Abstract
Among the diverse nucleic acid probes, molecular beacons (MBs) have shown their excellent potential in a variety of basic researches and practical applications. Their excellent selectivity, sensitivity, and detection without separation have led them to be particularly useful in real-time intracellular monitoring of gene expression, development of biosensors, and clinical diagnostics. This paper will focus on the properties of various MBs and discuss their potential applications.
Keywords: Molecular beacon, DNA, RNA, probe, biosensor, molecular diagnostics
Introduction
Molecular beacons (MBs) are single-stranded nucleic acid probes composed of three different functional domains: a stem, a loop, and a fluorophore/quencher pair (Figure 1) [1]. Fluorophore/quencher pairs, the signaling elements, produce the on/off signals depending on the conformational state of MBs. When a donor fluorophore is in its excited state, the energy is transferred via a non-radiative long-range dipole-dipole coupling mechanism to an acceptor quencher in close proximity (typically <10nm), called fluorescence resonance energy transfer (FRET), named after the German scientist Theodor Förster. On the contrary, the fluorescence signal is freely produced when the quencher is in far distance. Stems made of four to seven base pairs function as lockers to maintain the closed hairpin structures and bring a quencher nearby a fluorophore in a few nanometers’ distance without hybridization to the complementary targets. Thus, the quencher turns off fluorescence with high quenching efficiency via FRET. Loops are the recognizing elements that, upon hybridization to their complementary targets, induce a conformational change of MBs. This change increases the physical distance between the fluorophore and the quencher, resulting in fluorescence. The unique on/off signal mechanism has been very useful in the field of detecting DNA and RNA targets.
Figure 1.
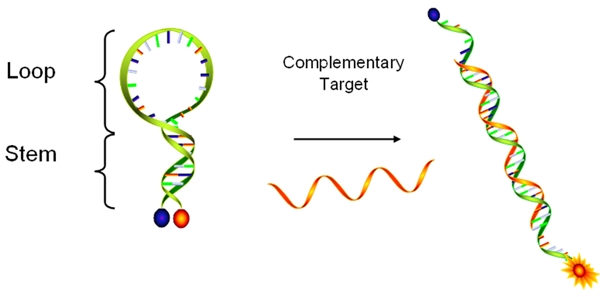
Diagram of a MB in a hybridized and unhybridized state. While the MB remains in free form without its target, it forms hairpin structure with quenched fluorescence. Once it binds to its target, the fluorophore restore the fluorescence signal due to the far distance from the quencher.
The unique hairpin structure and signaling mechanism endow the MBs with several advantages. First of all, the light-up signaling mechanism allows MBs to perform highly sensitive detection and monitoring of nucleic acids in real time. The unbound MBs stay with quenched fluorescence. Only hybridized MBs, whose amount is proportional to that of the target can produce high fluorescence signal. Such a detection-without-separation property is particularly useful in situations where it is either impossible or undesirable to extract the probe–target hybrids from an excess of the unbound probes. Another advantage of MBs is their relatively high signal-to-background ratio, providing higher sensitivity of detection. Upon hybridization to its target, a well designed MB can generate as high as 200-fold fluorescence enhancement under optimal conditions [2]. This provides the MBs with a significant advantage over other fluorescent probes in ultra-sensitive analysis. In addition to its sensitivity, MBs offer excellent selectivity. They are extraordinarily target-specific, and are able to differentiate as low as single-mismatched base pair sequences. The selectivity of MBs is a direct result of their hairpin conformation, as the stem hybrid acts as an activation energy barrier to the loop-target hybrid. The remarkable selectivity provided by the hairpin conformations has been demonstrated in a variety of biological environments where a number of different nucleic acid sequences and enormous amount of non-targets are present.
Due to these unique features in sensitivity and selectivity, the applications of MBs have been explored in many research fields since it was first created in 1996. Among all nucleic acid probes, MBs have demonstrated superior performances in real-time intracellular monitoring of gene expression, biosensor development and clinical diagnosis. We will focus on these areas in this paper.
MBs for Real-time RNA Monitoring in Living Cells
One of the primary advantages of MBs is their inherent capability of detection without separation. This advantage is critical for intracellular applications where any types of separation methods are unlikely to be applicable without damaging the living samples. For this reason, not only can MBs detect RNAs in the native environment, they can also visualize and tract the sub-cellular localization of RNAs in real time. To accomplish such a goal, designing MBs with excellent performance is critical.
The major concern in designing MBs for intracellular applications is the selection of an appropriate target region. This is especially critical since most regions of RNA targets remain in double strands. The selection of the target sites start with the prediction of possible RNA secondary structures. The target sites chosen are ideally single-stranded to assure that native mRNA structure would minimally compete with the proposed MB. In addition, it should be unique to represent the specific target. For the chosen regions, high affinity oligonucleotides of different lengths that complementary to the regions will then be used as the loop sequences of the MB. Each loop sequence is then flanked with two complementary arm sequences to generate a potential MB. Usually the stems are four to seven base pairs long and have a very high G and C content (75 to 100 percent) to ensure the hairpin conformation would not fluctuate in the living environment. However, it should not be too strong since it can prevent the binding of the MB to its target sequence, often resulting in low signal enhancement.
Currently the application of MBs for intracellular analysis is a field in its nascence (see Figure 2 for an example). Initial studies of MBs concentrated on the detection of the MB hybridization to RNA [3–5]. In 2003, Tyagi et al demonstrated that MBs could be used for the visualization of the distribution and transport of mRNA [6]. In this study, a MB for oskar mRNA was investigated in Drosophila melanogastar oocytes. To eliminate the background fluorescence from the MBs, a binary MB approach was developed, which utilized two MBs that targeted adjacent positions on the mRNA. Only when both MBs were hybridized to the mRNA sequence, a donor and acceptor fluorophore were brought within close proximity, allowing FRET to take place and generating a new signal that indicates hybridization of the MBs to the mRNA target. In addition to visualizing the mRNA distribution, they were able to track the migration of mRNA inside the cell and even into adjacent cells in the oocyte. Other studies have imaged MBs on viral mRNA inside the host cells to study the behavior of the viral mRNA [7]. This study not only investigated the localization of the mRNA inside cell but also utilized photobleaching of the fluorophore on the MB to study the diffusion of the MB-mRNA hybrids.
