Abstract
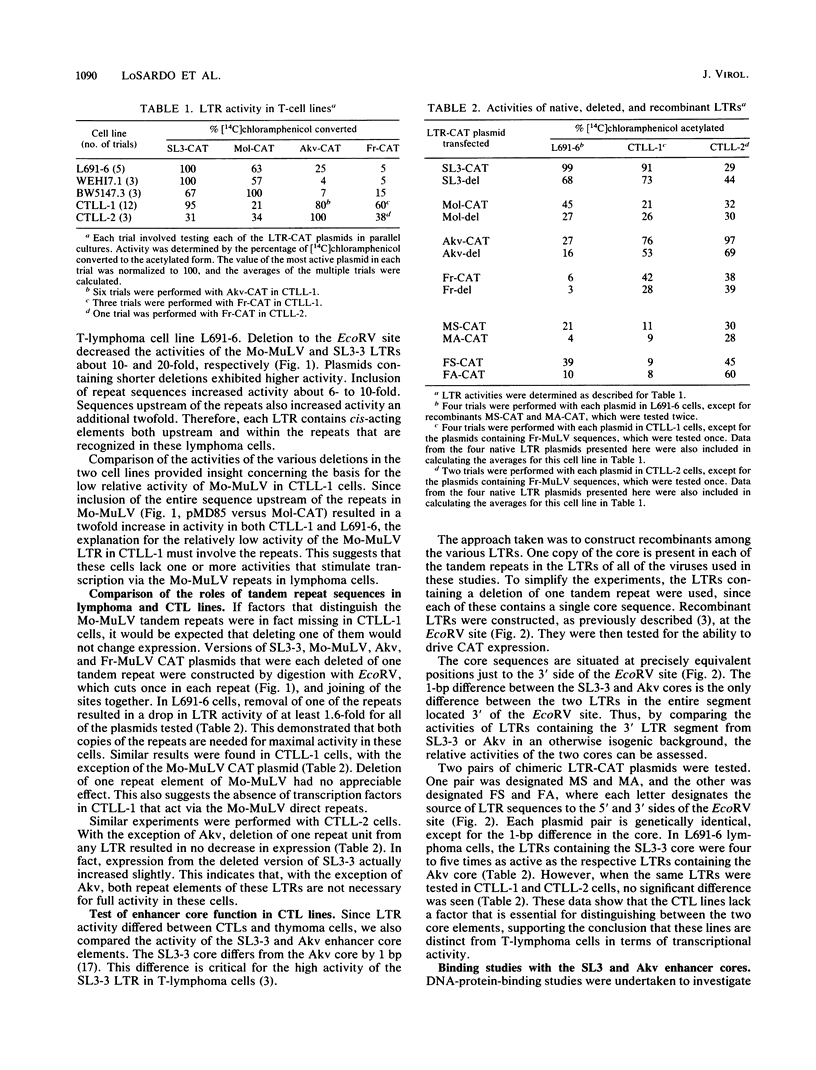
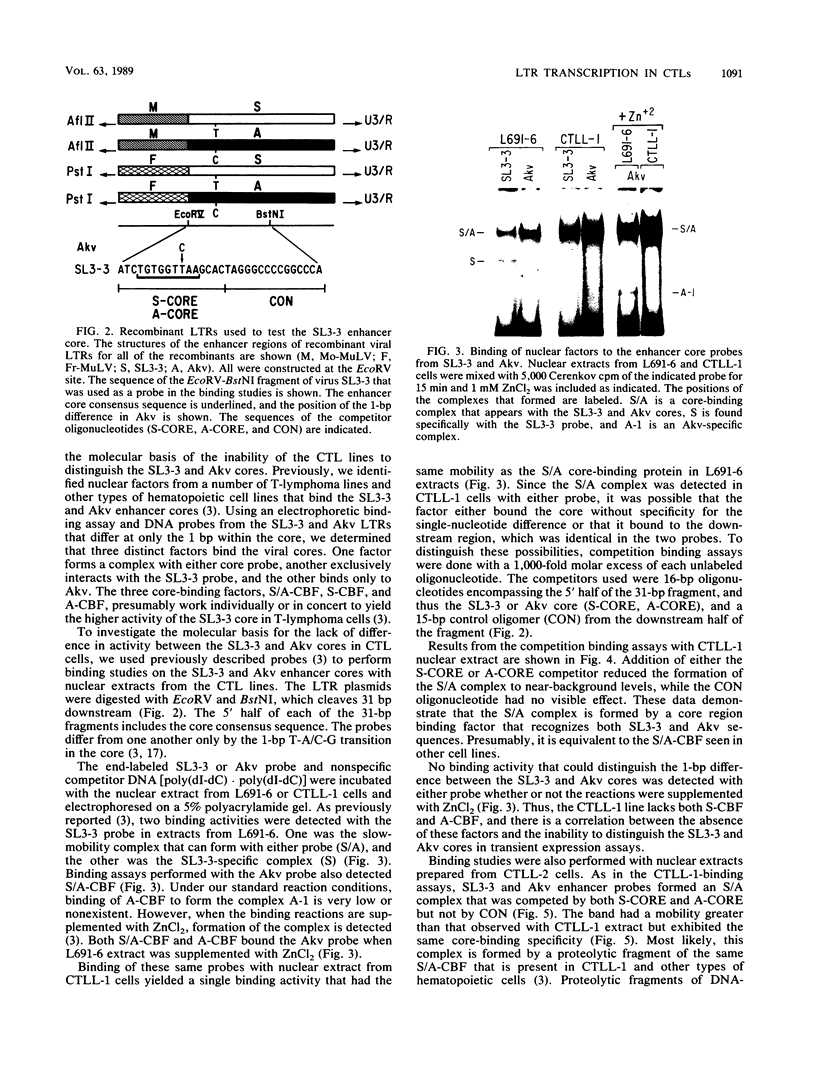
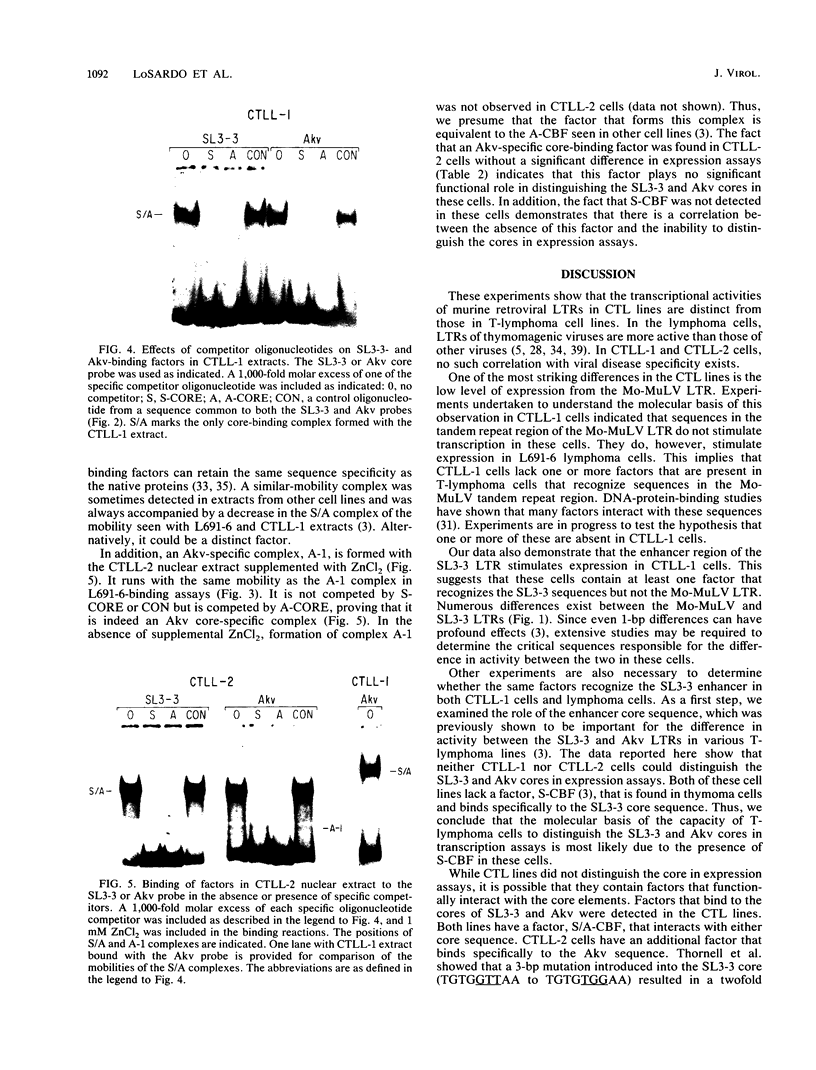
Transcriptional activities of the long terminal repeats (LTRs) of various murine leukemia viruses were tested in the cytotoxic T-cell lines CTLL-1 and CTLL-2. In contrast to T-lymphoma cells, in which the LTRs of T-lymphomagenic virus SL3-3 and Moloney murine leukemia virus are more active than those of other viruses, transcriptional activity in these mature, interleukin-2-dependent cells is not correlated with the specificity of viral leukemogenicity. Several approaches were used to investigate the molecular basis for LTR activity differences in lymphoma cells and mature cytotoxic T cells. Deletion analysis of the Moloney virus LTR showed that the direct repeats associated with enhancer activity have, at most, a slight effect on expression in CTLL-1 cells, whereas they stimulate expression six- to eightfold in T-lymphoma cells. This suggests that the mature T-cell line lacks one or more factors present in T-lymphoma cells that function to augment transcription from the Moloney murine leukemia virus LTR. We also used recombinant viral LTRs to investigate the role of the enhancer core element of SL3-3 in CTLL-1 and CTLL-2 cells. A one-base-pair difference between the core sequences of SL3-3 and nonleukemogenic Akv virus, which is important for SL3-3 activity in T-lymphoma cells, had no effect in these cells. The inability to distinguish the single-base-pair difference in expression assays was correlated with the absence of binding of a cellular factor, S-CBF, to the SL3-3 enhancer core in extracts of CTLL-1 and CTLL-2 nuclei. These studies may have implications for identification of the target cells for viral leukemogenesis, as well as for tracing of changes in the transcriptional machinery during T-lymphocyte differentiation.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Chandrasekaran R., Puigjaner L. C., Walker J. K., Hall I. H., Birdsall D. L., Ratliff R. L. Wrinkled DNA. Nucleic Acids Res. 1983 Mar 11;11(5):1457–1474. doi: 10.1093/nar/11.5.