Abstract
S10-spc-α is a 17.5 kb cluster of 32 genes encoding ribosomal proteins. This locus has an unusual composition and organization in Leptospira interrogans. We demonstrate the highly conserved nature of this region among diverse Leptospira and show its utility as a phylogenetically informative region. Comparative analyses were performed by PCR using primer sets covering the whole locus. Correctly sized fragments were obtained by PCR from all L. interrogans strains tested for each primer set indicating that this locus is well conserved in this species. Few differences were detected in amplification profiles between different pathogenic species, indicating that the S10-spc-α locus is conserved among pathogenic Leptospira. In contrast, PCR analysis of this locus using DNA from saprophytic Leptospira species and species with an intermediate pathogenic capacity generated varied results. Sequence alignment of the S10-spc-α locus from two pathogenic species, L. interrogans and L. borgpetersenii, with the corresponding locus from the saprophyte L. biflexa serovar Patoc showed that genetic organization of this locus is well conserved within Leptospira. Multilocus sequence typing (MLST) of four conserved regions resulted in the construction of well-defined phylogenetic trees that help resolve questions about the interrelationships of pathogenic Leptospira. Based on the results of secY sequence analysis, we found that reliable species identification of pathogenic Leptospira is possible by comparative analysis of a 245 bp region commonly used as a target for diagnostic PCR for leptospirosis. Comparative analysis of Leptospira strains revealed that strain H6 previously classified as L. inadai actually belongs to the pathogenic species L. interrogans and that L. meyeri strain ICF phylogenetically co-localized with the pathogenic clusters. These findings demonstrate that the S10-spc-α locus is highly conserved throughout the genus and may be more useful in comparing evolution of the genus than loci studied previously.
Introduction
Leptospirosis is one of the most widespread zoonotic diseases in the world and is caused by pathogenic spirochetes within the genus Leptospira. Spirochetes belong to an ancient branch of eubacteria, with Leptospira representing its deepest division [1]. Leptospira are genetically diverse bacteria. Genetic classification of this genus is based on DNA homology and divides pathogenic Leptospira into seven main species: L. interrogans, L. borgpetersenii, L. weilii, L. noguchii, L. santarosai, L. kirschneri and L. alexanderi [2]–[4]. In addition, there are currently eleven recognized species with a saprophytic or intermediate pathogenic status, including the saprophytic species L. biflexa and L. meyeri, and L. fainei and L. inadai exemplifying species with an intermediate status [5]–[9]. Whole genome sequencing of L. interrogans serovars Lai and Copenhageni and two strains of L. borgpetersenii serovar Hardjo has revealed the occurrence of frequent gene rearrangements and fragmentation, perhaps indicating a rapid adaptation to new environments by pathogenic Leptospira [10]–[12]. It has been proposed that genome reduction detected in L. borgpetersenii reflects lower environmental survivability corresponding to limited potential for indirect transmission [10], in contrast to L. interrogans, a species that frequently passes through surface water between mammalian hosts [13].
We previously characterized the S10-spc-α ribosomal protein cluster of L. interrogans serovar Lai [14]. The cluster consists of 17.5 kb comprising 32 genes that, with the exception of fus, tuf, secY, adk and infA, code for ribosomal proteins. The secY gene codes for preprotein translocase, a gene that has diagnostic value and potential for resolving taxonomic questions in Leptospira [5], [14]. Genetic organization of ribosomal proteins is highly conserved and a prototypical S10 locus may predate divergence of Archaea and Bacteria [15]. However, translocation of several genes throughout the S10-spc-α locus differentiates Gram-positive from Gram-negative bacteria [15]. The genetic organization of the L. interrogans S10-spc-α locus is unique, as it contains all genes found in the Escherichia coli locus, and all genes except map that are found in the Bacillus subtilis locus [14]. The L. interrogans S10-spc-α locus is not typical of other spirochetes; several genes found in the S10-spc-α locus of L. interrogans are translocated to different portions of the Borrelia burgdorferi and Treponema pallidum genomes [14]. Considering the high plasticity of the Leptospira genome [10], [11], [16], it is unclear if genetic organization of the S10-spc-α locus is conserved amongst Leptospira, or if the genetic organization shared among Borrelia and Treponema may occur among some Leptospira species, and predate divergence of Leptospira from other spirochete genera.
In this study, we examined genetic organization and content of the S10-spc-α locus in Leptospira, and report that this locus is highly conserved throughout the genus. These data suggest that maintenance of the S10-spc-α operon structure is essential regardless of the extent of other rearrangements that have occurred during Leptospira evolution. Comparative sequence analysis of four segments of the S10-spc-α locus provides new information on phylogenetic relationships between pathogenic Leptospira.
Results
Amplification of the S10-spc-α locus of L. interrogans
Correctly sized fragments as deduced from the positions of the primer pairs on the locus (Table 1) were obtained from all six L. interrogans strains (Lai, M20, RGA, Hond Utrecht IV, Pomona and Hardjoprajitno) for each of the 40 primer pairs tested. These data indicate that the S10-spc-α locus is well conserved in L. interrogans (Table S3). Remarkably, the amplification pattern of L. inadai serovar Malaya strain H6 was identical to that of L. interrogans, a finding that we note below indicates this strain was incorrectly classified previously as L. inadai.