Figure 2.
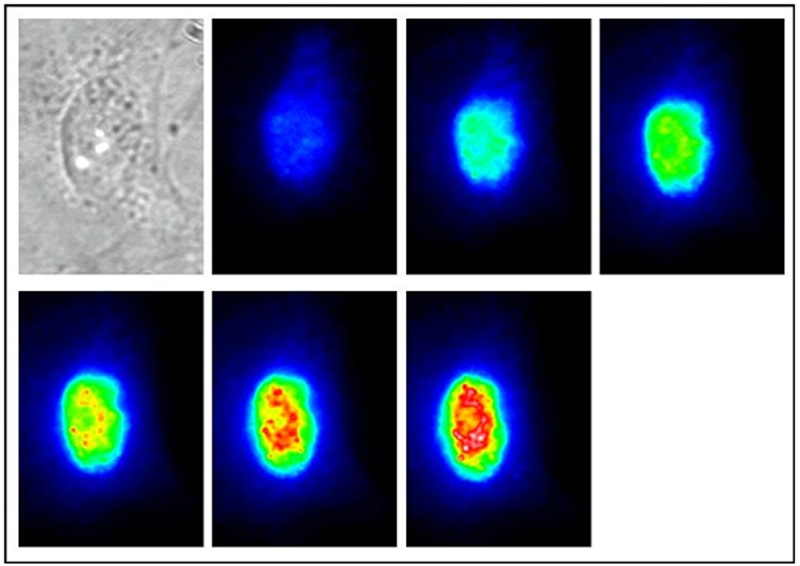
A transmission and the consecutive fluorescent images of a PtK2 cell injected with a β-1 andrenergic mRNA MB at 3-minute intervals for 18 minutes.
In 2005, Bao et al expanded the application of MBs in mRNA visualization by showing the colocalization of mRNA and intracellular organelles in human dermal fibroblasts [8]. In this study, MBs were used in conjunction with a fluorescent mitochondrial stain. Since the fluorescence from the MBs and the stain could be spectrally resolved, they were able to demonstrate that the mRNA of both glyceraldehyde 3-phosphate dehydrogenase and K-ras were specifically localized within the mitochondria. Their observation was confirmed by several control experiments, including the use of negative control MBs, fluorescence in situ hybridization (FISH) and detection of colocalization of 28S ribosomal RNA with the rough endoplasmic reticulum. The authors suggested that the observation of subcellular associations of mRNA with organelles such as mitochondria might provide new insight into the transport, dynamics, and functions of mRNA and mRNA-protein interactions.
Instead of focusing on localization and distribution, levels of mRNA expression have also been studied inside living cells using MBs. The binary MB approach was explored to determine the relative expression levels of K-ras and survivin mRNAs in human dermal fibroblasts [4]. According to the report, the expression level of K-ras mRNA was 2.25 fold higher, which was comparable to the ratio of 1.95 using RT-PCR. Recently, the stochasticity of manganese superoxide dismutase (MnSOD) mRNA expression in human breast carcinoma cells was studied using MBs with an internal standard reference probe to allow ratiometric analysis (Figure 3) [9]. In this work, the MnSOD expression in three different cell groups was studied and compared to each other and the expression of β-actin mRNA. The groups of cells studied included cells at basal expression levels, cells treated with Lipopolysaccharide (LPS), and cells transfected with a plasmid that overexpressed a cDNA clone of MnSOD. By using ratiometric analysis, many of experimental and instrumental variations were compensated, and the results could be directly compared among different cells. The study showed that the stochasticity of gene expression between the basal, LPS-treated, and the transfected cells was different for MnSOD while there was little to no difference in β-actin mRNA among the three groups. This represents a novel means to directly examine the stochasticity of transcription of MnSOD and other genes implicated in the regulation of cellular phenotype.
Figure 3.
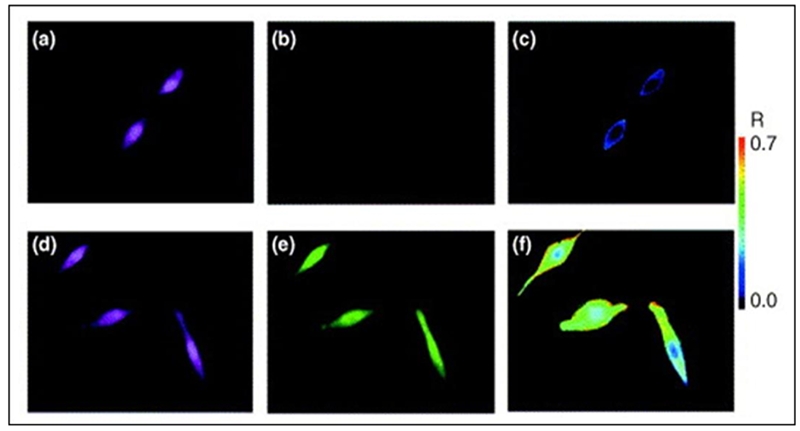
Intracellular imaging of single cells using MB probes. A ratiometric approach has been used to minimize experimental variations and enable more reliable data collection. On the top row are the cellular responses for ‘closed’ MBs. On the bottom row are the cellular responses for ‘open’ MBs. (a) and (d) are the fluorescence emission images of a reference probe. (b) and (e) correspond to fluorescence emission images of the MB probe response. (c) and (f) are representative ratiometric images of the MB responses by dividing the image from the MB by the image of the reference probe.
