Abstract
SNARE proteins mediate fusion of intracellular eukaryotic membranes and their α-helical transmembrane domains are known to contribute to lipid bilayer mixing. Synthetic transmembrane domain peptides were previously shown to mimic the function of SNARE proteins in that they trigger liposome fusion in a sequence-specific fashion. Here, we performed a detailed investigation of the conformational dynamics of the transmembrane helices of the presynaptic SNAREs synaptobrevin II and syntaxin 1a. To this end, we recorded deuterium/hydrogen-exchange kinetics in isotropic solution as well as in the membrane-embedded state. In solution, the exchange kinetics of each peptide can be described by three different classes of amide deuteriums that exchange with different rate constants. These are likely to originate from exchange at different domains of the helices. Interestingly, the rate constants of each class vary with the TMD sequence. Thus, the exchange rate is position-specific and sequence-specific. Further, the rate constants correlate with the previously determined membrane fusogenicities. In membranes, exchange is retarded and a significant proportion of amide hydrogens are protected from exchange. We conclude that the conformational dynamics of SNARE TMD helices is mechanistically linked to their ability to drive lipid mixing.
INTRODUCTION
It is becoming increasingly apparent that integral membrane proteins exhibit considerable conformational dynamics. For example, α-helical transmembrane domains (TMDs) may change their relative orientation with respect to each other, depending on the functional state of the respective protein. Thus, helix-helix packing angles may change or helices may rotate relative to each other (1–4). In addition to these rigid-body motions, transmembrane helices are dynamic entities by themselves in that they exhibit functionally relevant bending motions at hinge regions (5). At an even more subtle level, the helix backbones themselves exhibit small-scale vibrational dynamics on a picosecond timescale (6). Conformational flexibility of transmembrane helices can even lead to irreversible refolding to insoluble amyloid that is associated with severe pathological phenotypes as exemplified by the Aβ42 peptide involved in Alzheimer's disease and other TMDs (7,8). Little is currently known about the potential biological significance of these vibrational TMD motions. It seems clear, however, that a functional role of helix dynamics is subject to evolutionary fine-tuning if it depends on primary structure.
The single C-terminal transmembrane domain (TMD) of soluble n-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins has previously been implied to play a role in membrane fusion. SNAREs drive the fusion of most intracellular eukaryotic membranes, exemplified by fusion of synaptic vesicles with the presynaptic plasma membrane (9). SNARE-mediated membrane fusion is preceded by formation of stable complexes by their soluble coiled-coil domains, which is thought to mediate docking of cognate membranes (10). Docking is, however, insufficient for fusion as complete bilayer mixing depends on the presence of TMDs (11,12). These TMDs form α-helices in the membrane (13–15) that are capable of self-interaction (15–20). Recent evidence suggests that SNARE TMDs support outer leaflet mixing, i.e., initiation of fusion, as well as progression from a hemifused to a fully fused state (9,14,16,21).
That SNARE TMDs contribute to lipid mixing is supported by the finding that synthetic peptides harboring their hydrophobic cores drive liposome-liposome fusion in vitro (21,22). Since these TMD-peptides are devoid of soluble domains that could mediate membrane apposition, it appears that isolated TMDs increase the likelihood by which randomly colliding liposomes enter fusion. The fusogenicity of these TMD mimics has been related to their ability to adopt different secondary structures. Circular dichroism (CD) spectroscopy indicated that they formed mixtures of α-helical and β-sheet structures in inverse micelles; further, mutations that decreased fusogenicity were found to increase α-helicity (22).
Here, we extend these previous studies by examining the sequence-specific conformational dynamics of SNARE TMD helices in terms of local and transient unfolding of their backbones. We determined the kinetics of hydrogen/deuterium exchange (H/D-exchange) reactions where amide hydrogens that are transiently not engaged in hydrogen bonding due to local unfolding successively exchange for solvent deuterium (23–25). Exchange was monitored in isotropic solution and in the membrane-embedded state and the obtained kinetics are related to the previously determined fusogenicities of these TMDs.
EXPERIMENTAL PROCEDURES
Peptide synthesis
Peptides were synthesized by Boc chemistry (PSL, Heidelberg, Germany) and were >90% pure as judged by mass spectrometry. Concentrations were determined via UV spectroscopy using extinction coefficients of 5600 M−1 cm−1 for syb-L8, L16, and syx; and 6800 M−1 cm−1 for syb-wt and syb-multA, respectively.
Preparation of liposomes
Liposomes (final lipid concentration 3 mM) were made by lyophilizing mixtures of dimyristoyl phosphatidylcholine (DMPC) and peptides in 2-methyl-2-propanol, followed by hydration by shaking at 37°C for 1 h in 10 mM NH4Ac, pH 7.4 for mass spectrometry or in 10 mM Tris/HCl, pH 7.4, 10 mM NaCl for CD spectroscopy. The hydrated mixtures were subjected to 10 freeze/thaw cycles in liquid nitrogen and extruded through a polycarbonate membrane (50 nm pore size, 20 passages at 50°C) with a Mini-Extruder (Avanti, Greensboro, NC). Nominal peptide/lipid (P/L)-ratios were 0.01 (mol/mol). For CD spectroscopy, peptide concentrations were determined experimentally from Trp-fluorescence of the peptides as described (26). The fraction of liposome-bound peptides was determined by floating liposomes on sucrose gradients as described (27). Briefly, 300 μl of the liposome preparations were mixed with 700 μl 60% (v/v) sucrose and overlaid with 2.5 ml of 30% (v/v) sucrose and 0.5 ml 20 mM Tris/HCl, pH 7.4, 150 mM NaCl, 0.1 mM Na-EDTA. Upon centrifugation (320,000 × g, 20 h, 20°C) in an Optima LE-80K ultracentrifuge (Beckman Coulter, Fullerton, CA), the liposomes migrate to the interface between 30% sucrose and the Tris buffer while free peptides remain in the bottom fraction (27). Here, all of our peptides quantitatively comigrated with the lipid fraction which ascertains complete association with the liposomes.
Circular dichroism (CD) spectroscopy
For CD spectroscopy in TFE/buffer mixtures, peptides originally dissolved in 100% 2,2,2-trifluoroethanol (TFE) were brought to the respective buffer/TFE ratios by adding 20 mM NH4Ac, pH 7.4 while maintaining a peptide concentration of 50 μM. For CD spectroscopy of liposomes, peptide concentrations were determined as described above. CD spectra were obtained using a model No. J-710 CD spectrometer (JASCO, Oklahoma City, OK) from 190 nm to 260 nm in a 1.0 (solution) or 0.5 mm (liposomes) cuvette at 20°C using a response of 2 s, a scan speed of 100 nm/min, and a sensitivity of 100 mdeg/cm. Spectra were the signal-averaged accumulation of 10–16 scans with the baselines (corresponding to buffer in case of peptides in isotropic solution or corresponding to liposomes without peptide, respectively) subtracted. Mean molar ellipticities were calculated and secondary structure contents estimated by deconvoluting the spectra using the program CDNN (28). The experiments were carried out in triplicate.