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker P. E., Gillis S., Smith K. A. Monoclonal cytolytic T-cell lines. J Exp Med. 1979 Jan 1;149(1):273–278. doi: 10.1084/jem.149.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boral A. L., Okenquist S. A., Lenz J. Identification of the SL3-3 virus enhancer core as a T-lymphoma cell-specific element. J Virol. 1989 Jan;63(1):76–84. doi: 10.1128/jvi.63.1.76-84.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhnlein E., Lowenthal J. W., Siekevitz M., Ballard D. W., Franza B. R., Greene W. C. The same inducible nuclear proteins regulates mitogen activation of both the interleukin-2 receptor-alpha gene and type 1 HIV. Cell. 1988 Jun 3;53(5):827–836. doi: 10.1016/0092-8674(88)90099-2. [DOI] [PubMed] [Google Scholar]
- Bösze Z., Thiesen H. J., Charnay P. A transcriptional enhancer with specificity for erythroid cells is located in the long terminal repeat of the Friend murine leukemia virus. EMBO J. 1986 Jul;5(7):1615–1623. doi: 10.1002/j.1460-2075.1986.tb04404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celander D., Haseltine W. A. Tissue-specific transcription preference as a determinant of cell tropism and leukaemogenic potential of murine retroviruses. Nature. 1984 Nov 8;312(5990):159–162. doi: 10.1038/312159a0. [DOI] [PubMed] [Google Scholar]
- Chatis P. A., Holland C. A., Hartley J. W., Rowe W. P., Hopkins N. Role for the 3' end of the genome in determining disease specificity of Friend and Moloney murine leukemia viruses. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4408–4411. doi: 10.1073/pnas.80.14.4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesGroseillers L., Jolicoeur P. The tandem direct repeats within the long terminal repeat of murine leukemia viruses are the primary determinant of their leukemogenic potential. J Virol. 1984 Dec;52(3):945–952. doi: 10.1128/jvi.52.3.945-952.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesGroseillers L., Rassart E., Jolicoeur P. Thymotropism of murine leukemia virus is conferred by its long terminal repeat. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4203–4207. doi: 10.1073/pnas.80.14.4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond L. E., Berman J. W., Pellicer A. Differential expression of surface markers on thymic lymphomas induced by two carcinogenic agents in different mouse strains. Cell Immunol. 1987 Jun;107(1):115–120. doi: 10.1016/0008-8749(87)90271-1. [DOI] [PubMed] [Google Scholar]
- Gillis S., Smith K. A. Long term culture of tumour-specific cytotoxic T cells. Nature. 1977 Jul 14;268(5616):154–156. doi: 10.1038/268154a0. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A. W., Bankhurst A. D., Mason S., Warner N. L. Differentiated functions expressed by cultured mouse lymphoma cells. II. Theta antigen, surface immunoglobulin and a receptor for antibody on cells of a thymoma cell line. J Immunol. 1973 Feb;110(2):431–438. [PubMed] [Google Scholar]
- Ishimoto A., Adachi A., Sakai K., Matsuyama M. Long terminal repeat of Friend-MCF virus contains the sequence responsible for erythroid leukemia. Virology. 1985 Feb;141(1):30–42. doi: 10.1016/0042-6822(85)90180-1. [DOI] [PubMed] [Google Scholar]
- Ishimoto A., Takimoto M., Adachi A., Kakuyama M., Kato S., Kakimi K., Fukuoka K., Ogiu T., Matsuyama M. Sequences responsible for erythroid and lymphoid leukemia in the long terminal repeats of Friend-mink cell focus-forming and Moloney murine leukemia viruses. J Virol. 1987 Jun;61(6):1861–1866. doi: 10.1128/jvi.61.6.1861-1866.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury G., Gruss P. Enhancer elements. Cell. 1983 Jun;33(2):313–314. doi: 10.1016/0092-8674(83)90410-5. [DOI] [PubMed] [Google Scholar]
- Lenz J., Celander D., Crowther R. L., Patarca R., Perkins D. W., Haseltine W. A. Determination of the leukaemogenicity of a murine retrovirus by sequences within the long terminal repeat. 1984 Mar 29-Apr 4Nature. 308(5958):467–470. doi: 10.1038/308467a0. [DOI] [PubMed] [Google Scholar]