While researches have demonstrated the utility of MBs in intracellular monitoring of mRNA, problems still exist. Most of the causes are either from the complexity of living environment or the inherent properties of MBs. For example, MBs are vulnerable to intracellular enzymatic degradation, such as RNase cleavage of MB-bound RNA targets and non-specific opening by single-strand binding proteins (SSBs) [10], causing false-positive and -negative signals. For example, it has been reported that unmodified phosphodiester oligonucleotides may possess a half-life as short as 15–20 min in living cells [11]. In addition, the high background signal from autofluorescence of MBs themselves decreases the resolution. To solve the first problem, non-standard nucleic acids have been explored to design MBs, such as 2-OMe-modified RNAs [12–16], peptide nucleic acids (PNAs) [17–20], lock nucleic acids (LNAs) [21, 22] to improve the resistance to enzymatic activities (Table 1). 2-OMe-modified MBs show higher affinity, increased specificity, faster hybridization kinetics, and protection of bound RNA targets from RNase H cleavage [3]. However, 2-OMe-modified MBs open non-specifically inside cells, possible due to protein bindings [15, 16]. PNAs have peptide backbones. PNAs are not degraded by nucleases but have a neutral charge, and hybrids with RNA are thermally more stable compared to DNA-RNA and RNA-RNA duplexes. Xi et al [20] reported that use of PNA-MBs instead of traditional fluorescent in situ hybridization probes or DNA-MBs would be better under a wide range of environmental conditions. However, PNAs have not been widely used mainly because PNAs have drawbacks, such as limited solubility causing aggregation in biological environment. Recently, Tan et el investigated a locked nucleic acid (LNA)-MBs and demonstrated the great potential of LNA-MBs [21, 22]. LNAs are a conformationally restricted nucleic acid analogue, in which the ribose ring is locked into a rigid C3′-endo or Northern-type conformation by a simple 2′-O, 4′-C methylene bridge [23, 24]. LNAs as well as LNA-MBs have many attractive properties [21–24], such as high binding affinity, excellent base mismatch discrimination capability, and decreased susceptibility to nuclease digestion (Figure 4). Unlike DNA-MBs, the high structural stability of LNA-MBs showed a significantly lower background after delivered into cancer cells. In living environment, the LNA-MB showed no increase in fluorescence over a period of an hour without its target, while the DNA-MB exhibited a dramatic increase in signal after thirty minutes due to structural degradation. On the contrary, the completely open conformation of each beacon generated similar levels of fluorescence inside the cells after each MB bound to the synthetic complement. The longer lifetime and high structural stability of the LNA-MBs suggest that LNA-MB could be a useful probe for intracellular analysis.
Table 1.
Properties of different MBs
| Name | Major properties |
|---|---|
| 2-OMe-modified MBs | Resistant to nuclease digestion |
| Higher affinity to targets | |
| Do not support RNase H | |
| Vulnerable to SSBs | |
| PNA MBs | Resistant to nuclease digestion |
| Higher affinity to targets | |
| Low solubility in aqueous environment | |
| LNA MBs | Resistant to nuclease digestion |
| Higher affinity to targets | |
| Better structural stability | |
| Do not support RNase H | |
| Low fluorescence background in a living cell | |
| Resistant to SSBs depending on LNA MB design | |
| QD-labeled MBs | Higher brightness |
| Multiple-QD excitation with single wavelength | |
| CP-linked MBs | Higher brightness |
| Can be quenched with a single quencher | |
| Gold-MBs | Excellent quenching efficiency |
| Better selectivity | |
| SQ MBs | Remarkable signal enhancement (320 times with triple quencher assembly) |
Figure 4.
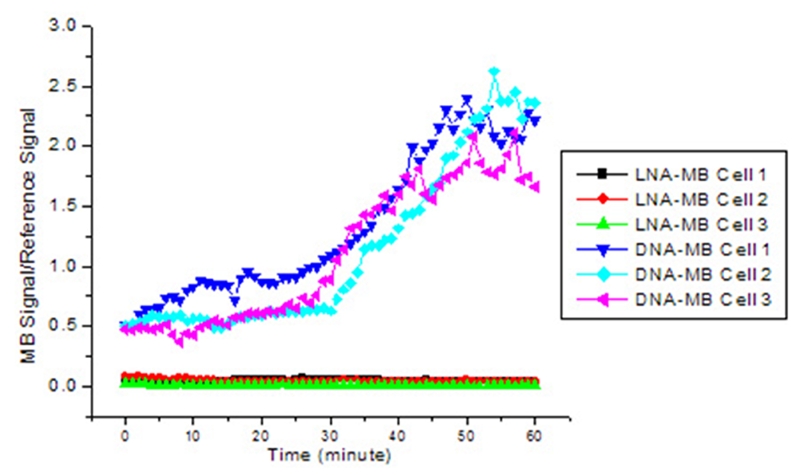
Background signal of LNA-MB and DNA-MB as a function of time after being injected into cells.
