Abstract
In the testis, the continuous production of sperm is maintained by a small population of stem cells called germ line stem cells (GSCs) in Drosophila, or spermatogonial stem cells (SSCs) in mammals. This stem cell population can self-renew or produce daughter cells that differentiate into mature sperm. In Drosophila, BMP signals inhibit GSC differentiation by blocking transcription of the gene, bag of marbles (bam). Once bam is activated, germ cells initiate differentiation. We identified a novel gene in mouse, Gm114, that shows homology to Drosophila bam. In male germ cells, expression of Gm114 begins at 12.5–13.5 days post coitum (dpc), the stage in mice when germ cells cease proliferation and begin differentiation into prospermatogonia. In adult testis, Gm114 is highly expressed in differentiated spermatocytes and spermatids but not in undifferentiated spermatogonia, strongly suggesting that, similar to Bam, GM114 plays an important role in mammalian germ line stem cell self-renewal and differentiation. Interestingly, deletion of the majority of the GM114 protein does not affect mouse viability or fertility. This suggests that either there is a function for the remaining N-terminal of GM114, or that there are alternative mechanisms in the mammalian system that control germ cell differentiation.
Keywords: testis, spermatogonial stem cells, Bam, GM114
Introduction
Mouse gonocytes undergo two or three rounds of mitosis in the genital ridge then enter sexually dimorphic patterns of differentiation at approximately 13.5 days post coitum (dpc) (McLaren, 2003). In the ovary, XX gonocytes directly enter meiosis and then arrest in meiotic prophase I. By contrast, in the testis, XY gonocytes are enclosed in testis cords where they enter mitotic arrest. After birth, gonocytes populate the perimeter of testis cords, where some of them are established as spermatogonial stem cells (SSCs) and undergo regulated proliferation thereafter.
The continuous production of mature sperm throughout life is maintained by this population of SSCs (de Rooij, 2001). SSCs are able to self-renew and to produce daughter cells that undergo several rounds of mitosis and enter meiosis to give rise to haploid spermatids, which subsequently are remodeled into spermatozoa. Growing evidence suggests that stem cell self-renewal and differentiation are heavily influenced by surrounding somatic cells, in a micro-environment often referred to as a stem cell niche (Chen et al., 2005; Meng et al., 2000). Sertoli cells are the supporting somatic cell type of the mammalian testis, and produce a number of factors, such as Glial cell line-derived Neurotrophic Factor (GDNF), which are important for stem cell self-renewal (Meng et al., 2000). Two transcription factors, Promyelocytic Leukemia Zinc-finger Factor (PLZF) and B cell CLL/lymphoma 6 member b (BCL6b), are expressed in SSCs and are required for SSC maintenance (Buaas et al., 2004; Costoya et al., 2004; Oatley et al., 2006). However, it is still unclear how the germ cell differentiation decision is made and how the differentiation process is initiated.
In Drosophila, GSC self renewal and differentiation has been relatively well defined (Chen and McKearin, 2005; Fuller and Spradling, 2007; Gilboa and Lehmann, 2004a; Szakmary et al., 2005; Zhao and Garbers, 2002). Drosophila GSCs are attached to a group of specialized somatic cells, referred to as hub cells in the testis and cap cells in the ovary. GSCs divide in an oriented division to produce two daughter cells. One daughter cell remains associated with the hub or cap cells and retains GSC potential. The other daughter cell moves away from the hub or cap cells and becomes a primary spermatogonium or cystoblast which undergoes four rounds of mitotic divisions with incomplete cytokinesis to yield 16 interconnected cells. In Drosophila, two TGF-β family members that are the orthologs of vertebrate bone morphogenetic proteins (Bmps), Decapentaplegic (Dpp) and Glass bottom boat (Gbb), are secreted from the somatic cell niche and are critical for maintaining GSCs in both the ovary and testis (Chen and McKearin, 2003; Shivdasani and Ingham, 2003). Dpp/Gbb are responsible for transcriptional repression of the differentiation factor bag of marbles (bam) in GSCs (Chen and McKearin, 2003; Kawase et al., 2004).
When germ cells are surrounded by somatic cells which secrete BMP signals, bam is repressed and GSCs are maintained. When germ cells move away from the stem cell niche and no longer receive the BMP signals, bam transcription is initiated, causing the germ cells to undergo differentiation. Mutations in bam result in an increase in the number of undifferentiated or less differentiated germ cells in both the ovary and testis, and a block in further differentiation (Gonczy et al., 1997; McKearin and Spradling, 1990). Ectopic expression of bam in GSCs results in a loss of the renewing stem cell population (Gilboa and Lehmann, 2004b; Ohlstein and McKearin, 1997; Schulz et al., 2004). All of these experiments implicate Bam as a key factor that regulates the decision between self-renewal and differentiation of Drosophila germ cells (Fuller and Spradling, 2007; Zhao and Garbers, 2002).
It has been reported that several Bmp family members, Bmp8a, Bmp8b, Bmp4 and Bmp7, are present in the adult mouse testis and required for spermatogenesis (Hu et al., 2004; Loveland and Hime, 2005; Zhao et al., 2001). Recently Bmp7 has been shown to affect germ cell survival during fetal stages (Ross, 2007). This raises the possibility that a similar pathway may regulate spermatogonial stem cell differentiation in mammals as in Drosophila. Here we have identified a novel murine gene, Gene model 114 (Gm114), which encodes a protein with homology to Drosophila Bam. We found that the expression pattern of Bam and GM114 was similar between Drosophila and mouse germ cells, suggesting that the pathway regulating GSC/SSC differentiation might be conserved during evolution. To further investigate the role of GM114, we generated mice carrying a deletion and frameshift eliminating the majority of the protein, including the entire region homologous to Drosophila Bam. In contrast to the dramatic phenotypes observed in the Drosophila bam mutant, Gm114 mutant mice are viable and fertile, and display no overt developmental defects.
