Abstract
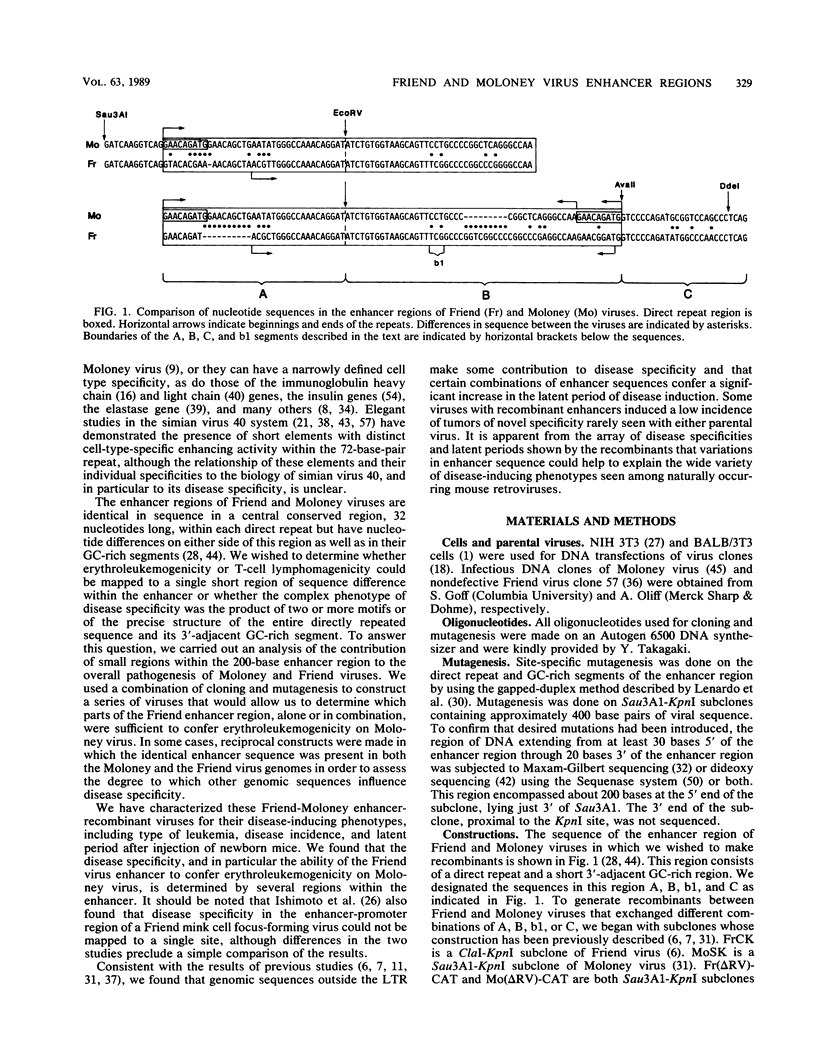
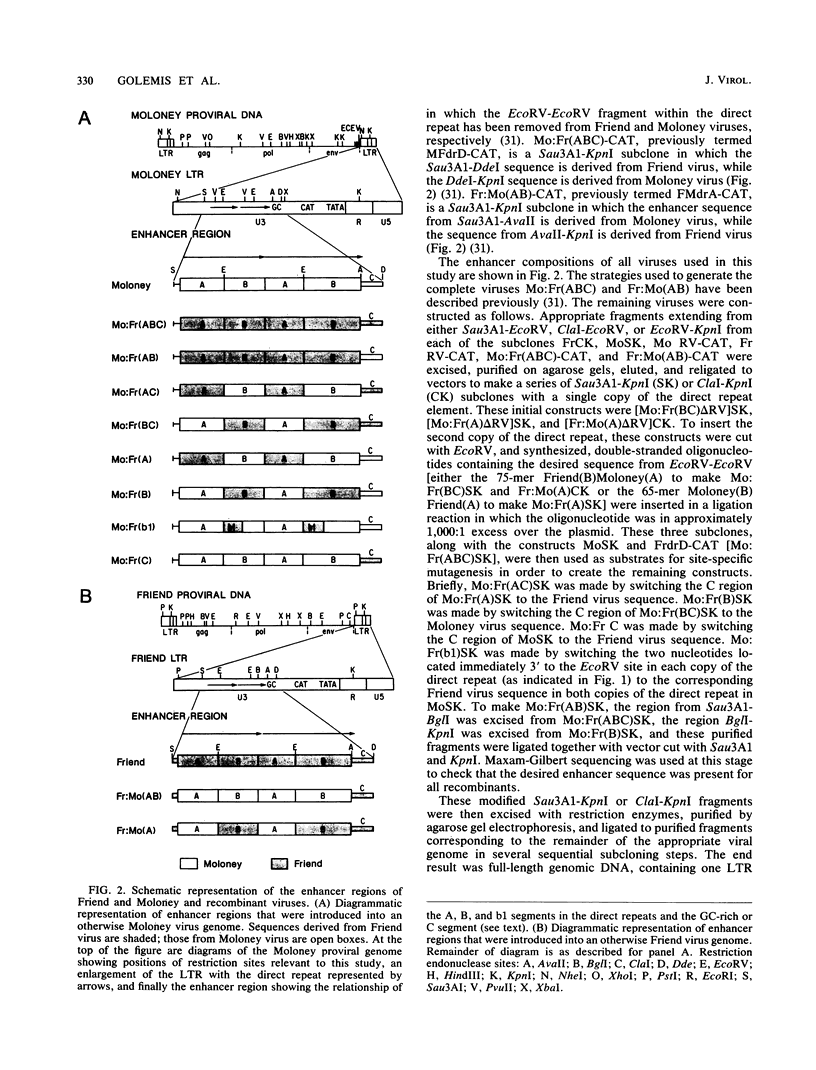
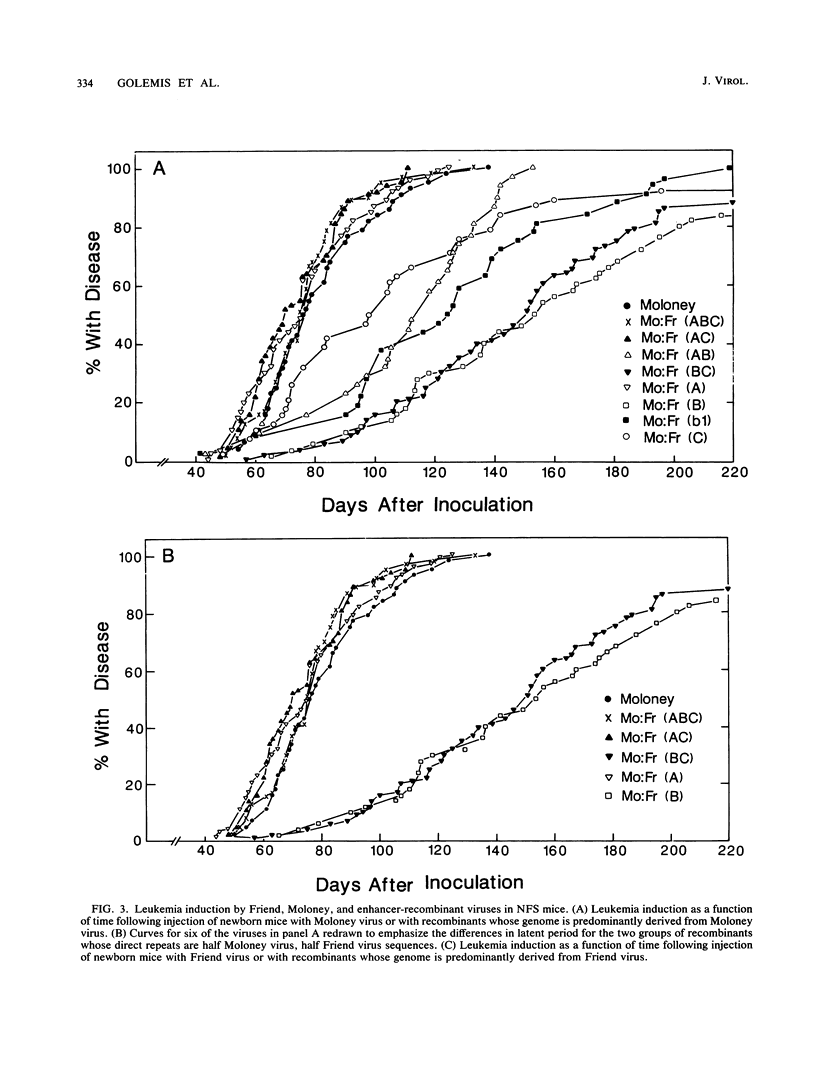
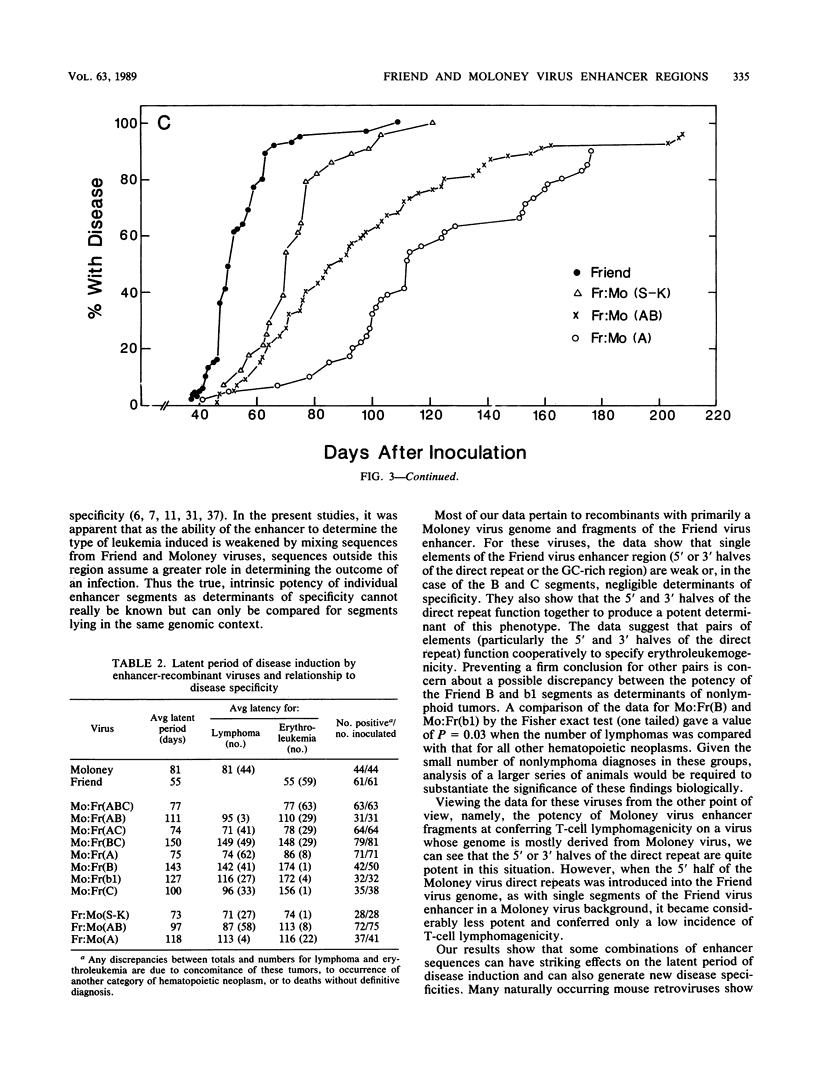
The nondefective Moloney and Friend murine leukemia viruses induce T-cell lymphomas and erythroleukemias, respectively, after being injected into newborn NFS mice. In previous studies, we showed that the distinct disease specificities of the two viruses could be switched by exchanging a small segment, about 200 nucleotides in length, encompassing their enhancer regions. This segment included the direct repeat sequence and an adjacent GC-rich region of about 20 nucleotides defined in studies of Moloney murine sarcoma virus enhancer-promoter function (L. A. Laimins, P. Gruss, R. Pozzatti, and G. Khoury, J. Virol. 49:183-189, 1984). The direct repeats of Friend and Moloney viruses are identical in a central core sequence of 32 nucleotides but have sequence differences on either side of this core as well as in their GC-rich segments. To determine whether disease specificity resides in part or in all of the direct repeat and GC-rich region, we constructed recombinants between Friend and Moloney viruses within this segment and tested them for their disease-inducing phenotypes. We found that disease specificity, in particular the ability of Friend virus sequence to confer erythroleukemogenicity on Moloney virus, is encoded throughout the region in at least three separable segments: the 5' and 3' halves of the direct repeat and the GC-rich segment. When just one of these segments (either both 5' halves of the direct repeat, both 3' halves, or just the GC-rich segment) from Friend virus was substituted into a Moloney virus genome, it conferred only a negligible or low incidence of erythroleukemia (less than or equal to 5% to between 10 and 15%). Any two segments together were considerably more potent (35 to 95% erythroleukemia), with the most effective pair being the two halves of the direct repeat. Individual segments and pairs of segments were considerably more potent determinants when they were matched with a genome of the same origin. Thus, although sequences outside the enhancer region are minor determinants of disease specificity when the enhancer is derived entirely from either Friend or Moloney virus, they can play a significant role when the enhancer is of mixed origin. Some recombinant enhancers conferred a long latent period of disease induction. This was particularly striking when the 5' halves of each copy of the direct repeat sequence were derived from Moloney virus and the 3' halves were derived from Friend virus.(ABSTRACT TRUNCATED AT 400 WORDS)
