes in chromosome number and/or the origin of structurally rearranged chromosomes may be associated with interspecific hybridization. The F1 hybrids are usually sterile and one component of sterility is due to adverse nucleo-cytoplasmic interactions (NCI) (Gill 1991; Maan 1991). During the process of evolutionary introgressive hybridization in subsequent generations, certain chromosomes that overcome adverse NCI and restore fertility are exclusively transmitted to the progeny and are often involved in complex chromosomal rearrangements (Gill 1991). In alloplasmic hybrid derivatives between T. aestivum and E. trachycaulus, all progeny plants in backcrosses to wheat carried either chromosome 1Ht or 1St or their short arms as telocentrics or in complex rearrangements with wheat chromosomes (Jiang and Gill 1993). The z5A plant was recovered in one such progeny. It was fully fertile and flowered 10 days earlier than the Chinese Spring wheat, the recurrent parent. In nature, the z5A plant will be a candidate for a founder population for the evolution of a new species.
The two genomes in alloplasmic hybrids between T. aestivum and E. trachycaulus exist in a common nucleus in an Elymus cytoplasm. Because of the incompatibility and genomic stress imposed by interspecific hybridization, certain intergenomic and/or chromosomal structural rearrangements might have occurred (Gill 1991), similar to those reported by Naranjo et al. (1987) and Jiang and Gill (1994). Nonhomologous recombination might be initiated to repair chromosomal DSBs by joining sequences with little or no homology. If chromosomes 1Ht and 5A were in close proximity at the time of DSB repair, DNA ends or other chromosome breaks on 1Ht and 5A might have been joined by nonhomologous end joining. Alternatively, because there were free DNA ends from chromosomal DSBs, these free ends might have enhanced random integration by the copy-join process of nonhomologous recombination to link new DNA sequences together (Merrihew et al. 1996). In our case, the sequences from the Elymus 1Ht chromosome were linked to those from wheat chromosome 5A because these two chromosomes were by chance physically close at the time.
The molecular and cytogenetic research has documented extensive chromosome changes, including inversions, translocations, and fusions/fissions leading to aneuploid changes and karyotype repatterning in the Brassica family (reviewed in Schubert 2007). It was suggested that aneuploid changes as well as extensive chromosome repatterning—including inversions, unequal translocations involving nonhomologous chromosomes, and elimination of minichromosomes—led to step changes in chromosome number from n = 8 in Arabidopsis lyrata to n = 5 in A. thaliana (Lysak et al. 2006). The experimental data on the origin of z5A allude to additional mechanisms. Results from our study indicated that the zebra chromosome arose by illegitimate recombination between nonhomologous chromosomes of T. aestivum and E. trachycaulus, resulting in a mosaic chromosome comprised of alternate blocks of T. aestivum and E. trachycaulus chromatin. Although our findings involved a polyploid species, it is possible that similar mechanisms might have been involved in diploid species.
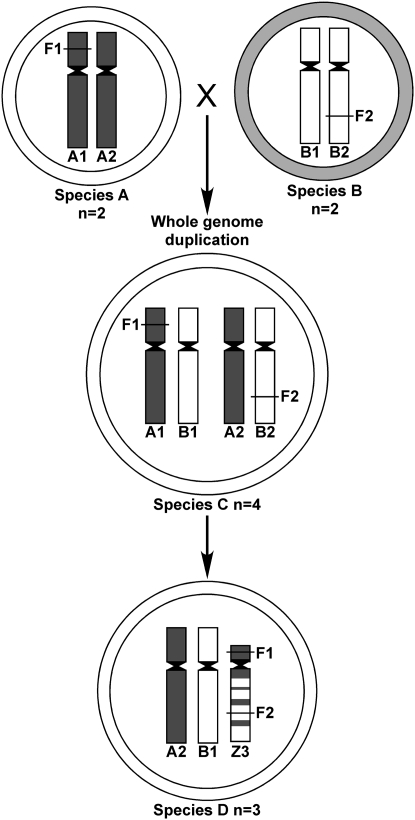
A hypothetical scheme showing possible evolutionary chromosomal changes during speciation is shown in Figure 4. We assume species A and species B with n = 2 co-evolved from a common ancestor and have differentiated cytoplasm and nuclear genomes. Species A has fertility factor F1 on A1 chromosome that is essential for restoring fertility to species-A-specific cytoplasm. Species B has another fertility factor, F2 on B2 chromosome, that is essential for the viability of the gametophyte or the sporophyte. Next, species A and B come in contact and undergo hybridization and a whole-genome duplication to form species C with n = 4 chromosomes. The whole-genome duplication is a polyploidy-associated mechanism that fixes hybrid vigor. However, hybridization is also mutagenic and may trigger chromosomal repatterning events. Genome duplication provides buffering for the formation of new chromosomes that otherwise may be deleterious because they involve segmental deletions and duplications. Most such events will be lethal; however, those that bring genetic factors F1 and F2 onto the novel chromosome Z3 will have strong selective advantage and may be fixed to form species D with n = 3 chromosomes. In our experimental hybrids, the z5A chromosome in the alloplasmic z5A plant had the Rf gene from E. trachycaulus that restored fertility. The group 5 long-arm chromosomes in the Triticeae are known to carry genes essential for the viability of the gametophyte (Endo and Gill 1996) and alien group 5 chromosomes show exclusive preferential transmission (Jiang and Gill 1998). This gene is the hypothetical gene F2 and because the z5A carried these two essential genes, the hybrid derivatives carrying z5A will be fertile and this chromosome will be fixed in the population.