Table 1. Primer pairs and positions in the S10-spc-α locus of L. interrogans serovar Lai.
| Fragment | Primer pair | Position | Genes | Fragment | Primer pair | Position | Genes |
| 1. | 737-745 | 843-1435 | urp | 35.* | 301-258 | 11601-12581 | rplE-rplF |
| 2. | 740-744 | 1305-1873 | urp | 36.* | 301-191 | 11601-12948 | rplE-rplF |
| 3. | 748-751 | 1759-2493 | fus | 37. | 314-191 | 12348-12948 | rpsH, rplF |
| 4. * | 752-751 | 2269-2493 | fus | 38. | 314-428c | 12348-14047 | rpsH-rpmD |
| 5. * | 752-729 | 2269-2832 | fus | 39. | 314-430c | 12348-14372 | rpsH-rplO |
| 6. * | 735-729 | 2406-2832 | fus | 40. | 802-R1c | 12735-13445 | rplF, rplR |
| 7. * | 735-743 | 2406-3304 | fus | 41.* | 802-428c | 12735-14047 | rplF-rpmD |
| 8. | 735-667 | 2406-4394 | fus, tuf | 42.* | 802-430c | 12735-14372 | rplF-rplO |
| 9. * | 743c-706 | 3304-3814 | fus | 43.* | 191c-428c | 12948-14047 | rplF-rpmD |
| 10.* | 743c-667 | 3304-4394 | fus, tuf | 44. | 191c-430c | 12948-14372 | rplF-rplO |
| 11.* | 800-660 | 3683-4327 | fus, tuf | 45.* | R1-428c | 13427-14047 | rplR-rpmD |
| 12. | 800-667 | 3683-4394 | fus, tuf | 46. | R1-430c | 13427-14372 | rplR-rplO |
| 13.* | 657-654 | 4350-5255 | tuf | 47.* | 428-430c | 14047-14372 | rpmD, rplO |
| 14. | 657-624c | 4350-5976 | tuf-rplC | 48. | 428-G2c | 14047-15468 | rpmD-secY |
| 15. | 659-648 | 4438-5465 | tuf | 49.* | 430-G2c | 14372-15468 | rplO, secY |
| 16.* | 732-624c | 5240-5976 | tuf-rplC | 50 | 634-635 | 14643-16387 | rplO-adk |
| 17.* | 647-618 | 5297-5806 | tuf, rpsJ | 51.* | 443-G2c | 15276-15468 | secY |
| 18.* | 647-624c | 5297-5976 | tuf-rplC | 52.* | 443-G1 | 15276-15752 | secY |
| 19. | 624-650 | 5976-6790 | rplC, rplD | 53.** | SecYII-SecYIV | 15289-15946 | secY |
| 20.* | 624-644 | 5976-7151 | rplC-rplW | 54. | G2-G1 | 15468-15752 | secY |
| 21. | 624-621c | 5976-7847 | rplC-rplB | 55.* | G2-444 | 15468-15970 | secY |
| 22.* | 651-644 | 6883-7151 | rplD, rplW | 56. | G2-429 | 15468-16353 | secY-adK |
| 23.* | 643-621c | 7138-7847 | rplW, rplB | 57.* | G2-400 | 15468-16640 | secY-infA |
| 24.* | 622-621c | 7689-7847 | rplB | 58.* | 260-458c | 16616-18104 | infA-rpsD |
| 25. | 621-625 | 7847-8504 | rplB, rpsS | 59.* | 458-507 | 18104-18696 | rpsD, rpoA |
| 26.* | 621-605c | 7847-9082 | rplB-rpsC | 60.* | 458-504 | 18104-19376 | rpsD, rpoA |
| 27.* | 605-460 | 9082-10196 | rpsC-rpmC | 61. | 450-479 | 18163-19264 | rpsD, rpoA |
| 28.* | 801-803c | 10105-10965 | rpmC-rplX | 62.* | 477-504 | 18584-19376 | rpoA |
| 29. | 801-301c | 10105-11601 | rpmC-rplE | 63. | 477-501c | 18584-19791 | rpoA, rplQ |
| 30. | 801-300 | 10105-12110 | rpmC-rpsH | 64. | 503-480 | 18862-19621 | rpoA, rplQ |
| 31.* | 310-309 | 10167-10672 | rpmC-rplN | 65.* | 478-501c | 19371-19791 | rpoA, rplQ |
| 32.* | 310-277 | 10167-11107 | rpmC-rplX | 66.* | 478-502 | 19371-20341 | rpoA, rplQ, |
| 33.* | 310-301c | 10167-11601 | rpmC-rplE | 67. | 501-502 | 19791-20341 | rplQ |
| 34.* | 301-300 | 11601-12110 | rplE-rpsH |
Fragments used in the phylogenetic analysis from the binary data.
Primer pair used to produced G1–G2 sequences from all pathogenic species.
Comparative PCR analysis of the S10-spc-α locus in pathogenic Leptospira
Amplification patterns of different L. borgpetersenii and L. kirschneri strains shared a high level of identity (one and two differences, respectively). However, marked strain differences were found within the species L. santarosai (8), L. noguchii (9), L. weilii (15) and L. alexanderi (14). Predictably, because genetic relatedness is used to differentiate Leptospira species, the amplification profiles varied depending on the species from which the template DNA was isolated (Table S3). These data show that strains composing these species likely have higher sequence variation within the S10-spc-α locus than that seen in L. interrogans. To confirm that failed PCR amplifications were due to sequence variation at or near the primer annealing sites, and not a disruption of gene synteny, a series of additional primers were designed that directed amplification from conserved sequences in adjacent genes through the regions in question. Amplification using these additional primer sets confirmed that all genes initially identified in the L. interrogans S10-spc-α locus were present throughout the same locus of all pathogenic Leptospira species. This conserved organization extends as far as fus, encoding EF-G at the 5′ end of the locus, through rpsD at the 3′ end of the locus. Thus the genetic organization of the S10-spc-α locus is conserved in all pathogenic Leptospira spp. with no signs of disruptions or translocations of genes within the locus.
Comparative PCR analysis of the S10-spc-α locus of non-pathogenic Leptospira
Attempts to perform PCR analysis of DNA from Leptospira species with saprophytic or intermediate (i.e. questionable) pathogenic status frequently failed to generate products or yielded anomalous sized amplicons. These data imply a marked divergence in the S10-spc-α sequence content from pathogenic Leptospira (Table S3). Interestingly, the amplification profile of L. meyeri strain ICF is consistent with a pathogenic status whereas the profile of L. meyeri strain Veldrat Semarang 173 is more similar to those of the saprophytic and intermediate species L. biflexa, L. fainei, and L. inadai.