To solve the problem of low signal to background ratio, there are two reported strategies. One is to improve the brightness of signaling elements. For example, replacement of the organic fluorophores with other brighter signaling materials, such as quantum dots and fluorescent polymers, has been reported. Quantum dots have very high extinction coefficients (millions cm−1M−1). The fluorescence intensity from a single quantum dot could be as high as that of 20 individual organic fluorophores. In fact, MBs with quantum dots have been reported [25]. Besides their advantage of offering higher brightness, the use of quantum dots may offer multiplexed imaging for RNA analysis due to their broad UV absorption and narrow emission spectra [25]. To avoid self-quenching arising from multiple substitutions, a polymer chain, called conjugated or conducting polymer (CP), has been utilized to replace the fluorophore in the beacon. These polymers are poly-unsaturated macromolecules in which all backbone atoms are sp or sp2 hybridized. They are known to exhibit photoluminescence with high quantum efficiency [26]. A unique and attractive property of fluorescent CPs is their fluorescence superquenching effect [27–29]; that is, the CPs are a hundred- to a million-fold more sensitive to fluorescence quenching compared to that of their low molecular weight analogues. The fluorescence superquenching is attributed to a combination of delocalization of the electronic excited state (exciton) and fast migration of the exciton along the CP chain. As a result, if the fluorescence of any single repeat unit is quenched, the entire polymer chain responds in the same fashion. An entire polymer chain of PPV with about 1000 repeating units has been shown to be quenched by a single methyl violet molecule [30]. When the MB is in its closed state, the CP will be brought close to the quencher, and it is expected that the fluorescence of the CP will be strongly suppressed. After the MB binds to the DNA targets, the fluorescence of the CP will be restored as a result of the increased distance between the CP and the quencher. After hybridization, the entire polymer will light up because the removal of quencher from the proximity of the CP. Compared to a traditional MB, this new design will greatly amplify the fluorescent signal obtained from target hybridization. The other strategy is to minimize the fluorescence background using metal quenchers or multiple assembled quenchers. For instance, metals such as gold have been exploited as a quencher in MB design. Dubertret et al [31] showed that 1.4 nm diameter gold nanoparticles quenched fluorescence as much as 100 times better than organic quencher DABCYL in MBs and had a higher quenching efficiency for dyes emitting near infrared region. Labeled with gold nanoparticle, MBs not only showed better sensitivity, but also improved selectivity in the detection of single base mismatch. A type of quencher assembly called superquenchers (SQs) for MB synthesis have showed remarkable signal enhancement [32]. The rationale is to assemble an array of quencher molecules to produce SQs for use in improving MB sensitivity. Pairing multiple quenchers to one fluorophore provides better quenching efficiency. This mainly comes from the improved absorption and the increased probability of dipole-dipole coupling between the quenchers and the fluorophore because of a collective quenching effect of these multiple quenchers. Thus, with the number of quenchers increased from one to three, MBs using fluorescein and DABCYL pair showed 14, 81, and 320 fold signal enhancement when hybridized to its target. This comparison clearly demonstrated that the SQ assembly greatly improves the signal to background ratio and thus, the analytical sensitivity of the fluorescent molecular probes.
MBs as Recognition Elements in Biosensor Development
MBs have also been shown to be outstanding recognition elements in the development of biosensors, especially arrays (Figure 5) [33, 34, 52]. MB arrays have demonstrated many advantages over conventional arrays. First, no target labeling is needed. Labeling is an important step in most microarray-based target preparation protocols. However, it is often time-consuming, labor-intensive and expensive. In addition, it can affect the levels of targets originally present in the sample. Secondly, no washing step is required. Due to the unique on-off signaling process, only MBs hybridized to their targets will produce fluorescence. Extra unbound MBs stay with quenched fluorescence. The high background in the conventional porous film microarrays is resolved. It will also help to miniaturize devices, such as lab-on-a-chip, design without having to consider washing problems in small volumes. Third, the hybridization process can be easily monitored in real time and more reliable results can be obtained from the hybridization dynamics curves. Fourth, the high specificity of MBs always ensures that mismatches can be easily discriminated. The presence of a hairpin structure maximizes the specificity of MB probes. Furthermore, unlike linear probes, MB probes are insensitive to mismatch type and position, which greatly simplifies probe design.
Figure 5.
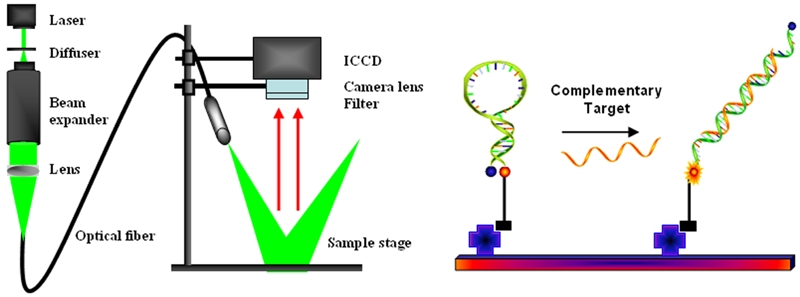
Schematic illustration of optical imaging setup (left panel) and MB arrays (right panel). MBs are immobilized using specific biomolecular interaction, such as streptavidin/biotin or covalent cross-linking.
There are many reports that MBs are the highly promising elements for microarrays using glass and fiber optics. In 1999, Tan et al reported that immobilized dual-labeled DNA probes on solid surface can still function as MBs to detect nucleic acid targets [33]. Under optimal condition, surface-immobilized MBs showed a linear response toward single strand DNA targets in the range of 5 to 10 nM with a detection limit of 2nM [35]. Using commercially available aldehyde-coated glass slides, the detection of Francisella tularensis has been accomplished with amine functionalized MB in a microarray format [36]. The research demonstrated that MBs have superior specificity toward complimentary oligonucleotide sequences to discriminate multiple and even single-nucleotide-mismatched sequences. In the case of using optical fibers as the surface, the performance of MBs appears to be better. Walt et el developed a randomly ordered fiber-optic gene array for rapid, parallel detection of unlabeled DNA targets with surface immobilized MBs [34]. They were able to detect genomic targets of cystic fibrosis with great selectivity, positional registration and fluorescence response monitoring of the microspheres by using an optical encoding scheme and an imaging fluorescence microscope system. The detection limit of the MBs immobilized on fiber optics is about 1.1 nM [37].
Although glass is the major material used in biosensor devices, attention has been paid to alternatives. Gold is a successful example [34–38]. It has particularly attractive molecular characteristics and has been extensively used in biotechnology and nanotechnology for signal transduction purposes. The interesting optical property of gold is that it can quench the fluorescence signal in very close proximity but enhance the fluorescence when it is moderately separated from the organic fluorophores. By immobilizing fluorophore-functionalized hairpin nucleic acids without quenching elements onto a gold surface, the fluorescence is quenched in the absence of targets due to the close proximity of the fluorophore to the gold surface. When the immobilized nucleic acids are hybridized with targets, they light up, and their fluorescence is enhanced. After hybridization with complementary targets, the fluorescence signal can increase more than 100 folds, and significant fluorescence intensity can be observed in less than 15 minutes.