Method and materials
Targeting vector construction and generation of mice carrying the Gm114 floxed allele
The conditional gene-targeting vector was constructed using a recombineering approach developed by Neal Copeland’s laboratory, essentially as described previously (Liu et al., 2003). Detailed recombineering protocols and information on how to receive the recombineering reagents can be found on the website http://recombineering.ncifcrf.gov. All primers were purchased from Integrated DNA Technologies (IDT). 129S7/AB2.2 mouse BAC DNA clone bMQ292a11 was donated from The Sanger Institute. BAC DNA was purified using a rapid alkaline lysis miniprep method. The BAC was transferred into the modified E. coli strain DY380 by electroporation. Two small DNA homologous arms were amplified from BAC clone bMQ292a11 with 2 pairs of primers: A 5’ AAT GCG GCC GCG TTG CTT TCT CTC TTG 3’; B 5’ CGG AAG CTT GGA AAA CAG GGC AAC AT 3’; Y 5’ GGC AAG CTT TCA GTT GCA CCA ACA 3’; Z 5’ GGC ACT AGT AGA AGA ACA CAT TCG ATC 3’. These two arms were cloned into the PL253 vector with SpeI and NotI sites. This retrieval vector was linearized by HindIII digestion, then gel purified and transformed into heat-shocked and electrocompetent DY380 cells containing the bMQ292a11 BAC clone. The genomic 10.2kb region was sub cloned into PL253 by homologous recombination and modified further in two targeting rounds using two vectors, PL452 and PL451, as described previously (Liu et al., 2003). First, to insert the single 5’ loxP site, a floxed neomycin/kanamycin cassette (from PL452) with homology to Gm114 intron 5 was generated. The primers used to amplify the two homology arms were as follows: C: 5’ CGC GTC GAC ACT TCT TGT CTT 3’; D: 5’ GGA ATT CTG AGC CTG GTC TTT G 3’; E: 5’ CGGGATCCCATAAACATACTAGTA 3’; F: 5’ AATAGCGGCCGCAGTGGTGACCTTCATAAT 3’. PCR conditions for amplification of all homology arms were 94°C for 30 s, 59°C for 60 s, and 72°C for 60 s for 30 cycles, using 1 µg BAC clone bMQ292a11 DNA as the template. Two homology arms were cloned into PL452, one after the other, with SalI/EcoRI (5’ arm) and BamHI/NotI (3’ arm) sites. The targeting cassette was released by SalI and NotI digestion and inserted into the retrieved Gm114 fragment by homologous recombination in DY380 cells. The neo cassette was then removed by transformation into arabinose-induced Cre-expressing EL350 cells to leave behind only a single loxP site. Next, using the same procedure, a Frt-neo-Frt-loxP cassette (from PL451) was inserted into intron 6. The primers used to amplify the homology arms for the 3’ targeting vector were as follows: G:
5’CCGCTCGAGACCTAACTCTTACCCT 3’; H:
5’CGGAATTCATTACCTCATTTAATGG 3’; I:
5’CGGGATCCACTTGGCTGAAGATA 3’; J:
5’TAAGCGGCCGCCCAAAATGTACCA 3’. The conditional targeting vector was then linearized by NotI digestion and electroporated into 129 Sv/Ev -derived embryonic stem (ES) cells, using standard procedures. G418 (200 µg/ml) resistant clones were analyzed by PCR with one primer from the neo cassette: 5’ GGGAGGATTGGGAAGACAAT 3’ and the other from the Gm114 locus but located outside the targeted region: 5’ CTCAGCACATCACCAGAGGA 3’. Selected clones were analyzed by Southern blot hybridization using BamHI or EcoRI digestion and a 0.5kb DNA fragment that lies outside the 3’ targeting region as a probe (see Fig. 5 and data not shown). Correctly targeted clones harboring both the single 5’ loxP site and the 3’ Frt-loxP-flanked neo cassette were injected into C57BL/6 blastocysts at the transgenic mouse facility at Duke University Medical Center using standard procedures.
Figure 5. Generation of Gm114−/− mice.
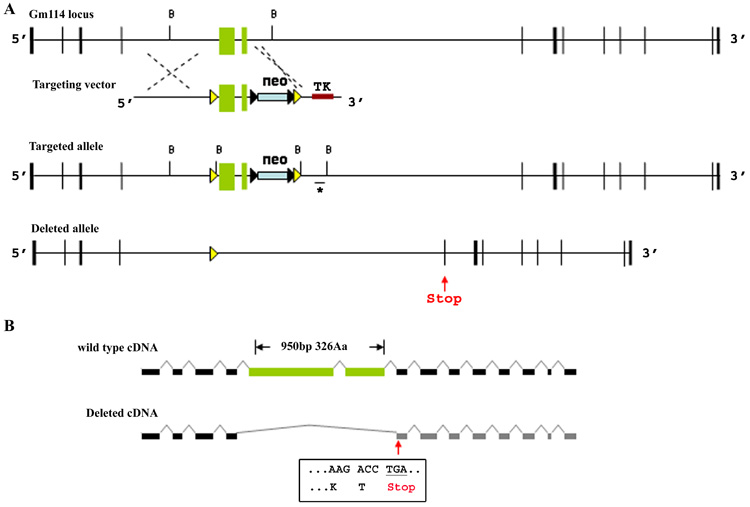
(A) Construct design to generate a conditional allele of Gm114. Two loxP sites (yellow triangles) were targeted into the Gm114 locus flanking exons 5 and 6 (green boxes) through homologous recombination. A neomycin cassette (blue box) flanked by frt sites (black triangles), and a thymidine kinase cassette (brown box) were used for positive and negative selection in ES cells. Southern blot and PCR analyses were used to identify targeted ES clones (data not shown). Southern blot analysis was performed using a BamHI restriction digest (cutting sites marked by B) and a specific DNA probe which is indicated by a line and asterisk underneath the targeted allele. (A, B) loxP recombination results in loss of 950bp of the coding region and generates a stop codon (red arrow) at beginning of exon 7 blocking translation of remaining exons. The DNA sequence in exon 7 and its encoded amino acid residues after the frame shift are shown in the box.
Generation of null mutants
After germ line transmission of the targeted conditional allele, Gm114loxP/+ mice were crossed to mice expressing Cre from the tissue non-specific alkaline phosphatase promoter (TNAP-cre) (Lomeli et al., 2000). This led to deletion of the conditional allele in the germ line and the generation of some offspring that were Gm114+/−.
Mouse strains and genotyping
Gm114+/− mice were maintained by intercrossing on a mixed genetic background (129/Sv; C57BL/6). TNAP-cre transgenic mice were generated as described by Lomeli et al.(2000) (Lomeli et al., 2000) and were kindly provided by Dr. Ting Xie in the Stowers Institute. Tail DNA was extracted using standard methods and genotyped by three-primer PCR analysis with the following primers: Gm17331 F, 5’ GAGGAAAGGTGCCTGGTCCAT 3’; Gm17690 R, 5’ CCAGTGGTGACCTTCATAATGAC 3’; and Gm21218 R, 5’ GCCCAAAATGTACCACACTG 3’. The wild-type band is 359 bp, the targeted allele band is 440 bp, and the deleted allele band is 602bp. PCR conditions were 94°C for 30 s, 61°C for 60 s, and 72°C for 60 s for 35 cycles. CD1 random-bred mice (Charles River) were used for immunocytochemistry and in situ hybridization.
5’ RACE
Total RNA was purified from dissected adult testes using the RNAeasy Mini Kit (QIAGEN, cat.74104). A 3’ primer, 5’ CGGGCCGGCTCTTTCTTCCCTGAC 3’, and a universal 5’ end primer from the SMART™ RACE cDNA amplification kit (BD Biosciences) were used to amplify 5’end cDNA. The PCR conditions were: 94°C for 30 s, 68°C for 30 s, and 72°C for 3min for 25 cycles.
Expression analysis
Total RNA was purified from dissected testes using the RNAeasy Mini Kit (QIAGEN, cat.74104). For RT-PCR analysis, first-strand synthesis was performed using the Superscript II kit (Invitrogen) and random hexamer primers. The resulting cDNA was analyzed by PCR using the following primers: 5’ CGGAATTCTCTGAAGGCAGAAAGTCTGCT 3’; 5’ CCGCTCGAGAGCTCATGTGTCATAAAAATC 3’. PCR conditions were 94°C for 15 s, 60°C for 30 s, and 72°C for 30 s for 30 cycles. The 1.3kb 3’ end cDNA was cloned and used as a probe for Northern analysis (see Fig. 2). Northern blot analysis was conducted according to standard procedures (Lerner and Nicchitta, 2006).