Recording of hydrogen/deuterium (H/D)- or deuterium/hydrogen (D/H)-exchange kinetics
Before D/H-exchange analysis in TFE/buffer solution, peptides were fully deuterated by incubation in 20% (v/v) dTFE, 10 mM ND4Ac, pD 7.4 at 95°C for 20 min before lyophilization and resolubilization in dTFE. To monitor D/H-exchange kinetics, the deuterated peptides (100 μM in 60% (v/v) dTFE, 10 mM ND4Ac, pD 7.4) were diluted 1:20 into 60% (v/v) TFE, 10 mM NH4Ac, pD 7.4 and incubated at the indicated temperature. After the indicated time periods, the exchange reactions were stopped by placing samples on ice and adding formic acid to a final concentration of 0.5% (v/v), which results in pH 2.5. In parallel, aliquots of each deuterated sample were diluted directly into 60% (v/v) TFE, 10 mM NH4Ac, pH 2.5 on ice. Under the latter condition, mainly deuteriums bound to N- and C-termini and to polar side-chain exchange. H/D-exchange reactions of peptides in liposomal membranes were started by diluting the liposome suspension 1:10 into 10 mM ND4Ac in D2O, pD 7.4 and stopped by addition of 50% (v/v) HCOOD to bring the pD to a value of 2.5 and placing the samples on ice.
Electrospray ionization mass spectrometry (ESI-MS)
ESI-MS was done by injecting 50 μl of the peptides in isotropic solution or embedded in liposomal membranes within ∼2 min of reaction stop into the micro electrospray ionization source of a model No. Q-Tof Ultima mass spectrometer (Waters, Milford, MA). Spectra were acquired in positive-ion mode (capillary voltage 2–3 kV; cone voltage 60 V) by accumulating 1-s scans over several minutes. Spectra were smoothed with the Savitzky-Golay algorithm over 80% of the peak areas (50 channels, five iterations) and centered (100 channels) and peak centers were taken for computation of mass differences. Data analysis was performed using MassLynx 4.0 Software (Waters). Only the spectra of the triply-charged peptide ions were evaluated. Peak-width analysis was done by determining peak width at 50% of maximal peak height (29).
Evaluation of exchange kinetics and curve fitting
For D/H-exchange done in TFE/buffer solution, the molecular masses of the triply charged ions were determined as described above and subjected to various corrections to account for ionization state, the mass of charge carriers, and dilution factors as follows. The number of deuteriums D on the exhaustively deuterated samples were determined according to D = 3(mD – mH) – 3, where mD and mH are the masses of the centered envelopes of the deuterated species and of nondeuterated reference samples, respectively. The number of deuteriums D as a function of time (D(t)) upon back-exchange in solution was determined by D(t) = 3(mD – mH) – 0.05 × Dmax, where Dmax is the calculated number of labile deuteriums on the respective peptide. The number of D upon forward exchange in liposomes was calculated by D(t) = 3(mD – mH) × 1.11 – 3. The forward (hydrogen/deuterium) exchange kinetics were converted to back (deuterium/hydrogen)-exchange kinetics by H(t) = Dmax − D(t).
To evaluate D/H-exchange kinetics of peptides in TFE/buffer solution, the data points corresponding to the corrected masses were approximated with the function D(t) = A × e−kA·t + B × e−kB·t + C × e−kC×t, where A, B, and C represent the numbers of amide deuteriums that exchange with the time constants kA, kB, and kC, respectively, and t is time (25,30). Since we are interested in the exchange kinetics of potentially hydrogen-bonded amide deuteriums, their theoretical number (19 D = D 22 N-D – 3 nonbonded N-D of an idealized α-helix of 23 residues) was taken as the initial data point (t = 0) for curve fitting. This appears to be the most accurate way to fit the kinetics since the numbers of deuteriums that exchanged under stop conditions (pH 2.5, on ice), corresponded to or even exceeded the calculated numbers of deuteriums bound to electronegative atoms. We thus deem it safe to assume that exclusively amide deuteriums remain at the first time point under exchange conditions (pH 7.4, 20°C or 37°C). For H/D-exchange of peptides in liposomal membranes, the data points were fit with D(t) = A·e−kA·t + B·e−kB·t + C. Fitting was done with Origin 7.5 software (OriginLab, Northampton, MA).
RESULTS
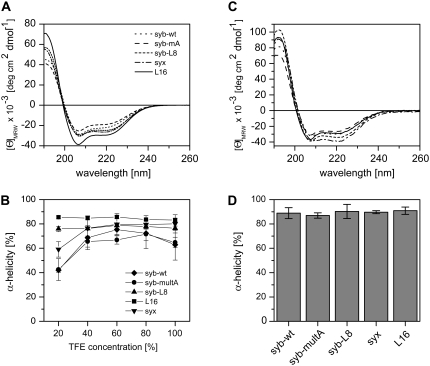
The objective of this study was to perform a detailed analysis of the conformational flexibility of the membrane-fusogenic TMDs from the synaptic SNARE proteins syntaxin 1A (syx) and synaptobrevin II (syb) by CD spectroscopy by recording H/D-exchange kinetics. In case of the syb TMD, we compared the wild-type TMD (syb-wt) to two mutant sequences. In syb-multA, three residues of the TMD-TMD interface are mutated for Ala (18) while syb-L8 has eight residues outside this interface exchanged for Leu. Both mutant TMDs have previously shown to be less fusogenic than syb-wt (22). For comparison, we analyzed an oligo-Leu (L16) sequence that functions as an artificial TMD (31) that is virtually nonfusogenic (22,26) (Fig. 1). In all peptides, the hydrophobic cores are flanked by Lys triplets and a Trp residue is incorporated for quantification.
FIGURE 1.
Peptide sequences. The mutated positions in syb-multA and syb-L8 are highlighted by bold-face type. L16 corresponds to a virtually nonfusogenic reference peptide. The hydrophobic residues are flanked by Lys triplets and a Trp residue was included for quantification.