In most biosensors, the sensitivity of MBs immobilized onto a solid surface is unfortunately never close to that of MBs in buffers. To improve MBs in biosensing applications, new MB probes were designed to enable a larger separation between the surface and the surface bound MBs by adding poly-T linker at one stem end of the MBs [39]. Using these MB probes, a DNA array has some, but very limited improvement in analytical sensitivity. Agarose-coated film has also been used for surface modifications with a much better performance of the surface-immobilized MBs [40]. One of the major causes for the decreased sensitivity is the slow hybridization kinetics of immobilized MBs [41]. Single MB molecules were imaged during the hybridization to individual complementary DNA probes. Among the 400 MB probes that were analyzed, 349 of them (87.5%) hybridized quickly and showed an abrupt fluorescence increase, while 51 probes (12.5%) reacted slowly, resulting in a gradual fluorescence increase. This ratio stayed about the same with varying concentrations of cDNA in MB hybridization on the liquid/surface interface. Statistical data of the 51 single-molecule hybridization images showed that there were multiple steps in hybridization process. These results helped us to better understand DNA hybridization processes using single molecule techniques, which will improve biosensor and biochip development.
MBs in Clinical Diagnosis
The outstanding properties of MBs have led to their applications in clinical diagnosis. The combination of MBs and PCR amplifications provides a rapid and accurate identification of pathogens and detection of gene mutations in human diseases [2, 42, 43]. The principle is shown in Figure 6. During the annealing step of PCR, MBs hybridize to the complementary targets and fluorescence signal is increased. Meanwhile, excess MBs remain in the closed conformations with quenched fluorescence. Therefore, the intensity of the fluorescence signal is directly proportional to the concentration of targets. Further, during the extension stage, the MBs hybridized to targets melt and thus do not interfere with polymerization. Multiple targets can be simultaneously detected using a ‘cocktail’ of MBs, and the relative differences of the targets can be determined. Since detection of fluorescence signal does not require sample separation during PCR amplification, sealed tubes are used to eliminate subsequent handling, reducing the risk of contamination. In addition, by-products, such as primer dimers and false amplicons, are unlikely detected because MBs can only be fluorescent in the presence of complementary targets.
Figure 6.
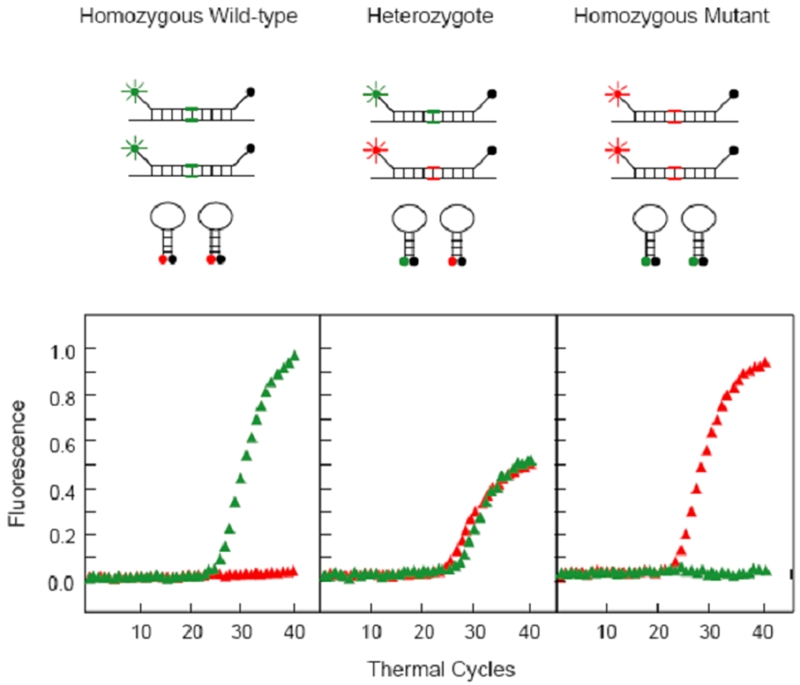
Principle of spectral genotyping using MBs by detection of a SNP in codon 325 of the estrogen receptor gene. In individuals homozygous for the major (C) allele, only the FAM-labeled MB hybridizes to the amplicons and thus is fluorescent, whereas the TET-labeled MB remains closed. In individuals homozygous for the minor(G)-allele, only the TET-labeled MB is fluorescent. In heterozygote individuals, both MBs hybridize to the amplicons, but since the total amounts of target available are theoretically the same, the levels of fluorescence from both fluorophores are decreased, compared with the fluorescence levels in the presence of a homozygous sample [2].
This type of assay generally requires fine tuning in designing MBs. First, proper selection of the reporter and quencher is important to increase the signal-to-background ratio. Second, it requires the use of a fluorometric thermocycler to monitor the fluorescence emission during the PCR reaction. Third, the melting temperature of the stem is about 10ºC above the annealing temperature to ensure that unbound MBs remain in the stem–loop conformation and, for the most part, are not open due to thermal fluctuations. Finally, it is important that the target-binding domain of the MB has a melting temperature slightly above the annealing temperature for optimal results.
There are many successful examples of using MBs as molecular probes to identify specific pathogens. MBs have been used to differentiate fungal pathogens with similar phenotypic and genotypic characteristics, such as Candida dubliniensis and Candida Albicans [44, 45]. Under well optimized condition, the assay was 100% sensitive and specific. Such accurate determination of highly similar pathogenic organisms could advance strategies for proper disease treatments. In another example, with nucleic acid sequence-based amplification (NASBA) of target molecules, MBs correctly identified West Nile, St Louis encephalitis and hepatitis B viruses with superior sensitivity and specificity [46, 47]. This type of assay can be easily incorporated into clinical laboratory testing, such as blood safety screening for transfusion medicine. In addition, MBs have also been found to be useful as a fast and reliable tool for the timely detection of food- and water-borne pathogens, such as Salmonella [48].