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aaronson S. A., Todaro G. J. Development of 3T3-like lines from Balb-c mouse embryo cultures: transformation susceptibility to SV40. J Cell Physiol. 1968 Oct;72(2):141–148. doi: 10.1002/jcp.1040720208. [DOI] [PubMed] [Google Scholar]
- Baltimore D. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature. 1970 Jun 27;226(5252):1209–1211. doi: 10.1038/2261209a0. [DOI] [PubMed] [Google Scholar]
- Bösze Z., Thiesen H. J., Charnay P. A transcriptional enhancer with specificity for erythroid cells is located in the long terminal repeat of the Friend murine leukemia virus. EMBO J. 1986 Jul;5(7):1615–1623. doi: 10.1002/j.1460-2075.1986.tb04404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celander D., Haseltine W. A. Tissue-specific transcription preference as a determinant of cell tropism and leukaemogenic potential of murine retroviruses. Nature. 1984 Nov 8;312(5990):159–162. doi: 10.1038/312159a0. [DOI] [PubMed] [Google Scholar]
- Celander D., Hsu B. L., Haseltine W. A. Regulatory elements within the murine leukemia virus enhancer regions mediate glucocorticoid responsiveness. J Virol. 1988 Apr;62(4):1314–1322. doi: 10.1128/jvi.62.4.1314-1322.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatis P. A., Holland C. A., Hartley J. W., Rowe W. P., Hopkins N. Role for the 3' end of the genome in determining disease specificity of Friend and Moloney murine leukemia viruses. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4408–4411. doi: 10.1073/pnas.80.14.4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatis P. A., Holland C. A., Silver J. E., Frederickson T. N., Hopkins N., Hartley J. W. A 3' end fragment encompassing the transcriptional enhancers of nondefective Friend virus confers erythroleukemogenicity on Moloney leukemia virus. J Virol. 1984 Oct;52(1):248–254. doi: 10.1128/jvi.52.1.248-254.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Company M., Errede B. Cell-type-dependent gene activation by yeast transposon Ty1 involves multiple regulatory determinants. Mol Cell Biol. 1987 Sep;7(9):3205–3211. doi: 10.1128/mcb.7.9.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone R. D., Weber-Benarous A., Baorto D., Mulligan R. C. Regulated expression of a complete human beta-globin gene encoded by a transmissible retrovirus vector. Mol Cell Biol. 1987 Feb;7(2):887–897. doi: 10.1128/mcb.7.2.