Figure 2. Structure of the Gm114 locus and transcript in testis.
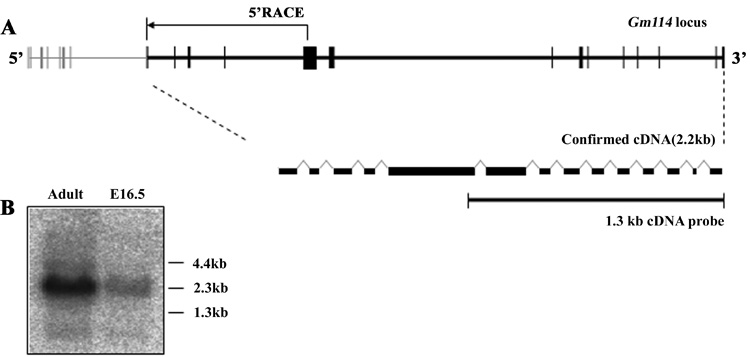
(A) Structure of predicted and confirmed Gm114 locus. 5’ RACE analysis mapped the start site of the Gm114 transcript, which is located at the predicted exon 8 (gray boxes represent predicted exons, black boxes represent confirmed exons). The arrow represents the result of 5’ RACE analysis using an internal 3’ primer located in the predicted 12th exon. The Gm114 gene is composed of 14 exons and is spliced into a 2.2 kb mRNA. (B) Northern blot analysis of adult and embryonic (16.5 dpc) testes total mRNA using a 1.3kb Gm114 cDNA probe (shown in A) confirmed that the major Gm114 transcript is slightly larger than 2kb.
Histology
Testes were dissected and fixed overnight in Bouin’s fixative (Polysciences, Inc, cat.16045) at 4°C. Samples were dehydrated and embedded in paraffin. 8µm sections were cut, stained using standard hematoxylin-eosin (H&E) procedures, mounted, and photographed using a Zeiss Axioplan 2 microscope.
Generation of an antibody against GM114
The 1.3kb 3’end of the Gm114 cDNA between BamHI and EcoRI sites was cloned into pGex6p-1 (Amersham Biosciences) and expressed in BL21-CodonPlus cells (Stratagene). Recombinant protein was purified from cell lysis supernatants using sequential Ni Sepharose, Q Sepharose, and Glutathione Sepharose columns (Amersham Biosciences). The GST tag was cleaved using Precision Protease (Amersham Biosciences) and depleted using glutathione resin. All purification steps were analyzed on a SDS-PAGE gradient gel stained with Coomassie Brilliant Blue. Three rabbits were challenged with purified GM114 protein and their serum was collected. Serum was further purified using a GM114 protein coupled CNBr Sepharose (Amersham Biosciences) affinity column. Serum and purified antibody were tested by Western blot using standard procedures.
In situ hybridization and immunocytochemistry
Testes were dissected and fixed overnight in 4% paraformaldehyde-phosphate-buffered saline (PFA-PBS). In situ hybridization was performed on whole-mount tissue or paraformaldehyde-fixed/OCT embedded cryosections, as described (Kim et al., 2006). The 0.5kb 5’ end of the Gm114 cDNA was used as an RNA probe template. Probes were synthesized using the DIG RNA labeling kit (Roche, Cat.1175025) following the manufacturer’s instructions. Fluorescent immunocytochemistry was performed on cryosections. Antibodies (Rabbit anti-GM114, 1:500; rat anti-PECAM (Pharmingen, San Diego, California, United States; 1:500); rabbit anti-Laminin (the kind gift of Harold Erikson, 1:500)) were added to the blocking solution and were incubated rocking at 4°C overnight. Samples were rinsed three times for 30 min in PBT (PBS and 0.1% Triton X-100) with 5% BSA and 0.1% heat-inactivated goat serum, and incubated overnight at 4°C in blocking solution with Cy2-, Cy3- or Cy5- conjugated secondary antibodies (1:500; Jackson Immunoresearch). Samples were washed three times for 30 min in PBT then mounted in DABCO and imaged on a Zeiss LSM420 confocal microscope.
Results
Identification of putative orthologs of Drosophila bam
Although the Drosophila bam gene has been studied since 1990 (McKearin and Spradling, 1990), no ortholog in mammals had previously been found (Zhao and Garbers, 2002). In 2004, we identified a mouse gene, Gene model 114 (Gm114), which shows significant homology to bam (Fig. 1A). The entire coding sequence of the Drosophila Bam protein was used to search the non-redundant database of GenBank coding sequences (NCBI) with the Position-Specific Iterated BLAST algorithm (PSI-BLAST) (Altschul et al., 1997). The top result was a predicted murine protein described as “similar to uncharacterized hypothalamic protein HT013” (XP_130476.4), which displayed 23% identity and 41% similarity with amino acids (aa) 41 through 197 of Bam. This gene was later given the designation of Gm114 by NCBI. No other murine sequences were identified that had significant identity over such a large region of the protein. Further sequence comparison using different alignment tools and working with smaller portions of the protein sequences identified additional regions of identity and similarity. In an optimal alignment of the two proteins (Fig. 1A, 1B), there is 21.5% identity and 32.7% similarity between Drosophila Bam and GM114 amino acid sequences. This is a relatively weak identity; however, the fact that there is some degree of homology over such a large region suggested that GM114 was a candidate Bam ortholog. While no other homologous sequences were identified in mouse, other vertebrate proteins with homology to GM114 were identified (Fig. 1C). For example, predicted HT013 in human is highly homologous to GM114 (66% identity). All these ortholog candidates are uncharacterized genes.
Figure 1. Identification of Gm114, a candidate ortholog of the Drosophila bag of marbles (bam) gene.