Secondary structure of TMD-peptides in solution and in a lipid membrane
SNARE TMDs form membrane-spanning α-helices (13–15). Here, we first determined the dependence of TMD secondary structure on solvent composition by CD spectroscopy to assess global helix stability and to establish solution conditions for subsequent D/H-exchange experiments. CD spectra were recorded in various mixtures of aqueous buffer and 2,2,2-trifluoroethanol (TFE). TFE is thought to stabilize helical structures indirectly by partial desolvation, i.e., destabilization, of the unfolded state (32), which is similar to the situation in the apolar region of a membrane. Starting from peptides in pure TFE, we gradually added aqueous buffer to obtain TFE/buffer mixtures ranging from 20% to 80% (v/v) TFE that contained the peptides at identical concentrations (50 μM). At 40–100% TFE, the spectra exhibit the characteristic line shapes diagnostic of α-helices with minima at 208 nm and 222 nm (Fig. 2 A) and their quantitative evaluation indicates α-helix contents from ∼65% to ∼85% (Fig. 2 B). L16 generally tends to be somewhat more helical than the SNARE TMDs. Interestingly, α-helicities of syb-wt, syx, and syb-multA, but not of syb-L8 and L16, decrease markedly when TFE concentration is reduced to 20%. Thus, syb-wt, syb-multA, and syx helices appear to be less stable than syb-L8 and L16 helices. Second, CD spectra were recorded from peptides that were integrated into the membranes of small unilamellar liposomes that were prepared from peptides and dimyristoyl phosphatidylcholine (DMPC). Flotation of the liposomes in sucrose gradients confirmed quantitative association of peptides with lipid (results not shown). CD spectra recorded from these proteoliposomes again indicate α-helical structures (Fig. 2 C). Deconvolution of the spectra suggests ∼90% α-helicity in each case (Fig. 2 D).
FIGURE 2.
Secondary structure of TMD-peptides determined by CD spectroscopy. (A) Spectra recorded in 60% (v/v) TFE, 20 mM NH4Ac, pH 7.4 at 50 μM peptide. (B) Dependence of α-helicity on TFE concentration. Note the partial unfolding of syx, syb-wt, and syb-multA at 20% TFE. (C) Spectra of peptides reconstituted into liposomal membranes made of DMPC at a P/L-ratio of 0.01. (D) α-Helix contents of membrane-embedded peptides. All spectra were corrected for the background signals seen with pure TFE/buffer (A and B) or pure liposomes (C and D). All values represent means of three independent measurements ± SD. Secondary structure contents were stable for several days.
Conformational flexibility from deuterium/hydrogen exchange kinetics in isotropic solution
One can roughly distinguish between two types of labile, i.e., exchangeable, hydrogens bound to a peptide. Hydrogens bound to electronegative amino-acid side-chain atoms exchange very rapidly. Exchange of amide hydrogens is slower and blocked if they are hydrogen-bonded within secondary structures elements. Transient unfolding of secondary structure allows for a gradual exchange of amide hydrogens for deuterium or vice versa. Exchange rates thus provide a measure of conformational flexibility (24,33,34).
Here, exchange kinetics was recorded at 60% TFE where all TMD peptides are largely helical to mimic the situation in a membrane. The advantage of working in isotropic solution is that all amide hydrogens are expected to be equally accessible to solvent. Exchange rates are therefore not influenced by shielding in a membrane (see also below).
First, the peptides were exhaustively deuterated, which resulted in >95% exchange of the labile hydrogens of all peptides (Dobs in Table 1).
TABLE 1.
D/H-exchange kinetics in isotropic solution
| Peptide | Dmax* | Dobs† | D0‡ | D24§ | kA [min−1]¶ | kB [min−1]‖ | kC [min−1]** |
|---|---|---|---|---|---|---|---|
| syx | 41 | 38.8 ± 1.8 | 15.9 ± 0.3 | ND†† | —‡‡ | 0.688 ± 0.483 | 0.0306 ± 0.0028 |
| syb-wt | 40 | 39.0 ± 0.3 | 15.5 ± 1.1 | 0.2 ± 0.4 | 12.6 ± 6.9 | 0.365 ± 0.114 | 0.0109 ± 0.0008 |
| syb-multA | 39 | 38.4 ± 0.1 | 15.9 ± 0.7 | 0.2 ± 0.1 | 9.0 ± 4.2 | 0.556 ± 0.213 | 0.0104 ± 0.0032 |
| syb-L8 | 39 | 38.4 ± 0.2 | 17.0 ± 0.6 | 0.5 ± 0.5 | 1.8 ± 0.35 | 0.033 ± 0.004 | 0.0019 ± 0.0006 |
| L16 | 38 | 36.9 ± 0.5 | 16.5 ± 0.1 | 1.6 ± 1.6 | 1.6 ± 0.11 | 0.015 ± 0.001 | 0.0011 ± 0.0005 |
Dmax is the calculated number of labile hydrogens/deuteriums.
Dobs is the number of deuteriums on the peptides upon exhaustive deuteration as determined from the mass increases of the triply charged ions.
D0 is the number of deuteriums that do not exchange under stop conditions (pH 2.5, on ice).
D24 is the number of deuteriums that remain after 24 h incubation under exchange conditions (2 min at pH 7.4, 20°C).
kA is the exchange rate constant of deuterium population A (= 9) exchanging with fast kinetics.
kB is the exchange rate constant of deuterium population B (= 4) exchanging with intermediate kinetics.
kC is the exchange rate constant of deuterium population C (= 6) exchanging with slow kinetics.
ND is Not Determined.
The kA value of syx is at least as high as that of syb, but could not be precisely calculated due to improper curve fitting at the earliest time points.
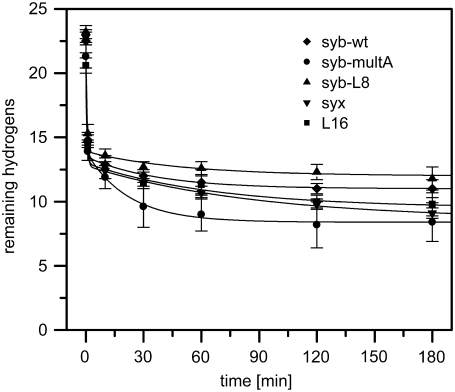
Second, aliquots of the fully deuterated peptides were incubated for 2 min under stop conditions (pH 2.5, samples on ice) at a final concentration of 5 μM. Under stop conditions, exchange rates are minimal and only deuteriums bound to N- and C-termini and to polar amino-acid side chains are expected to back-exchange for hydrogen (35,36). Exchange was monitored by determining the molecular masses of the triply charged peptide ions using ESI-MS. The triply-charged ions were consistently the most abundant species in our mass spectra and therefore evaluated for all subsequent analyses. An exemplary mass spectrum of the syb-wt TMD (Fig. 3 A) reveals that the isotopic envelope of the triply charged ion shifts from m/z 914.1 to m/z 905.6 under stop conditions to yield a peptide with 14.2 remaining deuteriums (mean = 15.5 D, see D0 in Table 1). Similar results were seen with all other peptides under stop conditions (∼16–17 D remaining, Table 1). The numbers of remaining deuteriums are invariably below the calculated number of potentially hydrogen-bonded amide deuteriums in an idealized 23-residue α-helix (= 19, see Experimental Procedures for explanation).