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFranco D., Yamamoto K. R. Two different factors act separately or together to specify functionally distinct activities at a single transcriptional enhancer. Mol Cell Biol. 1986 Apr;6(4):993–1001. doi: 10.1128/mcb.6.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesGroseillers L., Jolicoeur P. Mapping the viral sequences conferring leukemogenicity and disease specificity in Moloney and amphotropic murine leukemia viruses. J Virol. 1984 Nov;52(2):448–456. doi: 10.1128/jvi.52.2.448-456.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesGroseillers L., Rassart E., Jolicoeur P. Thymotropism of murine leukemia virus is conferred by its long terminal repeat. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4203–4207. doi: 10.1073/pnas.80.14.4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans L. H., Morrey J. D. Tissue-specific replication of Friend and Moloney murine leukemia viruses in infected mice. J Virol. 1987 May;61(5):1350–1357. doi: 10.1128/jvi.61.5.1350-1357.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies S. D., Morrison S. L., Oi V. T., Tonegawa S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983 Jul;33(3):717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Hanecak R., Mittal S., Davis B. R., Fan H. Generation of infectious Moloney murine leukemia viruses with deletions in the U3 portion of the long terminal repeat. Mol Cell Biol. 1986 Dec;6(12):4634–4640. doi: 10.1128/mcb.6.12.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbomel P., Bourachot B., Yaniv M. Two distinct enhancers with different cell specificities coexist in the regulatory region of polyoma. Cell. 1984 Dec;39(3 Pt 2):653–662. doi: 10.1016/0092-8674(84)90472-0. [DOI] [PubMed] [Google Scholar]
- Herr W., Clarke J. The SV40 enhancer is composed of multiple functional elements that can compensate for one another. Cell. 1986 May 9;45(3):461–470. doi: 10.1016/0092-8674(86)90332-6. [DOI] [PubMed] [Google Scholar]
- Hilberg F., Stocking C., Ostertag W., Grez M. Functional analysis of a retroviral host-range mutant: altered long terminal repeat sequences allow expression in embryonal carcinoma cells. Proc Natl Acad Sci U S A. 1987 Aug;84(15):5232–5236. doi: 10.1073/pnas.84.15.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Holland C. A., Anklesaria P., Sakakeeny M. A., Greenberger J. S. Enhancer sequences of a retroviral vector determine expression of a gene in multipotent hematopoietic progenitors and committed erythroid cells. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8662–8666. doi: 10.1073/pnas.84.23.8662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland C. A., Wozney J., Chatis P. A., Hopkins N., Hartley J. W. Construction of recombinants between molecular clones of murine retrovirus MCF 247 and Akv: determinant of an in vitro host range property that maps in the long terminal repeat. J Virol. 1985 Jan;53(1):152–157. doi: 10.1128/jvi.53.1.152-157.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimoto A., Takimoto M., Adachi A., Kakuyama M., Kato S., Kakimi K., Fukuoka K., Ogiu T., Matsuyama M. Sequences responsible for erythroid and lymphoid leukemia in the long terminal repeats of Friend-mink cell focus-forming and Moloney murine leukemia viruses. J Virol. 1987 Jun;61(6):1861–1866. doi: 10.1128/jvi.61.6.1861-1866.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jainchill J. L., Aaronson S. A., Todaro G. J. Murine sarcoma and leukemia viruses: assay using clonal lines of contact-inhibited mouse cells. J Virol. 1969 Nov;4(5):549–553. doi: 10.1128/jvi.4.5.549-553.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch W., Zimmermann W., Oliff A., Friedrich R. Molecular analysis of the envelope gene and long terminal repeat of Friend mink cell focus-inducing virus: implications for the functions of these sequences. J Virol. 1984 Mar;49(3):828–840. doi: 10.1128/jvi.49.3.828-840.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laimins L. A., Gruss P., Pozzatti R., Khoury G. Characterization of enhancer elements in the long terminal repeat of Moloney murine sarcoma virus. J Virol. 1984 Jan;49(1):183–189. doi: 10.1128/jvi.49.1.183-189.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenardo M., Pierce J. W., Baltimore D. Protein-binding sites in Ig gene enhancers determine transcriptional activity and inducibility. Science. 1987 Jun 19;236(4808):1573–1577. doi: 10.1126/science.3109035. [DOI] [PubMed] [Google Scholar]
- Li Y., Golemis E., Hartley J. W., Hopkins N. Disease specificity of nondefective Friend and Moloney murine leukemia viruses is controlled by a small number of nucleotides. J Virol. 1987 Mar;61(3):693–700. doi: 10.1128/jvi.61.3.693-700.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miksicek R., Heber A., Schmid W., Danesch U., Posseckert G., Beato M., Schütz G. Glucocorticoid responsiveness of the transcriptional enhancer of Moloney murine sarcoma virus. Cell. 1986 Jul 18;46(2):283–290. doi: 10.1016/0092-8674(86)90745-2. [DOI] [PubMed] [Google Scholar]
- Mosthaf L., Pawlita M., Gruss P. A viral enhancer element specifically active in human haematopoietic cells. Nature. 1985 Jun 13;315(6020):597–600. doi: 10.1038/315597a0. [DOI] [PubMed] [Google Scholar]