To determine if the genetic composition of this segment of the genome is different between saprophytic and pathogenic Leptospira, the corresponding regions of the L. biflexa, L. interrogans, and L. borgpetersenii genomes (GenBank accession numbers for L. interrogans, L. borgpetersenii and L. biflexa are AE016823, CP000348, CP000786, respectively) were aligned with BLAST and the results visualized by ACT (Fig. S1). These data show that saprophytic and pathogenic Leptospira have the same organization in the S10-spc-α locus, and the lack of successful PCR amplification is likely due to extensive sequence drift within the genus.
Phylogenetic analysis from binary data
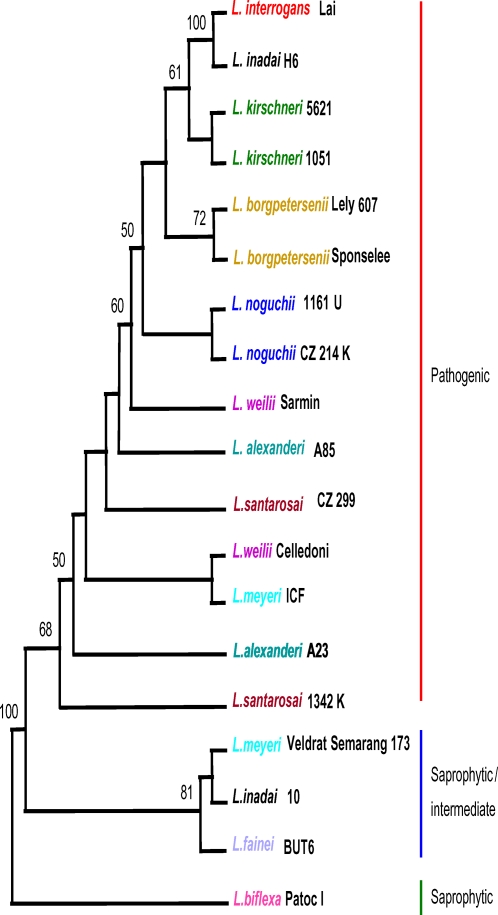
The parsimony criterion was used to infer phylogenetic relationships within Leptospira from binary data. The most parsimonious tree generated from these data shows two prominent well-supported clades: 1) a basal clade, with bootstrapping value of 81%, that includes L. fainei and L. inadai, two species with intermediate pathogenic status, and the saprophytic L. meyeri strain Veldrat Semarang 173; and 2) a sister clade, supported with a 68% bootstrap value, that contains pathogenic Leptospira species (Fig. 1). Within the pathogenic clade, relationships among L. alexanderi, L. santarosai, and L. weilii species, are poorly resolved. In contrast, L. interrogans, L. kirschneri, L. borgpetersenii, and L. noguchii are clustered in a well-supported clade where L. kirschneri and L. interrogans appear as closely related siblings.
Figure 1. Consensus tree based on PCR amplification data.
Majority-rule consensus tree elaborated under the parsimony criterion and based on binary data (absence/presence) coded from amplification patterns in the S10-spc-α locus for different Leptospira species. Numbers on nodes are bootstrap support after 100 replicates. Only bootstrap values above or equal to 50% are shown. Species included in the sequence analysis are coded in color. L. biflexa was used as the outgroup. CI = 0.346.
Surprisingly, there are two exceptions to the predicted distribution of strains. L. inadai strain H6 clusters with L. interrogans, and L. meyeri strain ICF branches within pathogenic species suggesting a pathogenic status for these two strains. As we note in the Discussion section, we believe strain H6 is incorrectly classified as L. inadai.
Multilocus sequence typing (MLST)
Phylogenetic analysis was done on four conserved loci within the S10-spc-α locus and compared to available Leptospira rrs sequence data (GenBank accession numbers EU365895-EU365966). DNA amplification of target sequences from the intermediate strains L. fainei strain BUT 6, L. inadai strain 10, and L. meyeri strain Veldrat Semarang 173 was not successful. Therefore, these strains were not included in the analysis. The sequences for the saprophytic strain Patoc I were deduced from its genome sequence [17]. None of the analyzed sequences are significantly deviated from neutral expectations (P>0.1). The shortest G1–G2 fragment (245 bp) showed the highest nucleotide diversity, π value of 0.14, whereas in the 300–301 fragment π was 0.09. Congruently, the mean divergence values (D) for pairwise comparisons ranged from 0.103 to 0.171 for the 300–301 and G1–G2 fragment, respectively. The lowest phylogenetic signal was obtained for 300–301 sequences. In contrast, the 621–625 and 624–650 fragments showed a phylogenetic signal slightly higher than the combined data set (Table 2).
Table 2. Nucleotide diversity, divergent estimations and parameters estimated from the sequences of 4 fragments in diverse Leptospira species.
| Fragment | Sites | Polymorphic Sites | Mean D | π | Θ per site | Tajima's D | P | -g1 |
| 300–301 | 469 | 179 | 0.1030 | 0.0891 | 0.1011 | −1.2016 | >0.1 | −0.5670 |
| 621–625 | 479 | 176 | 0.1190 | 0.1034 | 0.1306 | −0.8672 | >0.1 | −0.9404 |
| 624–650 | 491 | 226 | 0.1440 | 0.1205 | 0.1623 | −1.0739 | >0.1 | −1.0876 |
| G1–G2 | 245 | 91 | 0.1710 | 0.1434 | 0.1381 | 0.1606 | >0.1 | −0.5914 |
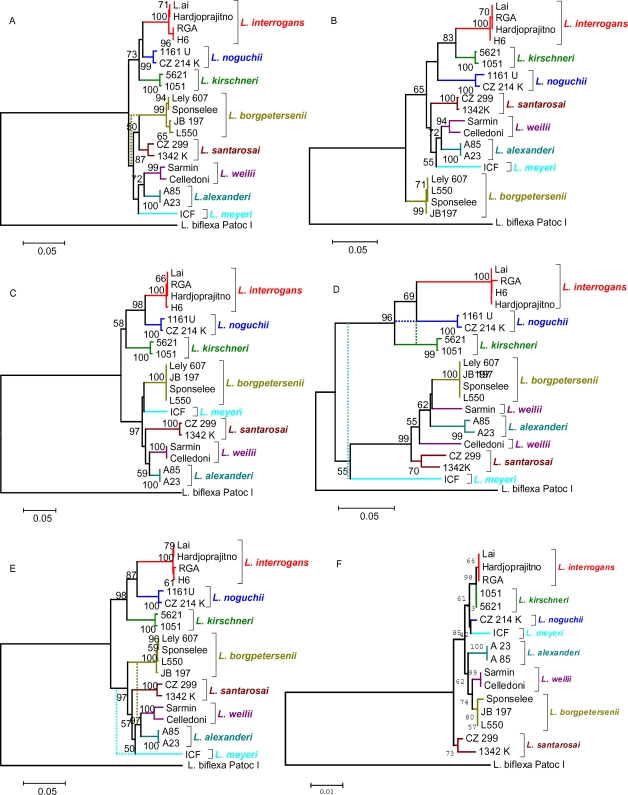
Distance and parsimony analysis yielded identical or similar topologies and bootstrapping values were comparable for the concordant nodes, although they were generally lower in parsimony trees. Alternative branching patterns in parsimony trees (with bootstrap value<50%) occurred in nodes showing the lowest bootstrap support in distance topologies.