FIGURE 3.
D/H-exchange kinetics in isotropic solution. Exhaustively deuterated peptides were assayed for back-exchange. (A) Exemplary mass spectra of the triply-charged syb-wt ion at different time points. The spectrum at t = 0 min was recorded after exchange under stop conditions (3 min incubation on ice at pH 2.5) where only very labile deuteriums exchange (see text for details). Low intensity isotopic envelopes at calculated masses ∼22 Da above the dominant envelopes likely originate from Na+-adducts. (B) A comparison of the exchange kinetics exhibited by syx, syb-wt, syb-multA, syb-L8, and L16. The data points at t = 0 correspond to the numbers of amide deuteriums seen after exchange under stop conditions. The data were fit with a three-term exponential function assuming D = 19 at t = 0 min (continuous lines; see Experimental Procedures for details). (C) Comparison of exchange rate constants that were calculated for deuterium classes whose size was equivalent for each peptide. No kA value syx is given for syx, whose kA is at least as high as that of syb but could not be precisely calculated due to improper curve fitting at the earliest time points. All values represent means ± SD of at least three independent measurements.
Third, D/H-exchange kinetics of amide deuteriums were determined under exchange conditions (pH 7.4, 20°C) for up to 24 h. As exemplified by the syb-wt spectra, the envelopes gradually shift with incubation time toward lower m/z values (Fig. 3 A). After 24 h incubation, back-exchange was virtually complete with all peptides (<2 Da mass difference from undeuterated species; D24 in Table 1). Fig. 3 B compares the exchange kinetics of all peptides recorded for 180 min. At a qualitative level, the numbers of amide deuteriums decrease nonlinearly with time. Interestingly, the kinetics depend on peptide sequence and exhibit the rank-order syx > syb-wt ≈ syb-multA > syb-L8 ≈ L16. For quantitative evaluation, the kinetics were fit with a triple exponential function (see Experimental Procedures). This fitting procedure subdivides each peptide's amide deuteriums into three classes (A, B, and C) that exchange with markedly different rate constants (kA, kB, kC).
The results show that syx, syb-wt, and syb-multA tend to contain more rapidly exchanging class A deuteriums (means: ∼ 4–7) and less slowly exchanging class C deuteriums (means: ∼4–8) than syb-L8 or L16 while similar numbers of class B deuteriums (means: ∼3–6) were detected with all peptides (Supplementary Material, Table S1). Exchange rate constants of class A deuteriums (Table S1) are similar, with the possible exception of syx, whose initial time-course could not be fit satisfactorily. Class B and C deuteriums exchange at rates that tend to follow the general rank-order syx > syb-wt ≈ syb-multA > syb-L8 > L16. To probe the effect of elevated temperature, we also recorded kinetics at 37°C where exchange was accelerated as expected. On average, we noted an approximately twofold increase of the numbers of class A deuteriums at the expense of class C deuteriums, an approximately twofold increase of kA, and a ∼5–14-fold increases of kC. The rank-orders of the rate constants were similar at 37°C and 20°C.
For a better comparison of the different exchange kinetics recorded at 20°C, we averaged the numbers of amide deuteriums within each class over all peptides, which resulted in A = 9, B = 4, and C = 6 deuteriums. Using these fixed class sizes, we then recalculated the rate constants (Table 1) that now fully reflect the kinetics. Depending on the TMD, kA ranges from 1.6 min−1 to 12.6 min−1 (corresponding to half-times t1/2 = 0.054 min to 0.43 min), kB ranges from 0.015 min−1 to 0.688 min−1 (t1/2 =1.89 min to 46.0 min), and kC ranges from 0.0011 min−1 to 0.0306 min−1 (t1/2 = 63.3 min to 628 min). Importantly, all rate constants follow the rank-order syx > syb-wt > syb-multA > syb-L8 > L16.
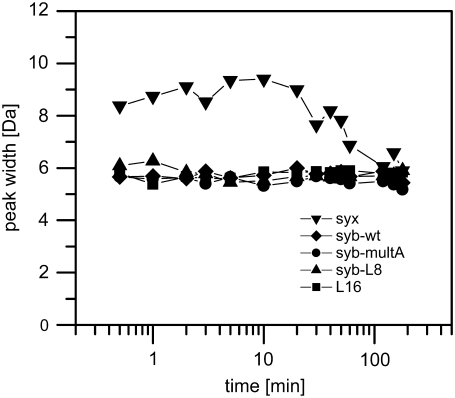
Next, we assessed whether transient TMD unfolding monitored by D/H-exchange is of a global or of a local nature by distinguishing between EX1 and EX2 kinetics. EX1 kinetics is seen if predominantly global unfolding results in simultaneous exposure of multiple amide groups and if the refolding rate is slower than the intrinsic rate of the exchange reaction. In this case, deuteriums exchange in a correlated fashion. If all amides exchange simultaneously, this results in a bimodal distribution of isotopic envelopes corresponding to completely exchanged and unexchanged species, respectively. The reverse holds true for ideal EX2 kinetics where uncorrelated exchange of individual deuteriums upon local unfolding yields gradually shifting envelopes. These may also broaden slightly due to the presence of intermittent mixtures of species containing slightly different numbers of hydrogen and deuterium atoms. If EX1 and EX2 kinetics are mixed, multiple isotopic envelopes that evolve in parallel may merge to significantly broadened peaks (34,37) at intermediate exchange periods and narrow down once exchange reaches completion. Slight or absent peak broadening is thus indicative of EX2 kinetics (29). Here, the gradual shifts of the envelopes observed for syb-wt (Fig. 3 A) and the other peptides (results not shown) suggest uncorrelated exchange (EX2 kinetics). To substantiate this point, we determined peak widths at different time points. The results show that the envelopes of syb-wt, syb-multA, syb-L8, and L16 remain at constant widths throughout the reaction supporting EX2 kinetics. The envelopes of syx are consistently somewhat broader and display slight intermediate broadening <2 Da (Fig. 4).
FIGURE 4.
Peak-width analysis. Peak widths at 50% of maximal peak height were determined from the deconvoluted mass spectra obtained in 60% (v/v) TFE, 10 mM NH4Ac, pH 7.4 at 20°C and plotted as a function of reaction time from t = 0.5 min to t = 3 h. Representative spectra were used in all cases except for syx, where lower signal/noise ratios resulted in a greater apparent variability of peak widths; syx peak widths were thus averaged from three independent spectra.
In sum, we draw the following conclusions:
The amide deuteriums of all TMD-peptides can be grouped into three major classes with markedly different exchange rate constants.