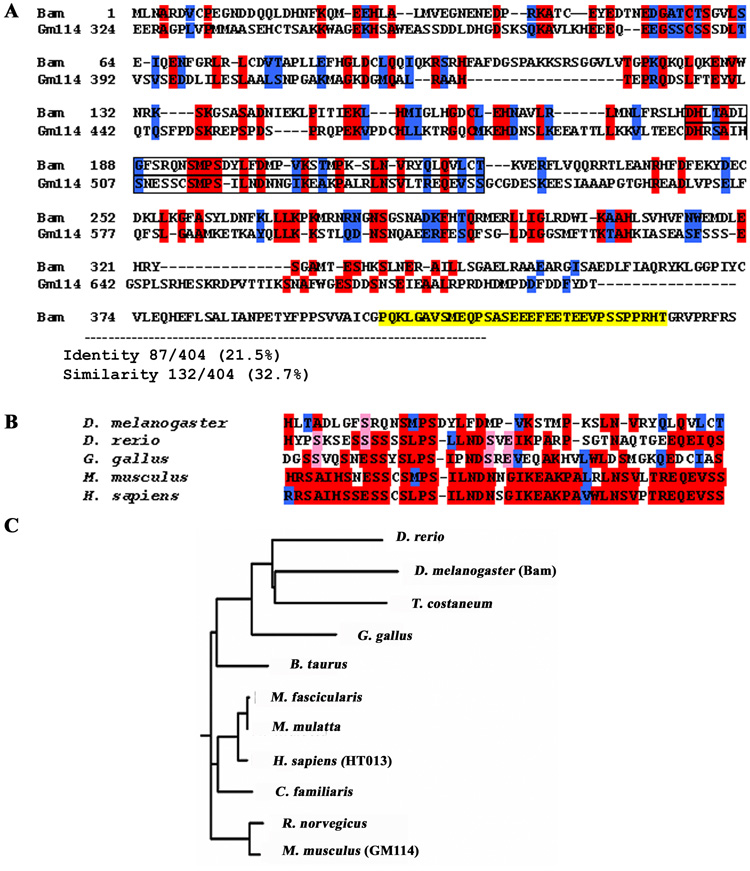
(A) BLAST analyses to identify candidate orthologs of the Drosophila protein. An uncharacterized mouse gene, termed Gene model 114 (Gm114) encodes a predicted protein with significant similarity to Bam (identical residues in red, conserved residues in blue, yellow is PEST sequence not contained in murine protein). Region of highest homology is boxed in (A) and aligned with other potential vertebrate orthologs identified in human, chicken, and zebrafish (B). (C) A phylogenetic tree of GM114 orthologs shows high conservation among mammalian species. Evolutionary distance was calculated and drawn using the Clustalw program at (http://myhits.isb-sib.ch/cgi-bin/clustalw). All putative orthologs are predicted genes with unknown function: Mus musculus (NP_001028470), Rattus norvegicus (ref|XP_230657.4|), Canis familiaris (ref|XP_848458.1|), Homo sapiens (gb|AAI05094.1|), Macaca mulatta (ref|XP_001093783.1|), Macaca fascicularis (dbj|BAE87931.1|), Bos taurus (ref|XP_591416.3|), Gallus gallus (ref|NP_001006394.1|), Tribolium castaneum (ref|XP_973854.1|), Drosophila melanogaster (NP_476800), Danio rerio (.ref|NP_001091860.1|).
Mapping the 5’ end of Gm114
Initially, the predicted sequence of Gm114 consisted of a 3.2kb coding region divided into 21 exons along 135kb of genomic sequence (Fig. 2A). To determine whether Gm114 was expressed in testis, we performed RT-PCR using primers positioned in exons 12 and 21. This experiment produced a 1.3kb band as expected (data not shown). To determine the size(s) of the transcript in testis, total mRNA from testes was analyzed by Northern blot using this 1.3kb specific cDNA as a probe (Fig. 2B). Gm114 is expressed in both adult and fetal 16.5 dpc testes. The majority of Gm114 transcripts were between 2–3 kb in size. We performed 5’RACE to map the start site of the transcript in adult testis (Fig. 2A). No sequence was amplified from the first 7 predicted exons. Instead our results indicated that the Gm114 transcript begins at the 153rd nucleotide of the predicted 8th exon. Consistent with this finding, a BRE and a TATA box consensus sequence are found at 35-30 nucleotides upstream from this start site. A continuous open reading frame starts with the ATG codon, 36 nucleotides downstream from the transcriptional start site. This transcript is 2152bp long and encodes 695aa in frame. In 2005 the GenBank sequence of Gm114 was updated, deleting the first 7 exons originally predicted, and confirming our 5’RACE results. All further information related to Gm114 exon and sequence numbers is presented according to this updated genomic structure.
The Gm114 mRNA expression pattern is consistent with a role in male germ cell development in mice
We examined Gm114 expression in gonads at different developmental stages by RNA in situ hybridization (Fig. 3). In mouse embryonic gonads, expression of Gm114 was specific to the XY gonad and was detected within testis cords. Expression gradually increased between 12.5 and 14.5 dpc (Fig. 3A) and was maintained in the neonatal and adult testis (Fig. 3B, 3C). Testis cords are comprised of Sertoli cells and germ cells. To determine which cells express Gm114, mRNA in situ hybridization was performed on KitW/Wv fetal gonads, which lack germ cells (Fig. 3D). Expression of Gm114 was not detected in the absence of germ cells. This suggested that Gm114 is either expressed in germ cells, or that its expression depends on the presence of germ cells. Gm114 is also detected in germ cells in the adult testis by RNA in situ hybridization (Fig. 3C). Very low or no expression of Gm114 was detected near the basal lamina of testis cords where SSCs and less differentiated spermatogonia reside in the mammalian testis. Expression levels increased through spermatocytes to round spermatids (Fig. 3C). This expression pattern suggested that Gm114 might be involved in male germ cell differentiation.
Figure 3. Expression pattern of Gm114 in the embryonic and adult testis suggests that Gm114 might be involved in male germ cell development.
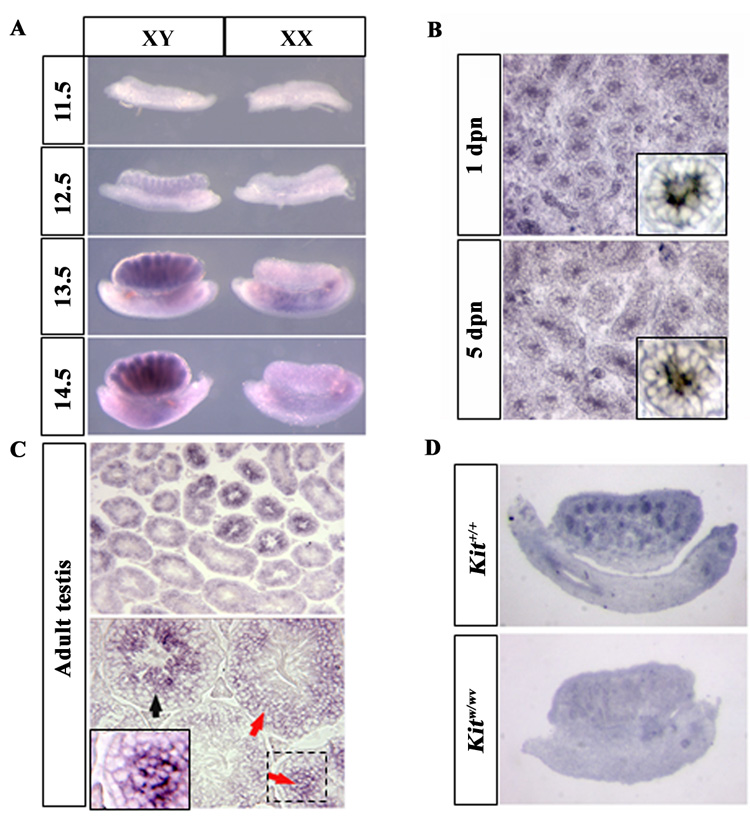
Expression of Gm114 was examined by mRNA in situ hybridization. (A) In the fetal gonad, expression of Gm114 is observed within testis cords and gradually increases between 12.5 and 14.5 dpc, when germ cells are arrested in mitosis and differentiate as prospermatogonia. Expression of Gm114 in germ cells is maintained during postnatal stages, as shown at day 1 and day 5 stages in (B, high magnification of a single cord shown in box). (C) In the adult testis, Gm114 is expressed in germ cells and expression increases throughout germ cell differentiation, with expression in spermatocytes and round spermatids (red arrows and high magnification of boxed region) and the highest expression in elongating spermatids (black arrow). (D) Gm114 expression was low or absent in testes cords of Kitw/wv mutant mice where germ cells are nearly completely absent.