In the composite tree (Fig. 2E), pathogenic strains were separated into two well-supported clades that are similar, but not identical to clades resolved in the binary tree. One clade consists of the sister sub-clades containing L. interrogans and L. noguchii, with L. kirschneri located in a basal position. This clade is consistently recovered in all topologies (Fig. 2), with the exception of the tree based on the 621–625 fragment (Fig. 2B) and parsimony topology generated from G1–G2 sequences (Fig. 2D), where L. kirschneri and L. noguchii swap their positions. The close relationships of these species are also apparent through comparative analysis using 16S rDNA sequence data (Fig. 2F) and independent binary data (Fig. 1). The second clade of the composite tree contains L. borgpetersenii, L. santarosai, L, alexanderi, L. weilii and L. meyeri strain ICF. Although, the branching pattern within this clade has lower support, the sibling relationship between L. alexanderi and L. weilii is well conserved. The relative positions of L. borgpetersenii, L. meyeri and L. santarosai are uncertain and vary depending on the data set and method of analysis (Fig. 2). In the tree inferred from the G1–G2 locus, the Celledoni and Sarmin strains of L. weilii are located in separate clades.
Figure 2. Phylogenetic trees based on Tamura-Nei distances and elaborated using the Neighbor-Joining method.
Distances were calculated from the 300–301 (A), 621–625 (B), 624–650 (C) and G1–G2 (D) sequence fragments within the S10-spc-α locus of pathogenic species of Leptospira. The total evidence was combined and analyzed under identical conditions (E). In addition, data available from 16S rDNA (rrs) sequences were used to obtain an alternative hypothesis for the relationships of diverse Leptospira strains (F). Dotted lines show alternative branching patterns, with bootstrapping values ≥50%, obtained in the consensus majority rule tree obtained by parsimony criterion. Numbers above branches represent the percentage of bootstrapping results (2000 replicates). Trees are drawn to scale as indicated by the bar depicted below each tree; bars represent the estimated distance in units of the number of base substitutions per site. The scale the 16S rRNA-based tree is expanded relative to other loci. L. biflexa was used as the outgroup.
The repeated findings that placed L. inadai strain H6 within the L. interrogans cluster, suggested that this strain is probably misclassified and belongs to L. interrogans. To rule out that an incorrect strain was used in our study, we repeated the sequence analysis with an H6 strain originating from the CDC collection used to establish the current taxonomic description of Leptospira [2]. Results with the CDC H6 reference strain were identical with results obtained with our strain excluding an error in our collection.
The S10-spc-α locus encodes ribosomal proteins that interact with rRNA, therefore ribosomal protein and rRNA sequences are expected to have parallel phylogenies. Because rrs is a well-accepted target for phylogenetic analysis we constructed a phylogenetic tree from available rrs sequence data. The rrs based phylogenetic tree is similar to the locus-deduced tree, showing close relationships between the species L. interrogans, L. kirschneri and L. noguchii. Both the clade support and genetic divergence among other Leptospira species based on rrs sequence data was lower than for S10-spc-α locus data alone, a finding consistent with a slower rate of sequence drift in rRNA than ribosomal protein genes.
Phylogeny of secY versus its G1–G2 domain
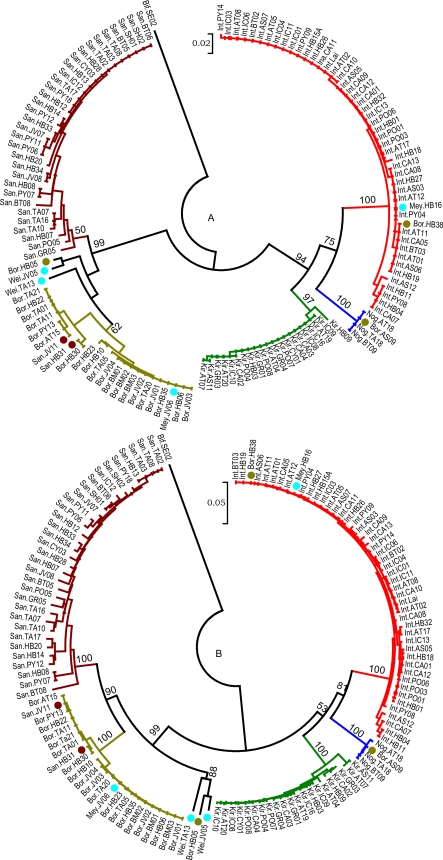
The 20-mer primers G1 and G2 amplify a 285 bp fragment of secY, and these primers were developed previously as a diagnostic PCR for the detection of Leptospira DNA [5]. A 245 bp fragment flanked by the G1–G2 primers has been shown previously to be a useful tool for discriminating between species [18]–[21]. This study provides an opportunity to broaden the evaluation of the G1–G2 domain by comparing the discriminative value of this domain with the majority of the secY sequence. Sequences for secY were obtained from 131 Leptospira strains (GenBank accession numbers EU357938–EU358070). The phylogenetic tree produced from secY sequence data was compared to a tree derived from the extracted sequences of the 245 bp fragment flanked by primers G1 and G2 (Fig. 3). These two trees are similar, resolve Leptospira species, and discriminate between strains. With few exceptions, all strains clustered with other members of the same species as determined by DNA-DNA hybridization analysis [4]. Because of a limitation presented by the original G2/G2 primer pair, it does not amplify DNA from L. kirschneri; two new primers were designed (SecYII and SecYIV) that flank the G1 and G2 annealing sites. These primers amplify secY sequences from all pathogenic strains (data not shown).