Comparing the different TMDs reveals that the deuteriums of all classes exchange with sequence-specific rates.
The kinetics suggest cumulative uncorrelated exchange reactions at locally unfolded helices.
Thus, our TMD-helices exhibit sequence-specific conformational flexibilities.
H/D-exchange kinetics in membranes
We also recorded H/D-exchange kinetics of our TMDs upon reconstitution in DMPC liposomes at a P/L-ratio of ∼0.01. Liposomes were directly introduced into the ion source of the mass spectrometer after reaction stop at the indicated incubation periods (38,39).
First, after recording H/D-exchange for 3 min under stop conditions ∼21–23 hydrogens remained unexchanged. In other words, ∼16–17 H exchanged under these conditions, which matches the numbers of hydrogens bound to electronegative atoms (calculated: 16–18 H, depending on the respective peptide). Apparently, therefore, very labile hydrogens exchange rapidly from membrane-embedded peptides, which is similar to the situation in solution. It is possible, however, that the hydrogens bound to the Trp, Cys, and/or Tyr side chains are protected from exchange by the membrane and that some exchanging hydrogens thus correspond to intrinsically non-hydrogen-bonded amides at the N-terminus.
Second, H/D-exchange kinetics were recorded under exchange conditions (37°C, pD 7.4) where amide hydrogens exchange. Similar to the situation in solution, we observed a gradual shift of the isotopic envelopes with incubation time. Further, peak width analysis does not indicate significant peak broadening and thus suggests uncorrelated exchange according to the EX2 mechanism (data not shown). Fig. 5 shows the kinetics in reverse to facilitate comparison to solution D/H-exchange kinetics (compare Fig. 3 B). It is evident that exchange is rapid within the first minutes.
FIGURE 5.
H/D-exchange kinetics in liposomal membranes. Peptides were reconstituted into liposomal membranes made of DMPC at P/L = 0.01 and assayed for their exchange kinetics. The data points at t = 0 correspond to the numbers of amide deuteriums seen after exchange for 3 min under stop conditions. The data were fit with a three-term exponential function (continuous lines; see Experimental Procedures for details). All values represent means ± SD of at least three independent measurements.
Thereafter, exchange slows down rapidly and levels off after 1–3 h. Curve fitting again subdivides each peptide's amide hydrogens into three classes. Class A hydrogens (means: ∼7–10) exchange with very similar rate constants (mean kA ∼2 min−1, t1/2 ∼0.35 min−1) while class B hydrogens (means: ∼3–6) display rate constants within a narrow range (mean kB ∼0.01 min−1 to ∼0.05 min−1, t1/2 ∼14 min−1 to ∼70 min−1). Class C (means: ∼8–11) reflects a virtually nonexchanging hydrogen population; even incubation for up to 24 h did not lead to further significant exchange (data not shown). It is of note that class B hydrogens of syb-multA appear to exchange significantly faster than those of the other TMDs (Fig. 5). When peptides were incorporated in liposomes made of palmitoyl-oleoyl-phosphatidylcholine, di-oleoyl-phosphatidylethanolamine; di-oleoyl-phosphatidylserine (3:1:1 molar ratio) similar results were obtained, except that, on average, approximately three fewer hydrogens exchanged upon 24 h. This is ascribed to the longer acyl chains of these lipids that are likely to shield larger parts of the TMDs (data not shown).
We conclude:
That the amide hydrogens of membrane-embedded TMDs can be grouped into two classes of exchangeable hydrogens with different exchange rate constants and one virtually nonexchangeable class that is apparently protected by the hydrophobic region of the membrane; and
That the exchangeable amide hydrogens of most TMDs exhibit similar exchange rate constants except syb-multA, for possible reasons discussed below.
DISCUSSION
Sequence- and position-specific conformational flexibility of SNARE TMD helices
We compared the D/H-exchange kinetics of different SNARE TMD helices in isotropic solution, where all amide deuteriums are equally exposed to solvent, and in membranes. The amide deuteriums of each TMD could be grouped into three classes (A, B, and C) that exchange with different kinetics. Which parts of the helices do the different classes correspond to? Since helices are thought to be more unstable at the termini (i.e., helix fraying) (40), it is likely that exchange of amide deuteriums is most rapid at the termini and slows down toward more central regions. Accordingly, we tentatively assign class A deuteriums to amide groups around the terminal Lys triplets and classes B and C to more central domains. Unfortunately, our attempts to directly map the different classes of deuterium by collision-induced dissociation of partially exchanged peptides using tandem mass spectrometry did not yield a consistent picture. This is likely due to strong deuterium scrambling, which was previously shown to blur the proton/deuterium distribution of Lys-tagged peptides (41).
Importantly, each class of amide deuteriums exhibits TMD-specific exchange kinetics. In particular, syx, syb-wt, and syb-multA TMDs exhibit significantly higher exchange rate constants than syb-L8 and L16. In other words, the kinetics of exchange does not only depend on the location of the amide within the helix but also on the respective primary structure. The differences are most pronounced between kB and kC values. It follows that conformational flexibility may show the largest TMD-specific variation for ∼10 central residues, which is equivalent to approximately three helical turns. Do the exchange kinetics originate from transient populations of fully unfolded helices or from local unfolding reactions?
Several observations argue in favor of local unfolding. First, exchange from a fully unfolded state is not expected to result in the observed classes of kinetically distinct amide deuteriums. Second, all TMDs investigated here exhibit similar helicities at 60% TFE where D/H-exchange was monitored. Third, ongoing D/H-exchange resulted in gradual shifts of the isotopic envelopes without significant peak broadening, which indicates uncorrelated exchange. We therefore deem it likely that exchange reflects vibrational motions of the helix backbone. These vibrations may proceed at picosecond timescales as suggested by molecular dynamics simulations of other TMD helices (6). When comparing D/H-exchange kinetics with the helix contents as measured by CD spectroscopy, only those TMDs with the faster exchange kinetics (syx, syb-wt, and syb-multA) exhibit significant global unfolding at 20% TFE. Thus, local helix unfolding at 60% TFE appears to be connected to global instability at 20% TFE.