- O'Donnell P. V., Woller R., Chu A. Stages in development of mink cell focus-inducing (MCF) virus-accelerated leukemia in AKR mice. J Exp Med. 1984 Sep 1;160(3):914–934. doi: 10.1084/jem.160.3.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliff A. I., Hager G. L., Chang E. H., Scolnick E. M., Chan H. W., Lowy D. R. Transfection of molecularly cloned Friend murine leukemia virus DNA yields a highly leukemogenic helper-independent type C virus. J Virol. 1980 Jan;33(1):475–486. doi: 10.1128/jvi.33.1.475-486.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliff A., Signorelli K., Collins L. The envelope gene and long terminal repeat sequences contribute to the pathogenic phenotype of helper-independent Friend viruses. J Virol. 1984 Sep;51(3):788–794. doi: 10.1128/jvi.51.3.788-794.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondek B., Shepard A., Herr W. Discrete elements within the SV40 enhancer region display different cell-specific enhancer activities. EMBO J. 1987 Apr;6(4):1017–1025. doi: 10.1002/j.1460-2075.1987.tb04854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz D. M., Palmiter R. D., Hammer R. E., Brinster R. L., Swift G. H., MacDonald R. J. Specific expression of an elastase-human growth hormone fusion gene in pancreatic acinar cells of transgenic mice. Nature. 1985 Feb 14;313(6003):600–602. doi: 10.1038/313600a0. [DOI] [PubMed] [Google Scholar]
- Rowe W. P., Pugh W. E., Hartley J. W. Plaque assay techniques for murine leukemia viruses. Virology. 1970 Dec;42(4):1136–1139. doi: 10.1016/0042-6822(70)90362-4. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirm S., Jiricny J., Schaffner W. The SV40 enhancer can be dissected into multiple segments, each with a different cell type specificity. Genes Dev. 1987 Mar;1(1):65–74. doi: 10.1101/gad.1.1.65. [DOI] [PubMed] [Google Scholar]
- Shinnick T. M., Lerner R. A., Sutcliffe J. G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981 Oct 15;293(5833):543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- Shoemaker C., Goff S., Gilboa E., Paskind M., Mitra S. W., Baltimore D. Structure of a cloned circular Moloney murine leukemia virus DNA molecule containing an inverted segment: implications for retrovirus integration. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3932–3936. doi: 10.1073/pnas.77.7.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short M. K., Okenquist S. A., Lenz J. Correlation of leukemogenic potential of murine retroviruses with transcriptional tissue preference of the viral long terminal repeats. J Virol. 