Figure 3. Circular phylogenetic trees based in Tamura-Nei distances and elaborated using Neighbor-Joining method.
Distances were calculated from G1–G2 (A) restricted sequences or the secY sequences (B), and are based on analysis of 131 strains of pathogenic species of Leptospira. Numbers above branches represent the percentage of bootstrapping results (2000 replicates). Only bootstrap values above or equal to 50% are shown. L. biflexa was used as the outgroup. Dots indicate strains with divergent positions compared to those from DNA-DNA reassociation analysis [2].
Discussion
Whole genome sequence analyses of different Leptospira species reveal extensive plasticity, including rearrangements, duplications, and disruptions of otherwise conserved segments of the genome [10]–[12], [17]. Previously, we demonstrated that L. interrogans strain Lai contained a large ribosomal protein locus spanning the S10, spc, and α loci identified in widely divergent eubacterial genera [14]. Notably, this entire locus is transcribed from either of two promoters upstream of fus, the first gene in the operon, and comprises one of the longest known prokaryotic transcripts [14]. In the present study, we show that genetic content and organization of the S10-spc-α locus is well conserved across the genus Leptospira, a finding that is somewhat remarkable given the extent of rearrangements that have disrupted synteny during Leptospira evolution. The conserved S10-spc-α organization includes the presence of the 5′ fus gene coding for elongation factor EF-G and the genes adk (adenylate kinase), infA (IF1), and rpsD (S4) located at the 3′ end of the locus, genes that are dispersed in the B. burgdorferi and T. pallidum genomes [14]. The genetic organization of the Leptospira S10-spc-α locus is unique among spirochetes [14], and the data presented in this work support phylogenetic evidence that suggests Leptospira are one of the oldest branches in spirochete evolution. Conservation of the Leptospira S10-spc-α locus is in stark contrast to the unique organization of rRNA genes, where the rrs, rrl, and rrn genes are not closely linked to each other, but are dispersed throughout the larger of two chromosomes comprising the Leptospira genome [16], [22]. Despite a lack of synteny for the ribosomal RNA genes, rRNA genes show limited sequence divergence. Generally, rRNA sequence conservation is a consequence of low tolerance to change due to structural constraints within the ribosome and a requirement to maintain specific binding sites for ribosomal proteins [23], [24].
PCR analysis of the S10-spc-α locus showed a number of regions that were more consistently amplified than other regions (Table S3), suggesting that either this locus has undergone rearrangements or that sequence drift affected the efficiency of primers to faithfully bind template from diverse species. Alignment of genomic sequences spanning the S10-spc-α locus showed that the genetic organization of this locus is conserved among pathogenic and saprophytic Leptospira (Fig. S1). Therefore, variable success in amplifying regions of the S10-spc-α locus from diverse Leptospira species is likely due to sequence drift; Leptospira species have substantial differences in sequence composition as shown by DNA∶DNA hybridization analysis [2]. Additionally, the PCR primers were designed primarily from the available genomic sequences of two pathogenic Leptospira serovars, and our results may be biased due to the divergence between pathogenic and saprophytic species. The binary PCR data positioned Leptospira species into two clades; one clade contained only pathogenic species, while the other contained both saprophytic species and species with intermediate pathogenic potential. One important aspect of our findings is confirmation that L. fainei, L. inadai, and L. meyeri, known to present a group of Leptospira with intermediate pathogenic potential, form a distinct cluster separate from true pathogenic species, suggesting the presence of three distinct lines of evolution within this genus.
We selected four loci within the S10-spc-α locus that were consistently amplified from Leptospira species in initial studies to perform phylogeny studies. Phylogenetic trees deduced from the separate loci as well as from the concatenated sequence were similar, and resulted in trees each having two clades, results similar to those obtained from the binary PCR data. The clades contained branches that, with few exceptions, reflected species designations based on rrs sequence analysis [25], MLST analysis [26], multilocus enzyme electrophoresis (MLEE) [27], and DNA homology data [2].
Three anomalies were found during comparison of the binary and sequence-based phylogenetic trees. First, the two strains of species L. meyeri were separated into different branches. Strain ICF was positioned in the pathogenic clusters whereas Veldrat Semarang 173 appeared in the saprophytic/intermediate pathogen cluster. This is consistent with previous reports that ICF is a pathogenic strain and Veldrat Semarang 173 is a saprophytic one [5], [28]. The findings of this work imply that L. meyeri is composed of strains with different pathogenic potential. A second anomaly detected in this work affects the classification of strain H6. Strain H6 was designated a member of L. inadai based on DNA∶DNA reassociation analysis [2], but MLEE data contradicted this finding [27]. To exclude the possibility that the discrepancy in previous studies, and in our work, was due to contamination, we analyzed strain H6 from both our collections and the reference collection at CDC used to develop the current species designations using DNA hybridization data [2], and found both strains had identical sequences to L. interrogans. Consequently, we recommend that strain H6 be reclassified as L. interrogans. The third anomaly involves L. weilii strains Celledoni and Sarmin. These two strains are separated into separate clades in the G1–G2 sequence-based tree (Fig. 2E). However, these two strains share the same clade based on analyses using binary data or sequence data. We believe that gene duplication and recombination events might have facilitated horizontal transfer of all or part of secY (corresponding to the G1–G2 region). It should be noted that genes contained in the S10-spc-α locus are duplicated in L. borgpetersenii strain L550, but are found as unique copy genes in all other sequenced Leptospira genomes, including L. borgpetersenii strain JB197 [10]. Thus, duplication of this locus and subsequent DNA acquisition via horizontal genetic transfer could facilitate stable integration of divergent secY genes.