Due to the excellent selectivity in distinguishing single-base mismatched target, MBs have been applied in the detection of point mutations and allele discrimination. Variation in DNA sequence plays a critical role in disease development. Single nucleotide polymorphisms (SNPs) [49] are being used as genetic markers, primarily for whole-genome scanning as an indication of interesting regions contributing to the traits under investigation or in more focused candidate gene association studies. Currently, around 10 million SNPs are stored in public databases. In diploid organisms such as humans, SNPs show biallelic polymorphisms. Base substitutions occur about once per 300∼1000 base pairs, with the minor allele occurring at least 1% in a population. Since SNP or a group of SNPs has been identified as a disease-associated marker, selective detection of SNP(s) can be extremely useful in disease diagnosis, genetic counseling, prognostification and treatment stratification with better prediction.
Blom et el reported that MBs can detect the point mutation in the methylenetetra-hydrofolate reductase (MTHFR) gene, a mutation that causes neural tube defects and is associated with an increased risk for cardiovascular diseases [50]. With the optimal design of MBs to distinguish the wild type and mutant alleles, the difference in the level of MTHFR gene expression is significant after 30 cycles of PCR. The competitive nature of this assay results in high specificity. It has been demonstrated that the new assay can detect ten copies of a rare target in the presence of 100,000 copies of abundant target after PCR amplification [43]. The MBs also appear to be promising for epidemiological studies. For instance, a clinical assay was developed to detect the antifolate resistance-associated S108N point mutation of dihydrofolate reductase (DHFR) gene in Plasmodium falciparum [51]. DHFR S108N mutation, along with some other rare mutations, are the major molecular events leading to drug resistance. It is therefore very important to detect them for proper drug treatment. One hundred African clinical isolates have been tested by this new method. In comparison to the PCR-restriction fragment length polymorphism assay, it was found that the MB method was very fast and reliable. Since SNPs are extremely abundant in the human genome and are associated with various human deceases, a simple and accurate method to determine allele frequencies in pooled DNA samples could provide important information to map out genes that contribute to complex diseases and to determine the epidemiological patterns. It could also serve as a tool for a wider range of other genetic applications.
Conclusion
With the completion of the human genome project, the research interest has shifted to focus on utilizing the genetic information for prediction and discovery. In addition, the development of more sensitive, selective, and high-throughput analytical systems has been strongly demanded. Among many different types of analysis, MBs have shown some promising results. The remarkable selectivity has shown that they can specifically detect the desired target out of enormous number of non-specific sequences. Their unique property of detection without separation makes them even more attractive in the research areas where the purification step is almost impossible to perform or a major cause of low signal to background ratio. Their great sensitivity due to the excellent quenching efficiency is always advantageous. Thus, these benefits over other analytical methods have made MBs ideal for intracellular monitoring of nucleic acids and biosensor development. Gene-expression analysis under the native condition was able to be performed due to the unique signaling mechanism of MBs. This could be very important because it can dynamically show the cellular responses to external stimulus, such as drug treatment as well as differential gene expressions between normal and abnormal cells. Development of biosensors using MBs can accelerate the advance of high-throughput analytical methods with highly maintained selectivity to deal with the massive number of targets. In addition, no need to label targets can minimize possible experimental errors. With PCR amplification, MBs have also been widely used in clinical diagnosis, for instance, detection of human pathogens and identification of disease-associated alleles or SNPs. Therefore, the unlimited potential and success of MBs will continue to accelerate the determination of biological processes related to gene expression and the development of clinical diagnostic tests.
Acknowledgments
The authors want to thank all our coworkers whose work is reported here. The research was supported by NSF NIRT grant EF-0304569, HIH R01GM066137 and ONR grant N00014-07-1-0982.
References
- 1.Tyagi S, Kramer FR. Molecular beacons: Probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 2.Tyagi S, Bratu DP, Kramer FR. Multicolor molecular beacons for allele discrimination. Nat Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 3.Perlette J, Tan WH. Real-time monitoring of intracellular mRNA hybridization inside single living cells. Anal Chem. 2001;73:5544–5550. doi: 10.1021/ac010633b. [DOI] [PubMed] [Google Scholar]