Figure 4.—
Hypothetical scheme leading to changes in chromosome number after interspecific hybridization, whole-genome duplication, and chromosomal rearrangements. Species A and B with n = 2 chromosomes have differentiated cytoplasms (indicated by open and shaded outer circles) and nuclear genomes. Species A has a fertility factor F1 on chromosome A1 that is essential for restoring fertility to species-A-specific cytoplasm and species B has another fertility factor, F2 on chromosome B2, that is essential for gametophytic or sporophytic viability. Interspecific hybridization between species A and B followed by whole-genome duplication created genomic stress that resulted in chromosomal repatterning and the formation of the novel chromosome Z3 by nonhomologous recombination, harboring both fertility factors F1 and F2. Plants with such a chromosomal constitution have strong selective advantage and will form species D with n = 3 chromosomes.
Acknowledgments
We thank Robert A. McIntosh and W. John Raupp for critical reading of the manuscript, Duane Wilson for excellent assistance, Li Huang, Lili Qi, Chongmei Dong, Scott Jackson, and Jiming Jiang for beneficial discussions. This research was supported by the Kansas Wheat Commission and a special U. S. Department of Agriculture grant to the Wheat Genetic and Genomic Resources Center. This article is contribution no. 08-323-J from the Kansas Agricultural Experimental Station, Kansas State University, Manhattan, Kansas 66506-5502.
References
- Curtis, C. A., A. J. Lukaszewski and M. Chrzasiek, 1991. Metaphase-I pairing of deficient chromosomes and genetic mapping of deficiency breakpoints in wheat. Genome 34 553–560. [Google Scholar]
- Endo, T. R., and B. S. Gill, 1996. The deletion stocks of common wheat. J. Hered. 87 295–307. [Google Scholar]
- Faris, J. D., K. M. Haen and B. S. Gill, 2000. Saturation mapping of a gene-rich recombinant hot spot region in wheat. Genetics 154 823–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francki, M. G., W. A. Berzonsky, H. W. Ohm and J. M. Anderson, 2002. Physical location of a HSP70 gene homologue on the centromere of chromosome 1B of wheat (Triticum aestivum L.). Theor. Appl. Genet. 104 184–191. [DOI] [PubMed] [Google Scholar]
- Gill, B. S., 1991. Nucleo-cytoplasmic interaction (NCI) hypothesis of genome evolution and speciation in polyploid plants. Nuclear and Organellar Genomes of Wheat Species. Proceedings of the Dr. H. Kihara Memorial International Symposium on Cytoplasmic Engineering in Wheat, edited by T. Sasakuma and T. Kinoshita. Kihara Memorial Yokohama Foundation for the Advancement of Life Science, Yokohama, Japan, pp. 48–53.
- Gill, B. S., B. Friebe and T. R. Endo, 1991. Standard karyotype and nomenclature system for description of chromosome bands and structural aberrations in wheat (Triticum aestivum). Genome 34 830–839. [Google Scholar]
- Gorbunova, V., and A. A. Levy, 1999. How plants make ends meet: DNA double-strand break repair. Trends Plant Sci. 4 263–269. [DOI] [PubMed] [Google Scholar]
- Huskins, C. L., 1946. Fatuoid, speltoid and related mutations of oats and wheat. Bot. Rev. 12 457–514. [Google Scholar]
- Jiang, J., 1993. Introgression of Elymus trachycaulus chromatin into common wheat. Ph.D. Thesis, Kansas State University, Manhattan, KS. [DOI] [PubMed]
- Jiang, J., and B. S. Gill, 1993. A ‘zebra’ chromosome arising from multiple translocations involving non-homologous chromosomes. Chromosoma 102 612–617. [DOI] [PubMed] [Google Scholar]
- Jiang, J., and B. S. Gill, 1994. Different species-specific chromosome translocations in Triticum timopheevii and T. turgidum support the diphyletic origin of polyploidy wheats. Chromosome Res. 2 59–64. [DOI] [PubMed] [Google Scholar]
- Jiang, J., and B. S. Gill, 1998. Preferential male transmission of an alien chromosome in wheat. J. Hered. 89 87–89. [Google Scholar]
- Jiang, J., K. L. D. Morris and B. S. Gill, 1994. Introgression of Elymus trachycaulus chromatin into common wheat. Chromosome Res. 2 3–13. [DOI] [PubMed] [Google Scholar]
- Jones, E., K. Rybka and A. J. Lukaszewski, 2002. The effect of a deficiency and a deletion on recombination in chromosome 1B in wheat. Theor. Appl. Genet. 104 1204–1208. [DOI] [PubMed] [Google Scholar]
- Lysak, M. A., A. Berr, A. Pecinka, R. Schmidt, K. McBreen et al., 2006. Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc. Natl. Acad. Sci. USA 103 5224–5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maan, S. S., 1991. Nucleo-cytoplasmic genetics of wheat. Nuclear and Organellar Genomes of Wheat Species. Proceedings of the Dr. H. Kihara Memorial International Symposium on Cytoplasmic Engineering in Wheat, edited by T. Sasakuma and T. Kinoshita. Kihara Memorial Yokohama Foundation for the Advancement of Life Science, Yokohama, Japan, pp. 175–194.