In the membrane, all TMDs were helical as ascertained by CD-spectroscopy, thus corroborating an earlier secondary structure analysis of the synaptobrevin II TMD done by Fourier transform infrared spectroscopy (13) and extending this result to the syntaxin 1A TMD. Examining H/D-exchange in membranes at 37°C gave a picture that is somewhat different from the one seen with dissolved TMDs, yet largely consistent with it. In the membrane, we can distinguish two classes of hydrogens (A, B) that exchange with different rate constants and one class (C) that is virtually unexchangeable. Compared to the rate constants obtained in solution at 37°C, kA and kB values of membrane-embedded TMDs drop on average ∼3-fold and ∼10-fold, respectively. Class A contains ∼7–10 hydrogens that are likely to correspond to amides around the terminal Lys-triplets located within the membrane headgroup region. Class B contains ∼3–6 hydrogens that may be located within that part of the membrane's acyl-chain region that is adjacent to the headgroups. This part is hydrophobic, yet partially permeated by water (42,43). The slow-down of class B kinetics upon membrane incorporation may have different reasons. First, the catalytically active hydroxyl ions are probably scarce; i.e., amides are partially shielded by the membrane. This may level the significantly different exchange kinetics of class B hydrogens that were seen in solution. Second, exchange rate constants may be affected by TMD-TMD interactions in the membrane. Indeed, membrane-embedding favors these interactions by high local peptide concentration, preorientation, and crowding effects (44). Accordingly, amide hydrogens that are hidden within helix-helix interfaces are likely to be protected from exchange. It is interesting in this context that membrane-spanning syb-multA class B hydrogens exchange more rapidly than those of the other TMDs, which is in line with the previous observation that syb-mult A self-interacts with lower affinity than syb or syx (17,18). We cannot exclude, however, that faster exchange of syb-multA class B hydrogens is also related to enhanced accessibility of those amides where bulky Leu and Ile residues of syb-wt were mutated to Ala. Approximately 8–11 virtually nonexchangeable hydrogens constitute class C; i.e., approximate three central turns of the helices appear to be completely shielded by the central part of the acyl-chain region.
The inability to achieve complete exchange with membrane-spanning helices is reminiscent of previous observations made with other TMDs. For example, recording H/D-exchange kinetics of TMD-peptides whose hydrophobic cores consist of Leu/Ala repeats and that are anchored by terminal Trp residues in DMPC membranes uncovered a fast population (t1/2 ≤ 3 min) of approximately six hydrogens that was assigned to the termini as well as to Trp side-chains, an intermediate population of ∼5–6 hydrogens (t1/2 = ∼101 to ∼103 min) that was ascribed to amides close to the helix termini, and a virtually nonexchangeable population of ∼10 hydrogens (t1/2 > 104 min) within their central parts. Collision-induced dissociation studies supported these assignments in revealing decreasing exchange rates toward the helix centers (39). Various studies done with natural membrane proteins or isolated TMDs also report only limited exchange in lipid bilayers. For example, only ∼24% of the HIV-1 virus Vpu protein TMD hydrogens exchanged in lipid bilayers (45), the fd coat protein TMD was protected from exchange in detergent micelles (46), only ∼24% of a peptide corresponding to the phospholemman TMD exchanged in lipid bilayers (47), the EmrE multidrug transporter amides proved mostly resistant to exchange (48), and only 45% of the full-length SliK potassium channel amides were prone to exchange (49). Notably, however, ∼90% of the lactose permease amides exchanged within 3 h (49) which is in line with the pronounced conformational flexibility of this polytopic transporter that may affect membrane structure to increase solvent accessibility (50).
TMD helix flexibility and membrane fusion
Do flexible TMD helices contribute to SNARE function? Previously, it was found that the ability of our TMD peptides to drive liposome-liposome fusion increases in the rank-order L16 < syb-L8 < syb-multA < syb-wt ≈ syx (22). Indeed, the different fusogenicities of syx, syb, syb-L8, and L16 TMDs are correlated to their respective conformational flexibilities seen here in solution. It is presently not clear how this apparent connection between flexibility and fusogenicity can be explained at a mechanistic level. We speculate that vibrational motions of the helix backbone may affect the conformation of the surrounding lipids and thus facilitate outer leaflet mixing of randomly colliding liposomes.
Why does the conformational flexibility of a helix depend on TMD primary structure? The β-branched amino acids Val and Ile are overrepresented in SNAREs as they collectively account for ∼50% of SNARE TMD sequences but only for ∼25% of unrelated TMDs (22). Ile and Val are known to destabilize helices. This destabilizing effect is ascribed to an increased entropy loss upon helix formation, since β-branched side chains exhibit a smaller number of side-chain conformations in helices versus random coils (51), which is due to steric clashes between their side chains and neighboring atoms of the helix (52,53). When applied to the TMD helices investigated in this study, it is not surprising that syb-L8 with its increased content of Leu as well as L16 exhibit decreased conformational flexibilities relative to syb-wt and syx. Consequently, fusogenicity would depend on the content in β-branched amino acids. Recent electron-spin resonance spectroscopy data support the idea of flexible SNARE TMDs. These experiments demonstrated significant motional dynamics of the oligo-Val stretch that constitutes the C-terminal half of the yeast SNARE Sso1p TMD that was embedded in a bilayer (15).
With syb-multA, only one Ile residue is mutated. Strong effects on flexibility are thus not to be expected in this case. Rather, the previously-noted (22) lower fusogenicity of syb-multA relative to syb-wt (22) may be due to a partial arrest at the hemifusion intermediate. In hemifusion, outer leaflets have mixed while inner leaflets stay separate (54,55). An impaired hemifusion-to-fusion transition has previously been related to reduced self-interaction of the vacuolar yeast SNARE Vam3p TMD (20,21). Indeed, our preliminary results suggest that liposome fusion by syb-multA results in partial arrest at hemifusion (W. Stelzer and D. Langosch, unpublished results).
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Acknowledgments
We thank Dr. Thomas Letzel for critical reading of the manuscript and his valuable comments.
This work was supported by the State of Bavaria and the Center of Integrative Protein Science Munich (CIPSM).
Holger Stalz's present address is Waters S.A.S., BP 608, 78056 Saint-Quentin, En Yvelines Cedex, France.
Editor: Peter Tieleman.