- 4.Santangelo PJ, Nix B, Tsourkas A, Bao G. Dual FRET molecular beacons for mRNA detection in living cells. Nucleic Acids Res. 2004;32:e57. doi: 10.1093/nar/gnh062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sokol DL, Zhang XL, Lu PZ, Gewitz AM. Real time detection of DNA RNA hybridization in living cells. Proc Natl Acad Sci USA. 1998;95:11538–11543. doi: 10.1073/pnas.95.20.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bratu DP, Cha BJ, Mhlanga MM, Kramer FR, Tyagi S. Visualizing the distribution and transport of mRNAs in living cells. Proc Natl Acad Sci USA. 2003;100:13308–13313. doi: 10.1073/pnas.2233244100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui ZQ, Zhang ZP, Zhang XE, Wen JK, Zhou YF, Xie WH. Visualizing the dynamic behavior of poliovirus plus-strand RNA in living host cells. Nucleic Acids Res. 2005;33:3245–3252. doi: 10.1093/nar/gki629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santangelo PJ, Nitin N, Bao G. Direct visualization of mRNA colocalization with mitochondria in living cells using molecular beacons. J Biomed Opt. 2005;10:44025. doi: 10.1117/1.2011402. [DOI] [PubMed] [Google Scholar]
- 9.Drake TJ, Medley CD, Sen A, Rogers RJ, Tan WH. Stochasticity of manganese superoxide dismutase mRNA expression in breast carcinoma cells by molecular beacon imaging. Chembiochem. 2005;6:2041–2047. doi: 10.1002/cbic.200500046. [DOI] [PubMed] [Google Scholar]
- 10.Fang XH, Li JJ, Tan WH. Using molecular beacons to probe molecular interactions between lactate dehydrogenase and single-stranded DNA. Anal Chem. 2000;72:3280–3285. doi: 10.1021/ac991434j. [DOI] [PubMed] [Google Scholar]
- 11.Uchiyama H, Hirano K, KashiwasakeJibu M, Taira K. Detection of undegraded oligonucleotides in vivo by fluorescence resonance energy transfer. J Biol Chem. 1996;271:380–384. doi: 10.1074/jbc.271.1.380. [DOI] [PubMed] [Google Scholar]
- 12.Kehlenbach RH. In vitro analysis of nuclear mRNA export using molecular beacons for target detection. Nucleic Acids Res. 2003;31:e64. doi: 10.1093/nar/gng063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marras SA, Gold B, Kramer FR, Smith I, Tyagi S. Real-time measurement of in vitro transcription. Nucleic Acids Res. 2004;32:e72. doi: 10.1093/nar/gnh068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mhlanga MM, Vargas DY, Fung CW, Kramer FR, Tyagi S. tRNA-linked molecular beacons for imaging mRNAs in the cytoplasm of living cells. Nucleic Acids Res. 2005;33:1902–1912. doi: 10.1093/nar/gki302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Molenaar C, Marras SA, Slats JCM, Truffert JC, Lemaitre M, Raap AK, Dirks RW, Tanke HJ. Linear 2 ′ O-Methyl RNA probes for the visualization of RNA in living cells. Nucleic Acids Res. 2001;29:e89. doi: 10.1093/nar/29.17.e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsourkas A, Behlke MA, Bao G. Hybridization of 2 ′-O-methyl and 2 ′-deoxy molecular beacons to RNA and DNA targets. Nucleic Acids Res. 2002;30:5168–5174. [PMC free article] [PubMed] [Google Scholar]
- 17.Kuhn H, Demidov VV, Gildea BD, Fiandaca MJ, Coull JC, Frank-Kamenetskii MD. PNA beacons for duplex DNA. Antisense Nucleic Acid Drug Dev. 2001;11:265–270. doi: 10.1089/108729001317022269. [DOI] [PubMed] [Google Scholar]
- 18.Petersen K, Vogel U, Rockenbauer E, Nielsen KV, Kolvraa S, Bolund L, Nexo B. Short PNA molecular beacons for real-time PCR allelic discrimination of single nucleotide poly-morphisms. Mol Cell Probes. 2004;18:117–122. doi: 10.1016/j.mcp.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 19.Seitz O. Solid-phase synthesis of doubly labeled peptide nucleic acids as probes for the real-time detection of hybridization. Angew Chem Int Ed Engl. 2000;39:3249–3252. doi: 10.1002/1521-3773(20000915)39:18<3249::aid-anie3249>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 20.Xi CW, Balberg M, Boppart SA, Raskin L. Use of DNA and peptide nucleic acid molecular beacons for detection and quantification of rRNA in solution and in whole cells. Appl Environ Microbiol. 2003;69:5673–5678. doi: 10.1128/AEM.69.9.5673-5678.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L, Yang CYJ, Medley CD, Benner SA, Tan WH. Locked nucleic acid molecular beacons. J Am Chem Soc. 2005;127:15664–15665. doi: 10.1021/ja052498g. [DOI] [PubMed] [Google Scholar]
- 22.Yang CJ, Wang L, Wu Y, Kim Y, Medley CD, Lin H, Tan W. Synthesis and investigation of deoxyribonucleic acid/locked nucleic acid chimeric molecular beacons. Nucleic Acids Res. 2007;35:4030–4041. doi: 10.1093/nar/gkm358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orum H, Wengel J. Locked nucleic acids: A promising molecular family for gene-function analysis and antisense drug development. Curr Opin Mol Ther. 2001;3:239–243. [PubMed] [Google Scholar]
- 24.Vester B, Wengel J. LNA (Locked nucleic acid): High-affinity targeting of complementary RNA and DNA. Biochemistry. 2004;43:13233–13241. doi: 10.1021/bi0485732. [DOI] [PubMed] [Google Scholar]
- 25.Kim JH, Morikis D, Ozkan M. Adaptation of inorganic quantum dots for stable molecular beacons. Sens Actuators B-Chem. 2004;102:315–319. [Google Scholar]
- 26.Hide F, Schwartz BJ, Diaz-Garcia MA, Heeger AJ. Conjugated polymers as solid-state laser materials. Synthetic Metals. 1997;91:35–40. [Google Scholar]
- 27.Kushon SA, Ley KD, Bradford K, Jones RM, McBranch D, Whitten D. Detection of DNA hybridization via fluorescent polymer superquenching. Langmuir. 2002;18:7245–7249. [Google Scholar]
- 28.Lu LD, Jones RM, McBranch D, Whitten D. Surface-enhanced superquenching of cyanine dyes as J-aggregates on Laponite clay nanoparticles. Langmuir. 2002;18:7706–7713. [Google Scholar]