- Li Y., Golemis E., Hartley J. W., Hopkins N. Disease specificity of nondefective Friend and Moloney murine leukemia viruses is controlled by a small number of nucleotides. J Virol. 1987 Mar;61(3):693–700. doi: 10.1128/jvi.61.3.693-700.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath M. S., Pillemer E., Kooistra D., Weissman I. L. The role of MuLV receptors on T-lymphoma cells in lymphoma cell proliferation. Contemp Top Immunobiol. 1980;11:157–184. doi: 10.1007/978-1-4684-3701-0_5. [DOI] [PubMed] [Google Scholar]
- Nabel G. J., Rice S. A., Knipe D. M., Baltimore D. Alternative mechanisms for activation of human immunodeficiency virus enhancer in T cells. Science. 1988 Mar 11;239(4845):1299–1302. doi: 10.1126/science.2830675. [DOI] [PubMed] [Google Scholar]
- O'Donnell P. V., Woller R., Chu A. Stages in development of mink cell focus-inducing (MCF) virus-accelerated leukemia in AKR mice. J Exp Med. 1984 Sep 1;160(3):914–934. doi: 10.1084/jem.160.3.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondek B., Gloss L., Herr W. The SV40 enhancer contains two distinct levels of organization. Nature. 1988 May 5;333(6168):40–45. doi: 10.1038/333040a0. [DOI] [PubMed] [Google Scholar]
- Ralph P. Retention of lymphocyte characteristics by myelomas and theta + -lymphomas: sensitivity to cortisol and phytohemagglutinin. J Immunol. 1973 Jun;110(6):1470–1475. [PubMed] [Google Scholar]
- Shaw J. P., Utz P. J., Durand D. B., Toole J. J., Emmel E. A., Crabtree G. R. Identification of a putative regulator of early T cell activation genes. Science. 1988 Jul 8;241(4862):202–205. doi: 10.1126/science.3260404. [DOI] [PubMed] [Google Scholar]
- Shinnick T. M., Lerner R. A., Sutcliffe J. G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981 Oct 15;293(5833):543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- Short M. K., Okenquist S. A., Lenz J. Correlation of leukemogenic potential of murine retroviruses with transcriptional tissue preference of the viral long terminal repeats. J Virol. 1987 Apr;61(4):1067–1072. doi: 10.1128/jvi.61.4.1067-1072.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siekevitz M., Josephs S. F., Dukovich M., Peffer N., Wong-Staal F., Greene W. C. Activation of the HIV-1 LTR by T cell mitogens and the trans-activator protein of HTLV-I. Science. 1987 Dec 11;238(4833):1575–1578. doi: 10.1126/science.2825351. [DOI] [PubMed] [Google Scholar]