GM114 is highly enriched in the cytoplasm of differentiated male germ cells
To complement our mRNA expression analysis we developed a polycolonal antibody against GM114 protein to study its expression and sub-cellular localization. By Western blot analysis of adult testis lysates, the antibody recognized a 76 kDa protein, the predicted size of full-length GM114 (see Fig. 6B). Like Bam, GM114 is a cytoplasmic protein at all stages tested (Fig. 4). Consistent with mRNA expression results, we detected GM114 protein in XY but not XX germ cells at 13.5 dpc (Fig. 4A, B). Expression of GM114 is maintained in XY germ cells throughout fetal stages and early postnatal stages (Fig. 4C, D). At about 7 days postnatal (dpn), when germ cells relocate from the center to the basal lamina of testis cords, GM114 is still detected in all germ cells at equivalent levels (Fig.4E, F). After the first wave of spermatogenesis is initiated around 10 dpn, GM114 expression was dramatically increased in early differentiated spermatocytes near the center of seminiferous tubules (Fig. 4G, H). Consistent with mRNA in situ results, when spermatogenesis proceeds further, GM114 was also found highly enriched in spermatids in later stages and in adult testes (Fig. 4I–K). However, undifferentiated and less differentiated spermatogonia that remain at the perimeter of the tubule expressed no or very low levels of GM114 (Fig.4H, stars in K and L) compared to the adjacent spermatocytes and spermatids. This pattern was confirmed by staining with GM114 and GCNA1, which marks spermatogonia and early spermatocytes in adult testes (Fig. 4L) (Enders and May, 1994). This protein expression pattern is similar to Bam’s pattern in Drosophila, strongly suggesting that GM114 is involved in the decision between self-renewal and differentiation in the mammalian germ line.
Figure 6. The testes of Gm114−/− mice develop normally and show no obvious difference from wild type littermates.
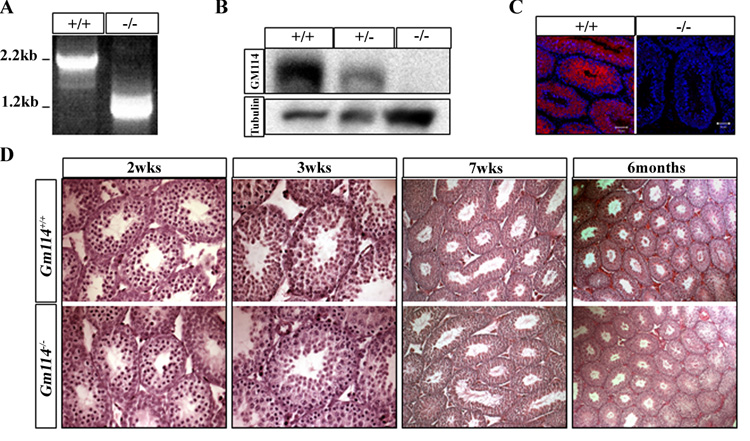
(A–C) GM114 expression is disrupted in mutant testes. Instead of a 2.2kb transcript, the deleted allele produces a 1.2kb transcript shown by RT-PCR (A). 76 kDa GM114 protein was not detected in mutant adult testes by (B) Western blot (γ-tubulin expression is shown as a loading control) or (C) fluorescent immunostaining (GM114 is red and DNA blue (propidium iodide)). The testis of Gm114 −/− mice developed normally and showed no obvious difference from wild type littermates (D). Hematoxylin-eosin (H&E) staining was performed on mutant and wild type testes at postnatal 2 weeks, 3 weeks, 7 weeks, and 6 months.
Figure 4. GM114 is a cytoplasmic protein expressed in male germ cells.
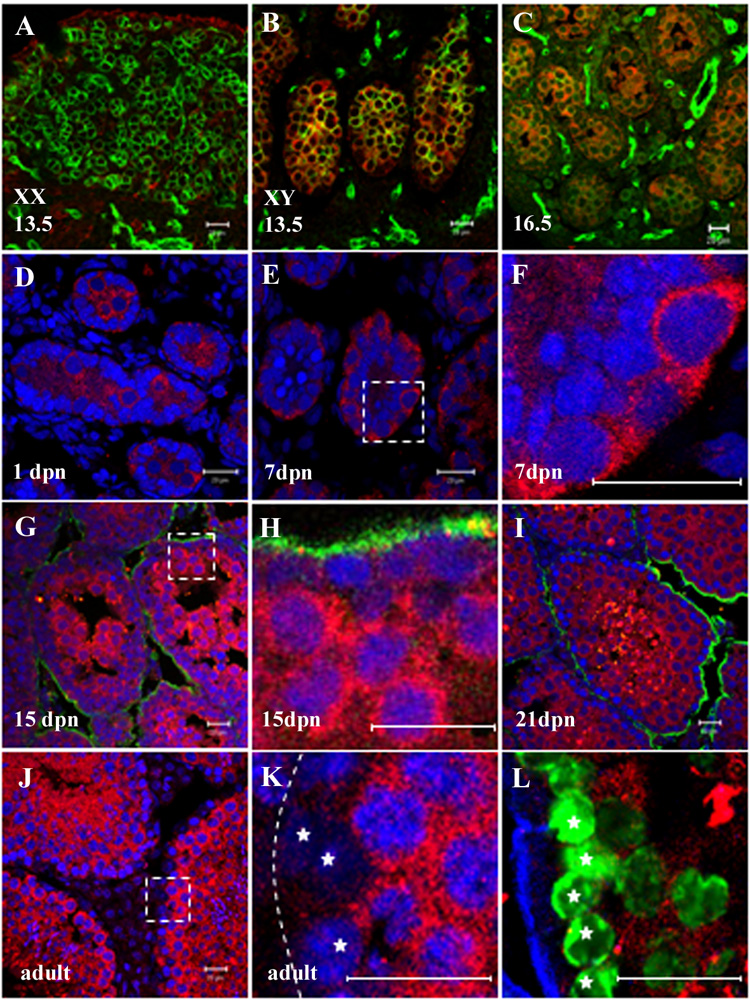
Fluorescent immunostaining was performed on gonads using a polyclonal antibody against GM114 (red) during fetal stages (A–C) and in postnatal and adult testes (D–L). (F), (H), and (K) are higher magnification of boxed regions in (E), (G) and (J). GM114 protein was detected in the cytoplasm of XY germ cells at 13.5 dpc (B) and16.5 dpc (C), but not in XX germ cells (A). A PECAM antibody labels germ cells and vascular endothelial cells (green in A–C). GM114 is maintained in all male germ cells from 1 to 7 dpn (D–F). At 15 dpn GM114 expression remains high in differentiated spermatocytes in the center of the seminiferous cords, but has declined in undifferentiated spermatogonia at the cord periphery (G, H). This pattern is seen at all later stages (J–L). Undifferentiated spermatogonia (stars in K and L, with low DNA density) have no or very low expression of GM114 compared to high expression in adjacent spermatocytes (high DNA density). The identity of undifferentiated spermatogoina at the periphery of the cord was confirmed by labeling with GCNA1 antibody (green in K) which marks spermatogonia and early spermatocytes (stars). DNA was stained with propidium iodide (blue in D–K). Basal lamina outlining testis cords is stained with an antibody against laminin (green in G–I; blue in L), or designated by a dashed line in K. All bars represent 20µm.