- 29.Lu LD, Jones RM, Bergstedt TS, McBranch D, Whitten D. Self-assembled “polymers” on nanoparticles: Superquenching and sensing applications. Abstracts of Papers of the American Chemical Society. 2002;223:D43. [Google Scholar]
- 30.Chen LH, McBranch DW, Wang HL, Helgeson R, Wudl F, Whitten DG. Highly sensitive biological and chemical sensors based on reversible fluorescence quenching in a conjugated polymer. Proc Natl Acad Sci USA. 1999;96:12287–12292. doi: 10.1073/pnas.96.22.12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dubertret B, Calame M, Libchaber AJ. Single-mismatch detection using gold-quenched fluorescent oligonucleotides. Nat Biotechnol. 2001;19:365–370. doi: 10.1038/86762. [DOI] [PubMed] [Google Scholar]
- 32.Yang CYJ, Lin H, Tan WH. Molecular assembly of superquenchers in signaling molecular interactions. J Am Chem Soc. 2005;127:12772–12773. doi: 10.1021/ja053482t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fang XH, Liu XJ, Schuster S, Tan WH. Designing a novel molecular beacon for surface-immobilized DNA hybridization studies. J Am Chem Soc. 1999;121:2921–2922. [Google Scholar]
- 34.Epstein JR, Leung AP, Lee KH, Walt DR. high-density, microsphere-based fiber optic DNA microarrays. Biosens Bioelectron. 2003;18:541–546. doi: 10.1016/s0956-5663(03)00021-6. [DOI] [PubMed] [Google Scholar]
- 35.Li J, Tan WH, Wang KM, Xiao D, Yang XH, He XX, Tang ZQ. Ultrasensitive optical DNA biosensor based on surface immobilization of molecular beacon by a bridge structure. Anal Sci. 2001;17:1149–1153. doi: 10.2116/analsci.17.1149. [DOI] [PubMed] [Google Scholar]
- 36.Ramachandran A, Flinchbaugh J, Ayoubi P, Olah GA, Malayer JR. Target discrimination by surface-immobilized molecular beacons designed to detect Francisella tularensis. Biosens Bioelectron. 2004;19:727–736. doi: 10.1016/j.bios.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 37.Liu XJ, Tan WH. A fiber-optic evanescent wave DNA biosensor based on novel molecular beacons. Anal Chem. 1999;71:5054–5059. doi: 10.1021/ac990561c. [DOI] [PubMed] [Google Scholar]
- 38.Du H, Strohsahl CM, Camera J, Miller BL, Krauss TD. Sensitivity and specificity of metal surface-immobilized “molecular beacon” biosensors. J Am Chem Soc. 2005;127:7932–7940. doi: 10.1021/ja042482a. [DOI] [PubMed] [Google Scholar]
- 39.Yao G, Tan WH. Molecular-beacon-based array for sensitive DNA analysis. Anal Biochem. 2004;331:216–223. doi: 10.1016/j.ab.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 40.Wang H, Li J, Liu HP, Liu QJ, Mei Q, Wang YJ, Zhu JJ, He NY, Lu ZH. Label-free hybridization detection of a single nucleotide mismatch by immobilization of molecular beacons on an agarose film. Nucleic Acids Res. 2002;30:e61. doi: 10.1093/nar/gnf061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao G, Fang XH, Yokota H, Yanagida T, Tan WH. Monitoring molecular beacon DNA probe hybridization at the single-molecule level. Chemistry. 2003;9:5686–5692. doi: 10.1002/chem.200304977. [DOI] [PubMed] [Google Scholar]
- 42.Tsourkas A, Bao G. Shedding light on health and disease using molecular beacons. Brief Funct Genomic Proteomic. 2003;1:372–384. doi: 10.1093/bfgp/1.4.372. [DOI] [PubMed] [Google Scholar]
- 43.Marras SAE, Kramer FR, Tyagi S. Multiplex detection of single-nucleotide variations using molecular beacons. Genet Anal. 1999;14:151–156. doi: 10.1016/s1050-3862(98)00018-7. [DOI] [PubMed] [Google Scholar]
- 44.Park S, Wong M, Marras SAE, Cross EW, Kiehn TE, Chaturvedi V, Tyagi S, Perlin DS. Rapid identification of Candida dubliniensis using a species-specific molecular beacon. J Clin Microbiol. 2000;38:2829–2836. doi: 10.1128/jcm.38.8.2829-2836.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sebti A, Kiehn TE, Perlin D, Chaturvedi V, Wong M, Doney A, Park S, Sepkowitz KA. Candida dubliniensis at a cancer center. Clin Infect Dis. 2001;32:1034–1038. doi: 10.1086/319599. [DOI] [PubMed] [Google Scholar]
- 46.Lanciotti RS, Kerst AJ. Nucleic acid sequence-based amplification assays for rapid detection of West Nile and St. Louis encephalitis viruses. J Clin Microbiol. 2001;39:4506–4513. doi: 10.1128/JCM.39.12.4506-4513.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yates S, Penning M, Goudsmit J, Frantzen I, van de Weijer B, van Strijp D, van Gemen B. Quantitative detection of hepatitis B virus DNA by real-time nucleic acid sequence-based amplification with molecular beacon detection. J Clin Microbiol. 2001;39:3656–3665. doi: 10.1128/JCM.39.10.3656-3665.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen W, Martinez G, Mulchandani A. Molecular beacons: A real-time polymerase chain reaction assay for detecting Salmonella. Anal Biochem. 2000;280:166–172. doi: 10.1006/abio.2000.4518. [DOI] [PubMed] [Google Scholar]
- 49.Brookes AJ. The essence of SNPs. Gene. 1999;234:177–186. doi: 10.1016/s0378-1119(99)00219-x. [DOI] [PubMed] [Google Scholar]
- 50.Giesendorf BAJ, Vet JAM, Tyagi S, Mensink EJMG, Trijbels FJM, Blom HJ. Molecular beacons: a new approach for semiautomated mutation analysis. Clin Chem. 1998;44:482–486. [PubMed] [Google Scholar]
- 51.Durand R, Eslahpazire J, Jafari S, Delabre JF, Marmorat-Khuong A, di Piazza JP, Le Bras J. Use of molecular beacons to detect an antifolate resistance-associated mutation in Plasmodium falciparum. Antimicrob Agents Chemother. 2000;44:3461–3464. doi: 10.1128/aac.44.12.3461-3464.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steemers FJ, Ferguson JA, Walt DR. Screening unlabeled DNA targets with randomly ordered fiber-optic gene arrays. Nat Biotechnol. 200;18:91–94. doi: 10.1038/72006. [DOI] [PubMed] [Google Scholar]