- Speck N. A., Baltimore D. Six distinct nuclear factors interact with the 75-base-pair repeat of the Moloney murine leukemia virus enhancer. Mol Cell Biol. 1987 Mar;7(3):1101–1110. doi: 10.1128/mcb.7.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Vigneron M., Matthes H., Wildeman A., Zenke M., Chambon P. Requirement of stereospecific alignments for initiation from the simian virus 40 early promoter. Nature. 1986 Jan 9;319(6049):121–126. doi: 10.1038/319121a0. [DOI] [PubMed] [Google Scholar]
- Thiesen H. J., Bösze Z., Henry L., Charnay P. A DNA element responsible for the different tissue specificities of Friend and Moloney retroviral enhancers. J Virol. 1988 Feb;62(2):614–618. doi: 10.1128/jvi.62.2.614-618.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornell A., Hallberg B., Grundström T. Differential protein binding in lymphocytes to a sequence in the enhancer of the mouse retrovirus SL3-3. Mol Cell Biol. 1988 Apr;8(4):1625–1637. doi: 10.1128/mcb.8.4.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villemur R., Rassart E., DesGroseillers L., Jolicoeur P. Molecular cloning of viral DNA from leukemogenic Gross passage A murine leukemia virus and nucleotide sequence of its long terminal repeat. J Virol. 1983 Feb;45(2):539–546. doi: 10.1128/jvi.45.2.539-546.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt M., Haggblom C., Swift S., Haas M. Envelope gene and long terminal repeat determine the different biological properties of Rauscher, Friend, and Moloney mink cell focus-inducing viruses. J Virol. 1985 Jul;55(1):184–192. doi: 10.1128/jvi.55.1.184-192.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiher H., König M., Gruss P. Multiple point mutations affecting the simian virus 40 enhancer. Science. 1983 Feb 11;219(4585):626–631. doi: 10.1126/science.6297005. [DOI] [PubMed] [Google Scholar]
- Yoshimura F. K., Davison B., Chaffin K. Murine leukemia virus long terminal repeat sequences can enhance gene activity in a cell-type-specific manner. Mol Cell Biol. 1985 Oct;5(10):2832–2835. doi: 10.1128/mcb.5.10.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]