References
- 1.Farrens, D. L., C. Altenbach, K. Yang, W. L. Hubbell, and H. G. Khorana. 1996. Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science. 274:768–770. [DOI] [PubMed] [Google Scholar]
- 2.Klingenberg, M. 2005. Ligand-protein interaction in biomembrane carriers. The induced transition fit of transport catalysis. Biochemistry. 44:8563–8570. [DOI] [PubMed] [Google Scholar]
- 3.Matthews, E. E., M. Zoonens, and D. M. Engelman. 2006. Dynamic helix interactions in transmembrane signaling. Cell. 127:447–450. [DOI] [PubMed] [Google Scholar]
- 4.Perozo, E., D. M. Cortes, P. Sompornpisut, A. Kloda, and B. Martinac. 2002. Open channel structure of MscL and the gating mechanism of mechanosensitive channels. Nature. 418:942–948. [DOI] [PubMed] [Google Scholar]
- 5.Cordes, F. S., J. N. Bright, and M. S. Sansom. 2002. Proline-induced distortions of transmembrane helices. J. Mol. Biol. 323:951–960. [DOI] [PubMed] [Google Scholar]
- 6.Mukherjee, P., I. Kass, I. T. Arkin, and M. T. Zanni. 2006. Structural disorder of the CD3ζ transmembrane domain studied with 2D IR spectroscopy and molecular dynamics simulations. J. Phys. Chem. B. 110:24740–24749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johansson, J. 2003. Molecular determinants for amyloid fibril formation: lessons from lung surfactant protein C. Swiss Med. Wkly. 133:275–282. [DOI] [PubMed] [Google Scholar]
- 8.Munter, L. M., P. Voigt, A. Harmeier, D. Kaden, K. E. Gottschalk, C. Weise, R. Pipkorn, M. Schaefer, D. Langosch, and G. Multhaup. 2007. GxxxG motifs within the amyloid precursor protein transmembrane sequence are critical for the etiology of Aβ42. EMBO J. 26:1702–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langosch, D., M. Hofmann, and C. Ungermann. 2007. The role of transmembrane domains in membrane fusion. Cell. Mol. Life Sci. 64:850–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weber, T., B. V. Zemelman, J. A. McNew, B. Westermann, M. Gmachl, F. Parlati, T. H. Sollner, and J. E. Rothman. 1998. SNAREpins: minimal machinery for membrane fusion. Cell. 92:759–772. [DOI] [PubMed] [Google Scholar]
- 11.Grote, E., M. Baba, Y. Ohsumi, and P. J. Novick. 2000. Geranylgeranylated SNAREs are dominant inhibitors of membrane fusion. J. Cell Biol. 151:453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rohde, J., L. Dietrich, D. Langosch, and C. Ungermann. 2003. The transmembrane domain of Vam3 affects the composition of cis- and trans-SNARE complexes to promote homotypic vacuole fusion. J. Biol. Chem. 278:1656–1662. [DOI] [PubMed] [Google Scholar]
- 13.Bowen, M., and A. T. Brunger. 2006. Conformation of the synaptobrevin transmembrane domain. Proc. Natl. Acad. Sci. USA. 103:8378–8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu, Y., F. Zhang, Z. Su, J. A. McNew, and Y. K. Shin. 2005. Hemifusion in SNARE-mediated membrane fusion. Nat. Struct. Mol. Biol. 12:417–422. [DOI] [PubMed] [Google Scholar]
- 15.Zhang, Y., and Y. K. Shin. 2006. Transmembrane organization of yeast syntaxin-analogue Sso1p. Biochemistry. 45:4173–4181. [DOI] [PubMed] [Google Scholar]
- 16.Giraudo, C. G., C. Hu, D. You, A. M. Slovic, E. V. Mosharov, D. Sulzer, T. J. Melia, and J. E. Rothman. 2005. SNAREs can promote complete fusion and hemifusion as alternative outcomes. J. Cell Biol. 170:249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laage, R., and D. Langosch. 1997. Dimerization of the synaptic vesicle protein synaptobrevin (vesicle-associated membrane protein) II depends on specific residues within the transmembrane segment. Eur. J. Biochem. 249:540–546. [DOI] [PubMed] [Google Scholar]
- 18.Laage, R., J. Rohde, B. Brosig, and D. Langosch. 2000. A conserved membrane-spanning amino acid motif drives homomeric and supports heteromeric assembly of presynaptic SNARE proteins. J. Biol. Chem. 275:17481–17487. [DOI] [PubMed] [Google Scholar]
- 19.Margittai, M., H. Otto, and R. Jahn. 1999. A stable interaction between syntaxin 1a and synaptobrevin 2 mediated by their transmembrane domains. FEBS Lett. 446:40–44. [DOI] [PubMed] [Google Scholar]
- 20.Roy, R., K. Peplowska, J. Rohde, C. Ungermann, and D. Langosch. 2006. Role of the Vam3p transmembrane segment in homodimerization and SNARE complex formation. Biochemistry. 45:7654–7660. [DOI] [PubMed] [Google Scholar]
- 21.Hofmann, M. W., K. Peplowska, J. Rohde, B. C. Poschner, C. Ungermann, and D. Langosch. 2006. Self-interaction of a SNARE transmembrane domain promotes the hemifusion-to-fusion transition. J. Mol. Biol. 364:1048–1060. [DOI] [PubMed] [Google Scholar]
- 22.Langosch, D., J. M. Crane, B. Brosig, A. Hellwig, L. K. Tamm, and J. Reed. 2001. Peptide mimics of SNARE transmembrane segments drive membrane fusion depending on their conformational plasticity. J. Mol. Biol. 311:709–721. [DOI] [PubMed] [Google Scholar]
- 23.Englander, J. J., C. Del Mar, W. Li, S. W. Englander, J. S. Kim, D. D. Stranz, Y. Hamuro, and V. L. Woods, Jr. 2003. Protein structure change studied by hydrogen-deuterium exchange, functional labeling, and mass spectrometry. Proc. Natl. Acad. Sci. USA. 100:7057–7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaltashov, I. A., and S. J. Eyles. 2002. Studies of biomolecular conformations and conformational dynamics by mass spectrometry. Mass Spectrom. Rev. 21:37–71. [DOI] [PubMed] [Google Scholar]
- 25.Yan, X., J. Watson, P. S. Ho, and M. L. Deinzer. 2004. Mass spectrometric approaches using electrospray ionization charge states and hydrogen-deuterium exchange for determining protein structures and their conformational changes. Mol. Cell. Proteomics. 3:10–23. [DOI] [PubMed] [Google Scholar]
- 26.Hofmann, M. W., K. Weise, J. Ollesch, P. Agrawal, H. Stalz, W. Stelzer, F. Hulsbergen, H. de Groot, K. Gerwert, J. Reed, and D. Langosch. 2004. De novo design of conformationally flexible transmembrane peptides driving membrane fusion. Proc. Natl. Acad. Sci. USA. 101:14776–14781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langosch, D., B. Brosig, and R. Pipkorn. 2001. Peptide mimics of the vesicular stomatitis virus G-protein transmembrane segment drive membrane fusion in vitro. J. Biol. Chem. 276:32016–32021. [DOI] [PubMed] [Google Scholar]
- 28.Bohm, G., R. Muhr, and R. Jaenicke. 1992. Quantitative analysis of protein far UV circular dichroism spectra by neural networks. Protein Eng. 5:191–195. [DOI] [PubMed] [Google Scholar]