In contrast to the expression of Drosophila bam in female cystoblast cells, we detected no expression of GM114 during female germ cell differentiation or in the adult ovary in mice (Fig. 4A and data not shown). These results are consistent with the fact in mammals no self-renewing germ line stem cell population has been identified in the embryonic or adult ovary after meiosis begins.
Generation of a Gm114 null allele by deletion of the two largest exons
To investigate the function of Gm114 in germ cells, we generated mice with a deletion in gene. According to its EST profile (UniGene’s EST ProfileViewer, NCBI), Gm114 is expressed in a range of other tissues, including brain, bone marrow, skin, spleen, and thymus. We also detected Gm114 expression in cultured embryonic stem (ES) cells and embryoid bodies (data not shown). To circumvent the possibility of embryonic lethality, we generated a conditional allele of Gm114 (Fig. 5A) To avoid the low recombination efficiency associated with loxP sites positioned far apart, and considering the absence of homology to Drosophila Bam in the first 4 exons, we chose to target the two central exons of Gm114. Exons 5 and 6 are the two largest exons of the gene and encode the region of the protein with homology to Drosophila Bam. One loxP site was inserted into the 4th intron and the other into the 6th intron of the Gm114 locus. The deletion of exons 5 and 6 would be predicted to disrupt Gm114 by eliminating 950bp of the 2085bp that comprise the coding sequence. Subsequent to this deletion, the allele could be transcribed into a truncated mRNA with exon 4 directly spliced to exon 7. This would result in a reading frame shift in exon 7, generating a stop codon at the beginning of exon 7 such that the remaining sequence could not be translated (Fig. 5B).
Gm114 is not required for germ cell development
Twenty targeted ES cell clones were selected by PCR and Southern blot analysis, and four of these were used to generate chimeras (data not shown). Four chimeras showed germ line transmission of the targeted allele and two resulting lines were used for further analyses. To generate a germ line specific deletion of Gm114, Gm114loxp/+ mice were crossed with tissue non-specific alkaline phosphatase cre (TNAP-cre) mice, which express Cre recombinase in germ cells (Lomeli et al., 2000). Gm114loxp/+; TNAP-cre mice were crossed with wild type mice and produced offspring carrying a Gm114 deletion allele, Gm114+/−. This result indicated that Cre recombinase successfully induced loxP-mediated excision in the Gm114loxp/+; TNAP-cre germ cell genome, and that haploid sperm carrying the Gm114 deletion allele could develop normally and fertilize oocytes. Gm114+/− mice were intercrossed and produced Gm114−/− homozygous offspring. To test whether the Gm114 transcript was disrupted as predicted, RT-PCR analysis was performed on mRNA from homozygous mutant and wild type testes at 21 dpn using primers specific for the 1st and the last exons (Fig. 6A). The full-length Gm114 transcript was detected in wild type gonads, while, as expected, a truncated form of Gm114 mRNA was present in Gm114−/− testes of a size consistent with deletion of exons 5 and 6. The mutant cDNA was cloned and 12 independent clones were sequenced and shown to be identical (data not shown). As predicted, this transcript aligns with the 5’ end of the wild type transcript, and contains a 950bp deletion with a stop codon generated at the beginning of exon 7 resulting from the reading frame shift (Fig. 5B). The remaining 5’ sequence encodes a 143aa protein, 1/5th of full size, with no homology to Drosophila Bam. To test whether the protein was disrupted in the mutant, expression of GM114 was analyzed by Western blot in wild type and Gm114−/− adult testes lysates (Fig. 6B). The 76 kDa GM114 protein was decreased in heterozygous testis samples and was absent in the homozygous mutants. The polyclonal anti-GM114 antibody was raised against the protein domain generated from exons 6 to 14, which includes sequence not directly deleted by loxP recombination but affected by the frameshift. Using this antibody we detected no new protein isoforms in mutant lysates (data not shown), suggesting that the deletion disrupts translation of the entire C-terminal end of the protein because of the new stop codon generated in exon 7. The GM114 protein also was not detected by immunocytochemistry in the mutant adult testis (Fig. 6C). These results indicate that the loxP deletion had the predicted result of deletion of half of the coding region of Gm114 and disruption of the reading frame of the remaining 3’ end sequence. Mice heterozygous for the Gm114 deletion developed normally and, when intercrossed, produced offspring at 1:2:1 Mendelian ratios (+/+ =30; +/− =85; −/− = 45). Gm114−/− mutants were the same size and weight as wild type littermates, and no obvious defects were discovered during fetal development. During postnatal stages, mutant mice also developed normally and showed no obvious differences from their littermates. We dissected mutant testes at various stages, from 1 dpn to 16 months of age. At all stages the testis had a normal size and weight, and showed normal spermatogenic development (Fig. 6D, Table 1). All stages of the seminiferous epithelium were represented and sperm were found in the epididymis. Homozygous mutant male bred normally and their litter sizes were indistinguishable from wild type even at 16 months of age.
Table 1.
Measures of testis function
| Testis / body weight (%) | Plugged female ** | Pregnant female | litter size (Ave.) | |
|---|---|---|---|---|
| Gm114+/+ | 0.23 (n=5) | 16 | 13 | 9.1 |
| Gm114−/− | 0.24 (n=4) * | 15 | 11 | 8.9 |
P=0.7724, no significant difference between the mutants and wild type adult mice;
5 wild type and 5 mutant adult male mice at 6 and 16 months of age. Each male mouse was set up in continuous mating with 3 wild type female mice. Mating statistics and fertility were also unaffected.
Discussion
In Drosophila, GSC is relatively well understood (Fuller and Spradling, 2007). Orthologs of many important factors involved in GSC self-renewal and differentiation have been found in mammals (Spassov and Jurecic, 2002; Tsuda et al., 2003; Zhao and Garbers, 2002). In this study, we discovered that the murine protein GM114 shares sequence similarity to Drosophila Bam, and also displays an expression pattern that consistant with a role in mouse spermatogonial differentiation. Based on this evidence, we suggest that Gm114 is an ortholog of Drosophila bam.