The S10-spc-α locus includes the secY gene encoding preprotein translocase. Primer pair G1–G2 is positioned within this gene and directs amplification of a 285 bp fragment from all pathogenic species except L. kirschneri [5]. Although it has been suggested that this small fragment has a high discriminating power making it useful for a quick speciation [18]–[21], data supporting that contention is fragmentary. One goal of this study was to determine if analysis of the G1–G2 region provided sufficient information for Leptospira spp. discrimination. Data generated in the present study is more comprehensive than previous reports; phylogenetic trees based on the G1–G2 segment are in accordance with rrs based trees, showing that analysis of this small fragment can be used to identify species.
Genetic analysis of the S10-spc-α locus contributes to a better understanding of Leptospira evolution. Trees generated from analysis of sequence data generated here provide analysis of more conserved loci than those studied previously [25], and may be more useful in comparing evolution of the genus. A conserved, yet distinct genetic organization of this locus provides additional support for the early divergence of Leptospira from other spirochetes. Finally, from a practical standpoint, we demonstrate that analysis of a 245 bp segment of secY is suitable for rapid identification of Leptospira species.
Materials and Methods
Bacterial strains and media
Leptospira strains used in this study were from the reference collections of the WHO/FAO/OIE Collaborating Center for Reference and Research on Leptospirosis at KIT Biomedical Research, Amsterdam, The Netherlands, and the USDA Leptospirosis Reference Center at the National Centers for Animal Health, Ames, USA (Table S1). Bacteria were propagated at 30°C in EMJH liquid media as described by Ellinghausen and McCullough [29] as modified by Johnson and Harris [30].
DNA extraction
Leptospira were grown to late log phase, harvested by centrifugation, and genomic DNA was extracted using a QIAamp DNA mini kit (Qiagen, Germany) following the manufacturer's instructions. DNA concentration was determined using a Nano-Drop-1000 spectrophotometer (ThermoFisher Scientific, USA) and by visual comparison with Smart Ladder SF (Eurogentec S.A., Belgium) after agarose gel electrophoresis in 1.5% agarose gels, stained with ethidium bromide according to standard procedures [31].
PCR analysis
Adjacent and overlapping fragments from the whole S10-spc-α locus were amplified by PCR from various Leptospira strains using primers listed in Table 1 and S2. Several primers were designed by cross-species alignment of available L. interrogans and L. borgpetersenii genome sequences [10]–[12] and access to the L. biflexa Patoc I genome sequence before publication (D. Bulach [17]). In addition, an iterative approach was used to develop primers useful for sequencing secY by identifying conserved regions suitable for amplification of adjacent variable regions across divergent species for which the genome sequences are yet unavailable. Primer sets were designed to produce a series of overlapping amplification products to ensure the presence and correct location of genes in the locus.
PCR amplifications were done on a PTC-100 Peltier Thermal Cycler (MJ Research, USA) using the following program: denaturation for 5 min. at 94°C, followed by 34 cycles consisting of annealing, 1 min at 52°C, primer extension, 2 min at 72°C, denaturation, 1 min at 94°C. PCR products were separated by agarose gel electrophoresis and visualized as described above.
Sequencing
PCR amplification products were purified by QIAquick PCR purification kits (Qiagen Corp.) prior to DNA sequencing. Nucleotide sequences were determined by dye termination reactions separated on ABI Prism 310 and ABI 3700 (Applied Biosystems, USA) DNA sequencers. Sequencing was done on both complementary and forward strands and repeated at least twice to obtain reliable sequence data. Sequence data were edited using Sequencher (Gene Codes Corp., USA).
Phylogenetic analysis: Binary Analysis of PCR data
The presence (1) or absence (0) of correctly amplified fragments within the S10-spc-α locus, for each of the analyzed species, was codified in a discrete binary 40-character matrix covering a complete set of 24 taxa, representing eleven Leptospira species (Table S1). The characters were weighted proportionally to fragment size and assumed sequence homology for fragments with identical estimated size. The data matrix was analyzed under parsimony criteria using the branch and bound algorithm; support for branches in the unrooted tree was estimated by bootstrapping (100 replicates) with the program PAUP* v. 4.0b10 [32]. The inferred phylogenetic relationships are based on both gene organization and sequence variation within the complete S10-spc-α locus. Phylogenetic signals contained in this data set were evaluated by g 1 estimation (g1 = −0.947). The negative skew of the distribution of three lengths, under parsimony criterion, is originated from trees with low scores based in highly informative data [33].
Phylogenetic analysis: Comparative Sequence Analysis
Sequence data from four loci within the S10-spc-α locus were obtained to conduct a distance and parsimony-based phylogenetic analysis of pathogenic Leptospira using MEGA4 and PAUP* v. 4.0b10, respectively. Nucleotide diversity and diverse sequence parameters were obtained with MEGA4 [34] and DNASP [35]. The hypothesis that all mutations are selectively neutral was tested using Tajima's D test [36] implemented in DNASP. The confidence limits of D (two-tailed test) was obtained assuming that D follows the beta distribution and the confidence limits given in equation 47 and Table 2, respectively in Tajima, 1989 [36]. Confidence intervals were also determined for Tajima's D by computer simulations using the coalescent algorithm. In distance analysis, midpoint rooted trees were obtained by the neighbor-joining method with Tamura-Nei distances [37], [38] and the cluster support was estimated by bootstrapping with 2000 replicates [39]. The gaps were ignored only when they are included in the two sequences compared, using the pairwise-deletion option. In parsimony analysis, a branch-and-bound search was used with 2000 bootstraps. The homogeneity of the four partitioned data sets was evaluated using the incongruence-length difference test [40] implemented in PAUP* v. 4.0b10. An initial tree was inferred from a data set that concatenated all the available sequenced fragments, i.e. data from the rplE, rpsN, rpsH (primer pair 301–300), rplB, rpsS (primer pair 621–625), rplC, rplD (primer pair 624–650) and secY (primer pairs G1–G2) (Table 1). This data set represents 1684 bp from each of nine Leptospira species and 19 representative strains, including L. biflexa strain Patoc I as an outgroup. Concurrently, sequence data from each of the individual fragments used in the concatenated set were analyzed separately using identical analysis methodology, to search for topological local incongruence responsible for the low support of particular nodes in the initial tree. In addition, two secY fragments (spanning the G1–G2 and SecY II–IV primer sets, respectively) were used for the reconstruction of phylogenetic relationships between 131 strains, using L. biflexa strain Patoc I as an outgroup. The secY sequences were stripped to a standard size of 1289 bp, whereas the sequences derived from G1–G2 were significantly shorter: 245 bp. Using this latter fragment, a comparison of monophyletic clustering and resolution of Leptospira species respect to the standard 16S rDNA data was done.