- 29.Weis, D. D., J. R. Engen, and I. J. Kass. 2006. Semi-automated data processing of hydrogen exchange mass spectra using HX-Express. J. Am. Soc. Mass Spectrom. 17:1700–1703. [DOI] [PubMed] [Google Scholar]
- 30.Englander, S. W., and N. R. Kallenbach. 1983. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q. Rev. Biophys. 16:521–655. [DOI] [PubMed] [Google Scholar]
- 31.Gurezka, R., R. Laage, B. Brosig, and D. Langosch. 1999. A heptad motif of leucine residues found in membrane proteins can drive self-assembly of artificial transmembrane segments. J. Biol. Chem. 274:9265–9270. [DOI] [PubMed] [Google Scholar]
- 32.Kentsis, A., and T. R. Sosnick. 1998. Trifluoroethanol promotes helix formation by destabilizing backbone exposure: desolvation rather than native hydrogen bonding defines the kinetic pathway of dimeric coiled coil folding. Biochemistry. 37:14613–14622. [DOI] [PubMed] [Google Scholar]
- 33.Englander, S. W. 2000. Protein folding intermediates and pathways studied by hydrogen exchange. Annu. Rev. Biophys. Biomol. Struct. 29:213–238. [DOI] [PubMed] [Google Scholar]
- 34.Ferraro, D. M., N. D. Lazo, and A. D. Robertson. 2004. EX1 hydrogen exchange and protein folding. Biochemistry. 43:587–594. [DOI] [PubMed] [Google Scholar]
- 35.Bai, Y., J. S. Milne, L. Mayne, and S. W. Englander. 1993. Primary structure effects on peptide group hydrogen exchange. Proteins. 17:75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dempsey, C. E. 2001. Hydrogen exchange in peptides and proteins using NMR spectroscopy. Prog. Nucl. Magn. Reson. Spectrosc. 39:135–170. [Google Scholar]
- 37.Xiao, H., J. K. Hoerner, S. J. Eyles, A. Dobo, E. Voigtman, A. I. Mel'cuk, and I. A. Kaltashov. 2005. Mapping protein energy landscapes with amide hydrogen exchange and mass spectrometry: I. A generalized model for a two-state protein and comparison with experiment. Protein Sci. 14:543–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Demmers, J. A., J. Haverkamp, A. J. Heck, R. E. Koeppe 2nd, and J. A. Killian. 2000. Electrospray ionization mass spectrometry as a tool to analyze hydrogen/deuterium exchange kinetics of transmembrane peptides in lipid bilayers. Proc. Natl. Acad. Sci. USA. 97:3189–3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Demmers, J. A., E. van Duijn, J. Haverkamp, D. V. Greathouse, R. E. Koeppe 2nd, A. J. Heck, and J. A. Killian. 2001. Interfacial positioning and stability of transmembrane peptides in lipid bilayers studied by combining hydrogen/deuterium exchange and mass spectrometry. J. Biol. Chem. 276:34501–34508. [DOI] [PubMed] [Google Scholar]
- 40.Wand, A. J., H. Roder, and S. W. Englander. 1986. Two-dimensional 1H NMR studies of cytochrome c: hydrogen exchange in the N-terminal helix. Biochemistry. 25:1107–1114. [DOI] [PubMed] [Google Scholar]
- 41.Demmers, J. A., D. T. Rijkers, J. Haverkamp, J. A. Killian, and A. J. Heck. 2002. Factors affecting gas-phase deuterium scrambling in peptide ions and their implications for protein structure determination. J. Am. Chem. Soc. 124:11191–11198. [DOI] [PubMed] [Google Scholar]
- 42.Marsh, D. 2001. Polarity and permeation profiles in lipid membranes. Proc. Natl. Acad. Sci. USA. 98:7777–7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Subczynski, W. K., A. Wisniewska, J. J. Yin, J. S. Hyde, and A. Kusumi. 1994. Hydrophobic barriers of lipid bilayer membranes formed by reduction of water penetration by alkyl chain unsaturation and cholesterol. Biochemistry. 33:7670–7681. [DOI] [PubMed] [Google Scholar]
- 44.Grasberger, B., A. P. Minton, C. DeLisi, and H. Metzger. 1986. Interaction between proteins localized in membranes. Proc. Natl. Acad. Sci. USA. 83:6258–6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kukol, A., and I. T. Arkin. 1999. Vpu transmembrane peptide structure obtained by site-specific Fourier transform infrared dichroism and global molecular dynamics searching. Biophys. J. 77:1594–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Veglia, G., A. C. Zeri, C. Ma, and S. J. Opella. 2002. Deuterium/hydrogen exchange factors measured by solution nuclear magnetic resonance spectroscopy as indicators of the structure and topology of membrane proteins. Biophys. J. 82:2176–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beevers, A. J., and A. Kukol. 2006. Secondary structure, orientation, and oligomerization of phospholemman, a cardiac transmembrane protein. Protein Sci. 15:1127–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arkin, I. T., W. P. Russ, M. Lebendiker, and S. Schuldiner. 1996. Determining the secondary structure and orientation of EmrE, a multi-drug transporter, indicates a transmembrane four-helix bundle. Biochemistry. 35:7233–7238. [DOI] [PubMed] [Google Scholar]
- 49.le Coutre, J., H. R. Kaback, C. K. Patel, L. Heginbotham, and C. Miller. 1998. Fourier transform infrared spectroscopy reveals a rigid α-helical assembly for the tetrameric Streptomyces lividans K+ channel. Proc. Natl. Acad. Sci. USA. 95:6114–6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abramson, J., H. R. Kaback, and S. Iwata. 2004. Structural comparison of lactose permease and the glycerol-3-phosphate antiporter: members of the major facilitator superfamily. Curr. Opin. Struct. Biol. 14:413–419. [DOI] [PubMed] [Google Scholar]
- 51.Chellgren, B. W., and T. P. Creamer. 2006. Side-chain entropy effects on protein secondary structure formation. Proteins. 62:411–420. [DOI] [PubMed] [Google Scholar]
- 52.Griffiths-Jones, S. R., G. J. Sharman, A. J. Maynard, and M. S. Searle. 1998. Modulation of intrinsic φ,ψ propensities of amino acids by neighboring residues in the coil regions of protein structures: NMR analysis and dissection of a β-hairpin peptide. J. Mol. Biol. 284:1597–1609. [DOI] [PubMed] [Google Scholar]
- 53.Street, A. G., and S. L. Mayo. 1999. Intrinsic β-sheet propensities result from van der Waals interactions between side chains and the local backbone. Proc. Natl. Acad. Sci. USA. 96:9074–9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chernomordik, L. V., and M. M. Kozlov. 2003. Protein-lipid interplay in fusion and fission of biological membranes. Annu. Rev. Biochem. 72:175–207. [DOI] [PubMed] [Google Scholar]
- 55.Tamm, L. K., J. Crane, and V. Kiessling. 2003. Membrane fusion: a structural perspective on the interplay of lipids and proteins. Curr. Opin. Struct. Biol. 13:453–466. [DOI] [PubMed] [Google Scholar]