During mouse embryonic development, male gonocytes enter mitotic arrest after 12.5 dpc and at least some of them commit to differentiation as prospermatogonia. Prospermatogonia lose the ability to give rise to pluripotent EG cells in culture, but acquire the potential to engraft and initiate spermatogenesis in adult recipient seminiferous tubules after transplantation (McLaren, 2003; Ohta et al., 2004). GM114 appears to be a transitional marker that is up-regulated in prospermatogonial cells, strongly suggesting a potential role in the regulation of male germ cell differentiation. Soon after birth some prospermatogonia migrate to the periphery of the testis cord and establish the SSC population. It is not known whether the definitive SSCs represent a pre-determined subset of the prospermatogonial cells in the fetal testis, or whether the entire population of prospermatogonial cells retains SSC potential, and the definitive SSCs are “selected” by their occupancy of the stem cell niche. Our data is consistent with the second possibility. XY germ cells that reach the basal lamina of testis cords at 10 dpn all initially express GM114, but those that settle into the SSC niche rapidly down-regulate GM114. This is strongly reminiscent of experiments in Drosophila suggesting that definitive GSCs are induced from a pluripotent population by signals from the niche (Fuller and Spradling, 2007).
In adult testes, Gm114 has a very similar expression pattern to bam in the Drosophila testis (Gonczy et al., 1997). It is not detectable in SSCs and their immediate progeny, but is highly enriched in differentiated spermatocytes and spermatids, suggesting a similar mechanism of GSC regulation between mouse and fly. There is no characteristic protein domain in either Bam or GM114, making it difficult to predict a biochemical function. It is possible that GM114 and Bam are components of the mRNA translational regulation pathway, since in Drosophila bam genetically interacts with several genes, such as benign gonial cell neoplasm (bgcn), pumilio, and nanos, which are all associated with mRNA translational regulation. Their orthologs are also found in mammals (Chen and McKearin, 2005; Ohlstein et al., 2000; Spassov and Jurecic, 2002; Szakmary et al., 2005; Tsuda et al., 2006; Tsuda et al., 2003; Zhao and Garbers, 2002). In mice, Nanos2 and GM114 have a similar expression pattern at fetal stages. However, after 5–7 dpn, when gonocytes move to the periphery of the tubules, the expression of Nanos2 restricts to a few spermatogonial cells along the basal lamina, and is maintained only in the SSCs of the adult testis (Tsuda et al., 2006; Tsuda et al., 2003). Pumilio2 has a similar expression pattern as Nanos2 in postnatal and adult testes (Xu et al., 2007). This reciprocal pattern between Nanos2/Pumilio and Gm114 suggests that they might have antagonistic functions in SSC self-renewal and differentiation. No genetic interaction among these genes has yet been reported in mouse.
In Drosophila, GSC maintenance requires BMP signals from the somatic environment to repress bam expression. In mammals, GDNF, a TGF-β family member secreted by Sertoli cells, has been shown to act through its receptor C-RET in spermatogonia to prevent germ cell differentiation (Meng et al., 2000; Naughton et al., 2006). Additionally, several Bmps and Smads are present in mouse testes (Loveland and Hime, 2005). PLZF, a transcriptional repressor in spermatogonia, was recently shown to be necessary for GSC maintenance (Buaas et al., 2004; Costoya et al., 2004). Although no direct downstream target of PLZF has been found, a group of genes was up-regulated in the PLZF null mutant. Gm114 expression increased both in PLZF mutants and in cultured SSCs after withdrawal of GDNF signaling (microarray analysis result, GEO accession number: GDS836 and GDS2112) (Costoya et al., 2004; Oatley et al., 2006). It will be interesting to know whether Gm114 is repressed in GSCs through one of these pathways.
Despite the homology between Gm114 and Drosophila bam and their provocative parallel expression patterns, we did not uncover a functional parallel between Gm114 and Drosophila bam. The remaining coding region at the 5’ end of GM114 can still be translated into a 143aa protein whose sequence is conserved among candidate mammalian orthologs. We cannot rule out the possibility that this small protein retains residual function through its N terminal fragment, which is only 1/5th the size of GM114 and exhibits no homology to Drosophila Bam. This would imply the evolution of a new functional domain in mammals unrelated to the Drosophila protein.
We think a more likely explanation is pathway redundancy in the mammalian system. Although we have identified no other Gm114 ortholog in mammals, functional redundancy may be achieved through robust compensatory pathways. For example, several genes involved in spermatogonial differentiation, such as Dazl (Ruggiu et al., 1997), Utp14b (Rohozinski and Bishop, 2004), Sohlh1 (Ballow et al., 2006) and Sox3 (Raverot et al., 2005), may compensate for loss of Gm114.
While the expression pattern of GM114 clearly distinguishes the stem cell component from differentiating spermatocytes, similar to Drosophila Bam, further studies will be necessary to identify GM114 partners and their interactions, and to clarify the role of GM114 in mammalian germ cell differentiation.
Acknowledgments
We are grateful for technical support from Dr. Vann Bennett (antibody); Dr. Christopher Nicchitta (Northern blot); Cheryl Bock and Dr. Brigid Hogan, (Gm114−/− mice); Dr. Neal Copeland and Dr. Jrgang Cheng (BAC recombineering); Dr. Mitch Eddy (Histological analysis); and Dr. Ting Xie for providing the TNAP-cre transgenic line. The BAC clone was kindly donated by The Sanger Institute. This work was funded by grants from NICHD (303 5136), the Lance Armstrong Foundation and the Duke University Medical Center Stem Cell Research Program. We are grateful for all discussion and comments from Capel lab members.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballow D, et al. Sohlh1 is essential for spermatogonial differentiation. Dev Biol. 2006;294:161–167. doi: 10.1016/j.ydbio.2006.02.027. [DOI] [PubMed] [Google Scholar]
- Buaas FW, et al. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet. 2004;36:647–652. doi: 10.1038/ng1366. [DOI] [PubMed] [Google Scholar]
- Chen C, et al. ERM is required for transcriptional control of the spermatogonial stem cell niche. Nature. 2005;436:1030–1034. doi: 10.1038/nature03894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, McKearin D. Dpp signaling silences bam transcription directly to establish asymmetric divisions of germline stem cells. Curr Biol. 2003;13:1786–1791. doi: 10.1016/j.cub.2003.09.033. [DOI] [PubMed] [Google Scholar]
- Chen D, McKearin D. Gene circuitry controlling a stem cell niche. Curr Biol. 2005;15:179–184. doi: 10.1016/j.cub.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Costoya JA, et al. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet. 2004;36:653–659. doi: 10.1038/ng1367. [DOI] [PubMed] [Google Scholar]