ACT Alignment
Alignment of S10-spc-α locus sequences from L. interrogans, L. borgpetersenii, and L. biflexa was done using BLASTN [41] with settings adjusted to identify regions having ≥80% sequence identity. Data were visualized using the Artemis Comparison Tool (ACT) [42].
Supporting Information
Alignment of the L. biflexa, L. interrogans and L. borgpetersenii S10-spc-a genome sequences. Regions of greater than 80% sequence identity are shown as blue (between L. interrogans and L. borgpetersenii) and red (L. biflexa and L. interrogans). White regions indicate segments where sequence identity drops below 80%. Regions of similarity were determined using Blastn under default settings except the -m 8 output option was used. The display was generated using ACT. Note that the orientation of these sequences shown in the figure is consistent with the genomic sequence data in GenBank and are inverted relative to the direction of transcription. GenBank accession numbers for the genomes of L. interrogans, L. borgpetersenii and L. biflexa are AE016823, CP000348, CP000786, respectively.
(2.24 MB DOC)
Leptospira strains used for the S10-spc-α locus study.
(0.25 MB DOC)
All primers used for the S10-spc-α locus analysis.
(0.09 MB DOC)
Amplification through the S10-spc-α operon of Leptospira spp. Positive and negative PCR scores for amplification reactions along the locus from various strains.
(0.05 MB DOC)
Acknowledgments
We thank Marc Suchard and David Haake for their continuous support and critical reading of the manuscript, David Alt and Richard Hornsby for expert technical support, and Mark Wilson for providing Leptospira cultures.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported by EC grant ICA4-CT-2001-10086, and by the U.S. Department of Agriculture.
References
- 1.Paster BJ, Dewhirst FE, Weisburg WG, Tordoff LA, Fraser GJ, et al. Phylogenetic analysis of the spirochetes. J Bacteriol. 1991;173:6101–6109. doi: 10.1128/jb.173.19.6101-6109.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brenner DJ, Kaufmann AF, Sulzer KR, Steigerwalt AG, Rogers FC, et al. Further determination of DNA relatedness between serogroups and serovars in the family Leptospiraceae with a proposal for Leptospira alexanderi sp. nov. and four new Leptospira genomospecies. Int J Syst Bacteriol 49 Pt. 1999;2:839–858. doi: 10.1099/00207713-49-2-839. [DOI] [PubMed] [Google Scholar]
- 3.Ramadass P, Jarvis B, Corner R, Penny D, Marshall R. Genetic characterization of pathogenic Leptospira species by DNA hybridization. Int J Syst Bacteriol. 1992;42:215–219. doi: 10.1099/00207713-42-2-215. [DOI] [PubMed] [Google Scholar]
- 4.Yasuda P, Steigerwalt A, Sulzer K, Kaufmann A, Rogers F, et al. Deoxyribonucleic acid relatedness between serogroups and serovars in the family Leptospiraceae with proposals for seven new Leptospira species. Int J Syst Bacteriol. 1987;37:407–415. doi: 10.1099/00207713-49-2-839. [DOI] [PubMed] [Google Scholar]
- 5.Gravekamp C, Van de Kemp H, Franzen M, Carrington D, Schoone GJ, et al. Detection of seven species of pathogenic leptospires by PCR using two sets of primers. J Gen Microbiol. 1993;139:1691–1700. doi: 10.1099/00221287-139-8-1691. [DOI] [PubMed] [Google Scholar]
- 6.Letocart M, Baranton G, Perolat P. Rapid identification of pathogenic Leptospira species (Leptospira interrogans, L. borgpetersenii, and L. kirschneri) with species-specific DNA probes produced by arbitrarily primed PCR. J Clin Microbiol. 1997;35:248–253. doi: 10.1128/jcm.35.1.248-253.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levett PN, Morey RE, Galloway RL, Steigerwalt AG. Leptospira broomii sp. nov., isolated from humans with leptospirosis. Int J Syst Evol Microbiol. 2006;56:671–673. doi: 10.1099/ijs.0.63783-0. [DOI] [PubMed] [Google Scholar]
- 8.Slack AT, Symonds ML, Dohnt MF, Smythe LD. Identification of pathogenic Leptospira species by conventional or real-time PCR and sequencing of the DNA gyrase subunit B encoding gene. BMC Microbiol. 2006;6:95. doi: 10.1186/1471-2180-6-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perolat P, Chappel RJ, Adler B, Baranton G, Bulach DM, et al. Leptospira fainei sp. nov., isolated from pigs in Australia. Int J Syst Bacteriol. 1998;48 Pt 3:851–858. doi: 10.1099/00207713-48-3-851. [DOI] [PubMed] [Google Scholar]
- 10.Bulach DM, Zuerner RL, Wilson P, Seemann T, McGrath A, et al. Genome reduction in Leptospira borgpetersenii reflects limited transmission potential. Proc Natl Acad Sci USA. 2006;103:14560–14565. doi: 10.1073/pnas.0603979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nascimento AL, Ko AI, Martins EA, Monteiro-Vitorello CB, Ho PL, et al. Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J Bacteriol. 2004;186:2164–2172. doi: 10.1128/JB.186.7.2164-2172.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren SX, Fu G, Jiang XG, Zeng R, Miao YG, et al. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature. 2003;422:888–893. doi: 10.1038/nature01597. [DOI] [PubMed] [Google Scholar]