- MacKey, J., 1954. Neutron and X-ray experiments in wheat and a revision of the speltoid problem. Hereditas 40 65–180. [Google Scholar]
- Merrihew, R. V., K. Marburger, S. L. Pennington, D. B. Roth and J. H. Wilson, 1996. High-frequency illegitimate integration of transfected DNA at preintegrated target sites in a mammalian genome. Mol. Cell. Biol. 16 10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, K. L. D., W. J. Raupp and B. S. Gill, 1990. Isolation of Ht genome chromosome additions from polyploid Elymus trachycaulus (StStHtHt) into common wheat (Triticum aestivum). Genome 33 16–22. [Google Scholar]
- Naranjo, T., P. G. Roca, P. G. Goicoechea and R. Giraldez, 1987. Arm homoeology of wheat and rye chromosomes. Genome 29 873–882. [Google Scholar]
- Pacher, M., W. Schmidt-Puchta and H. Puchta, 2007. Two unlinked double-strand breaks can induce reciprocal exchanges in plant genomes via homologous recombination and nonhomologous end joining. Genetics 175 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, L., B. Friebe and B. S. Gill, 2002. A strategy for enhancing recombination in proximal regions of chromosomes. Chromosome Res. 10 645–654. [DOI] [PubMed] [Google Scholar]
- Salse, J., S. Bolot, M. Throude, V. Jouffe, B. Piegu et al., 2008. Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution. Plant Cell 20 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert, I., 2007. Chromosome evolution. Curr. Opin. Plant Biol. 10 109–115. [DOI] [PubMed] [Google Scholar]
- Sears, E. R., 1952. Misdivision of univalents in common wheat. Chromosoma 4 535–550. [DOI] [PubMed] [Google Scholar]
- Sears, E. R., 1954. The aneuploids of common wheat. Mo. Agr. Exp. Sta. Res. Bull. 572 1–59. [Google Scholar]
- Sharma, H. C., and B. S. Gill, 1981. Wide hybridization. Ann. Wheat Newsl. 27 106. [Google Scholar]
- Sharma, H. C., and B. S. Gill, 1983. a New hybrids between Agropyron and wheat. 2. Production, morphology and cytogenetic analysis of F1 hybrids and backcross derivatives. Theor. Appl. Genet. 66 111–121. [DOI] [PubMed] [Google Scholar]
- Sharma, H. C., and B. S. Gill, 1983. b New hybrids between Agropyron and wheat. III. Backcross derivatives, effect of Agropyron cytoplasm and production of Agropyron addition lines, pp. 213–221 in Proceedings of the 6th International Wheat Genetics Symposium. Plant Germ-Plasm Institute, Kyoto, Japan.
- Simons, K. C., J. P. Fellers, H. N. Trick, Z. Zhang, Y. S. Tai et al., 2006. Molecular characterization of the major wheat domestication gene Q. Genetics 172 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins, G. L., Jr., 1950. Variation and Evolution in Plants. Columbia University Press, New York.
- Unrau, J., W. E. Smith and R. C. McGinnis, 1950. Spike density, speltoidy and compactoidy in hexaploid wheat. Can. J. Res. 28 273–276. [Google Scholar]
- Van Deynze, A. E., J. C. Nelson, M. E. Sorrells, S. R. McCouch, J. Dubcovsky et al., 1995. Molecular-genetic maps for group 1 chromosomes of Triteceae species and their relation to chromosomes in rice and oat. Genome 38 45–59. [DOI] [PubMed] [Google Scholar]
- Zhang, P., B. Friebe, A. J. Lukaszewski and B. S. Gill, 2001. The centromere structure in Robertsonian wheat-rye translocation chromosomes indicates that centric breakage-fusion can occur at different positions within the primary constriction. Chromosoma 110 335–344. [DOI] [PubMed] [Google Scholar]
- Zhang, P., W. Li, J. Fellers, B. Friebe and B. S. Gill, 2004. BAC-FISH in wheat identifies chromosome landmarks consisting of different types of transposable elements. Chromosoma 112 288–299. [DOI] [PubMed] [Google Scholar]