1987 Apr;61(4):1067–1072. doi: 10.1128/jvi.61.4.1067-1072.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Speck N. A., Baltimore D. Six distinct nuclear factors interact with the 75-base-pair repeat of the Moloney murine leukemia virus enhancer. Mol Cell Biol. 1987 Mar;7(3):1101–1110. doi: 10.1128/mcb.7.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocking C., Kollek R., Bergholz U., Ostertag W. Point mutations in the U3 region of the long terminal repeat of Moloney murine leukemia virus determine disease specificity of the myeloproliferative sarcoma virus. Virology. 1986 Aug;153(1):145–149. doi: 10.1016/0042-6822(86)90015-2. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiesen H. J., Bösze Z., Henry L., Charnay P. A DNA element responsible for the different tissue specificities of Friend and Moloney retroviral enhancers. J Virol. 1988 Feb;62(2):614–618. doi: 10.1128/jvi.62.2.614-618.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troxler D. H., Scolnick E. M. Rapid leukemia induced by cloned friend strain of replicating murine type-C virus. Association with induction of xenotropic-related RNA sequences contained in spleen focus-forming virus. Virology. 1978 Mar;85(1):17–27. doi: 10.1016/0042-6822(78)90408-7. [DOI] [PubMed] [Google Scholar]
- Vogt M., Haggblom C., Swift S., Haas M. Envelope gene and long terminal repeat determine the different biological properties of Rauscher, Friend, and Moloney mink cell focus-inducing viruses. J Virol. 1985 Jul;55(1):184–192. doi: 10.1128/jvi.55.1.184-192.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker M. D., Edlund T., Boulet A. M., Rutter W. J. Cell-specific expression controlled by the 5'-flanking region of insulin and chymotrypsin genes. Nature. 1983 Dec 8;306(5943):557–561. doi: 10.1038/306557a0. [DOI] [PubMed] [Google Scholar]
- Yoshimura F. K., Davison B., Chaffin K. Murine leukemia virus long terminal repeat sequences can enhance gene activity in a cell-type-specific manner. Mol Cell Biol. 1985 Oct;5(10):2832–2835. doi: 10.1128/mcb.5.10.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenke M., Grundström T., Matthes H., Wintzerith M., Schatz C., Wildeman A., Chambon P. Multiple sequence motifs are involved in SV40 enhancer function. EMBO J. 1986 Feb;5(2):387–397. doi: 10.1002/j.1460-2075.1986.tb04224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wachter R., Fiers W. Preparative two-dimensional polyacrylamide gel electrophoresis of 32 P-labeled RNA. Anal Biochem. 1972 Sep;49(1):184–197. doi: 10.1016/0003-2697(72)90257-6. [DOI] [PubMed] [Google Scholar]