- de Rooij DG. Proliferation and differentiation of spermatogonial stem cells. Reproduction. 2001;121:347–354. doi: 10.1530/rep.0.1210347. [DOI] [PubMed] [Google Scholar]
- Enders GC, May JJ., 2nd Developmentally regulated expression of a mouse germ cell nuclear antigen examined from embryonic day 11 to adult in male and female mice. Dev Biol. 1994;163:331–340. doi: 10.1006/dbio.1994.1152. [DOI] [PubMed] [Google Scholar]
- Fuller MT, Spradling AC. Male and female Drosophila germline stem cells: two versions of immortality. Science. 2007;316:402–404. doi: 10.1126/science.1140861. [DOI] [PubMed] [Google Scholar]
- Gilboa L, Lehmann R. How different is Venus from Mars? The genetics of germ-line stem cells in Drosophila females and males. Development. 2004a;131:4895–4905. doi: 10.1242/dev.01373. [DOI] [PubMed] [Google Scholar]
- Gilboa L, Lehmann R. Repression of primordial germ cell differentiation parallels germ line stem cell maintenance. Curr Biol. 2004b;14:981–986. doi: 10.1016/j.cub.2004.05.049. [DOI] [PubMed] [Google Scholar]
- Gonczy P, et al. bag-of-marbles and benign gonial cell neoplasm act in the germline to restrict proliferation during Drosophila spermatogenesis. Development. 1997;124:4361–4371. doi: 10.1242/dev.124.21.4361. [DOI] [PubMed] [Google Scholar]
- Hu J, et al. Developmental expression and function of Bmp4 in spermatogenesis and in maintaining epididymal integrity. Dev Biol. 2004;276:158–171. doi: 10.1016/j.ydbio.2004.08.034. [DOI] [PubMed] [Google Scholar]
- Kawase E, et al. Gbb/Bmp signaling is essential for maintaining germline stem cells and for repressing bam transcription in the Drosophila testis. Development. 2004;131:1365–1375. doi: 10.1242/dev.01025. [DOI] [PubMed] [Google Scholar]
- Kim Y, et al. Fgf9 and Wnt4 act as antagonistic signals to regulate mammalian sex determination. PLoS Biol. 2006;4:e187. doi: 10.1371/journal.pbio.0040187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner RS, Nicchitta CV. mRNA translation is compartmentalized to the endoplasmic reticulum following physiological inhibition of cap-dependent translation. Rna. 2006;12:775–789. doi: 10.1261/rna.2318906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, et al. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomeli H, et al. Targeted insertion of Cre recombinase into the TNAP gene: excision in primordial germ cells. Genesis. 2000;26:116–117. [PubMed] [Google Scholar]
- Loveland KL, Hime G. TGFbeta superfamily members in spermatogenesis: setting the stage for fertility in mouse and Drosophila. Cell Tissue Res. 2005;322:141–146. doi: 10.1007/s00441-005-0008-0. [DOI] [PubMed] [Google Scholar]
- McKearin DM, Spradling AC. bag-of-marbles: a Drosophila gene required to initiate both male and female gametogenesis. Genes Dev. 1990;4:2242–2251. doi: 10.1101/gad.4.12b.2242. [DOI] [PubMed] [Google Scholar]
- McLaren A. Primordial germ cells in the mouse. Dev Biol. 2003;262:1–15. doi: 10.1016/s0012-1606(03)00214-8. [DOI] [PubMed] [Google Scholar]
- Meng X, et al. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science. 2000;287:1489–1493. doi: 10.1126/science.287.5457.1489. [DOI] [PubMed] [Google Scholar]
- Naughton CK, et al. Glial cell-line derived neurotrophic factor-mediated RET signaling regulates spermatogonial stem cell fate. Biol Reprod. 2006;74:314–321. doi: 10.1095/biolreprod.105.047365. [DOI] [PubMed] [Google Scholar]
- Oatley JM, et al. Identifying genes important for spermatogonial stem cell self-renewal and survival. Proc Natl Acad Sci U S A. 2006;103:9524–9529. doi: 10.1073/pnas.0603332103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B, et al. The Drosophila cystoblast differentiation factor, benign gonial cell neoplasm, is related to DExH-box proteins and interacts genetically with bag-of-marbles. Genetics. 2000;155:1809–1819. doi: 10.1093/genetics/155.4.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B, McKearin D. Ectopic expression of the Drosophila Bam protein eliminates oogenic germline stem cells. Development. 1997;124:3651–3662. doi: 10.1242/dev.124.18.3651. [DOI] [PubMed] [Google Scholar]
- Ohta H, et al. Commitment of fetal male germ cells to spermatogonial stem cells during mouse embryonic development. Biol Reprod. 2004;70:1286–1291. doi: 10.1095/biolreprod.103.024612. [DOI] [PubMed] [Google Scholar]
- Raverot G, et al. Sox3 expression in undifferentiated spermatogonia is required for the progression of spermatogenesis. Dev Biol. 2005;283:215–225. doi: 10.1016/j.ydbio.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Rohozinski J, Bishop CE. The mouse juvenile spermatogonial depletion (jsd) phenotype is due to a mutation in the X-derived retrogene, mUtp14b. Proc Natl Acad Sci U S A. 2004;101:11695–11700. doi: 10.1073/pnas.0401130101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross A, Munger S, Capel B. Bmp7 Regulates Germ Cell Proliferation in Mouse Fetal Gonads. sexual development. 2007;1:11. doi: 10.1159/000100034. [DOI] [PubMed] [Google Scholar]
- Ruggiu M, et al. The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature. 1997;389:73–77. doi: 10.1038/37987. [DOI] [PubMed] [Google Scholar]
- Schulz C, et al. A misexpression screen reveals effects of bag-of-marbles and TGF beta class signaling on the Drosophila male germ-line stem cell lineage. Genetics. 2004;167:707–723. doi: 10.1534/genetics.103.023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivdasani AA, Ingham PW. Regulation of stem cell maintenance and transit amplifying cell proliferation by tgf-beta signaling in Drosophila spermatogenesis. Curr Biol. 2003;13:2065–2072. doi: 10.1016/j.cub.2003.10.063. [DOI] [PubMed] [Google Scholar]
- Spassov DS, Jurecic R. Cloning and comparative sequence analysis of PUM1 and PUM2 genes, human members of the Pumilio family of RNA-binding proteins. Gene. 2002;299:195–204. doi: 10.1016/s0378-1119(02)01060-0. [DOI] [PubMed] [Google Scholar]
- Szakmary A, et al. Regulatory relationship among piwi, pumilio, and bag-of-marbles in Drosophila germline stem cell self-renewal and differentiation. Curr Biol. 2005;15:171–178. doi: 10.1016/j.cub.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Tsuda M, et al. Implication of nanos2-3'UTR in the expression and function of nanos2. Mech Dev. 2006;123:440–449. doi: 10.1016/j.mod.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Tsuda M, et al. Conserved role of nanos proteins in germ cell development. Science. 2003;301:1239–1241. doi: 10.1126/science.1085222. [DOI] [PubMed] [Google Scholar]
- Xu EY, et al. A gene trap mutation of a murine homolog of the Drosophila stem cell factor Pumilio results in smaller testes but does not affect litter size or fertility. Mol Reprod Dev. 2007;74:912–921. doi: 10.1002/mrd.20687. [DOI] [PubMed] [Google Scholar]
- Zhao GQ, et al. Mutation in Bmp7 exacerbates the phenotype of Bmp8a mutants in spermatogenesis and epididymis. Dev Biol. 2001;240:212–222. doi: 10.1006/dbio.2001.0448. [DOI] [PubMed] [Google Scholar]
- Zhao GQ, Garbers DL. Male germ cell specification and differentiation. Dev Cell. 2002;2:537–547. doi: 10.1016/s1534-5807(02)00173-9. [DOI] [PubMed] [Google Scholar]