- 13.Faine S, Adler B, Bolin C, Perolat P. Melbourne, Australia: MediSci; 1999. Leptospira and Leptospirosis, 2nd ed. p. 272. [Google Scholar]
- 14.Zuerner RL, Hartskeerl RA, van de Kemp H, Bal AE. Characterization of the Leptospira interrogans S10-spc-alpha operon. FEMS Microbiol Lett. 2000;182:303–308. doi: 10.1111/j.1574-6968.2000.tb08912.x. [DOI] [PubMed] [Google Scholar]
- 15.Watanabe H, Mori H, Itoh T, Gojobori T. Genome plasticity as a paradigm of eubacteria evolution. J Mol Evol. 1997;44(Suppl 1):S57–64. doi: 10.1007/pl00000052. [DOI] [PubMed] [Google Scholar]
- 16.Zuerner RL, Herrmann JL, Saint Girons I. Comparison of genetic maps for two Leptospira interrogans serovars provides evidence for two chromosomes and intraspecies heterogeneity. J Bacteriol. 1993;175:5445–5451. doi: 10.1128/jb.175.17.5445-5451.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Picardeau M, Bulach DM, Bouchier C, Zuerner RL, Zidane N, et al. Genome Sequence of the Saprophyte Leptospira biflexa Provides Insights into the Evolution of Leptospira and the Pathogenesis of Leptospirosis. PLoS ONE. 2008;3:e1607. doi: 10.1371/journal.pone.0001607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De los A Valverede MA, Raminez JM, Montes de Octa LG, Goris MGA, Ahmed N. Arenal, a new Leptospira serovar of serogroup Javanica, isolated from a patient in Costa Rica. Infection, Genetics and Evolution. 2008 doi: 10.1016/j.meegid.2008.02.008. In press. [DOI] [PubMed] [Google Scholar]
- 19.Oliveira MA, Caballero OL, Vago AR, Harskeerl RA, Romanha AJ, et al. Low-stringency single specific primer PCR for identification of Leptospira. J Med Microbiol. 2003;52:127–135. doi: 10.1099/jmm.0.04923-0. [DOI] [PubMed] [Google Scholar]
- 20.Priya CG, Hoogendijk KT, Berg MVD, Rathinam SR, Ahmed A, et al. Field rats form a major infection source of leptospirosis in and around Madurai, India. J Postgrad Med. 2007;53:236–240. doi: 10.4103/0022-3859.37511. [DOI] [PubMed] [Google Scholar]
- 21.Rossetti CA, Liem M, Samartino LE, Hartskeerl RA. Buenos Aires, a new Leptospira serovar of serogroup Djasiman, isolated from an aborted dog fetus in Argentina. Vet Microbiol. 2005;107:241–248. doi: 10.1016/j.vetmic.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 22.Baril C, Herrmann JL, Richaud C, Margarita D, Girons IS. Scattering of the rRNA genes on the physical map of the circular chromosome of Leptospira interrogans serovar icterohaemorrhagiae. J Bacteriol. 1992;174:7566–7571. doi: 10.1128/jb.174.23.7566-7571.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El-Baradi TTAL, de Regt VCHF, Einerhand SWC, Teixido J, Planta RJ, et al. Ribosomal Proteins EL11 from Escherichia coli and L15 from Saccharomyces cerevisiae bind to the same site in both yeast 26S and Mouse 28S rRNA. J Mol Biol. 1987;195:909–917. doi: 10.1016/0022-2836(87)90494-3. [DOI] [PubMed] [Google Scholar]
- 24.Gourse RL, Thurlow DL, Gerbi SA, Zimmermann RA. Specific binding of a prokaryotic ribosomal protein to a eukaryotic ribosomal RNA: implications for evolution and autoregulation. Proc Natl Acad Sci USA. 1981;78:2722–2726. doi: 10.1073/pnas.78.5.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haake DA, Suchard MA, Kelley MM, Dundoo M, Alt DP, et al. Molecular evolution and mosaicism of leptospiral outer membrane proteins involves horizontal DNA transfer. J Bacteriol. 2004;186:2818–2828. doi: 10.1128/JB.186.9.2818-2828.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahmed N, Devi SM, Valverde M, Vijayachari P, Machang'u RS, et al. Multilocus sequence typing method for identification and genotypic classification of pathogenic Leptospira species. Ann Clin Microbiol Antimicrob. 2006;5:28. doi: 10.1186/1476-0711-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Letocart M, Boerlin P, Boerlin-Petzold F, Goudet J, Baranton G, et al. Genetic structure of the genus Leptospira by multilocus enzyme electrophoresis. Int J Syst Bacteriol. 1999;49:231–238. [Google Scholar]
- 28.Kositanont U, Rugsasuk S, Leelaporn A, Phulsuksombati D, Tantitanawat S, et al. Detection and differentiation between pathogenic and saprophytic Leptospira spp. by multiplex polymerase chain reaction. Diagn Microbiol Infect Dis. 2007;57:117–122. doi: 10.1016/j.diagmicrobio.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 29.Ellinghausen HC, Jr, McCullough WG. Nutrition of Leptospira Pomona and Growth of 13 Other Serotypes: Fractionation of Oleic Albumin Complex and a Medium of Bovine Albumin and Polysorbate 80. Am J Vet Res. 1965;26:45–51. [PubMed] [Google Scholar]
- 30.Johnson RC, Harris VG. Differentiation of pathogenic and saprophytic leptospires. I. Growth at low temperatures. J Bacteriol. 1967;94:27–31. doi: 10.1128/jb.94.1.27-31.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maniatis T, Fritsch EF, Sambrook J. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1983. Molecular Cloning: A Laboratory Manual. p. 545. [Google Scholar]
- 32.Swofford DL. Sunderland, MA: Sinauer Associates, Inc; 1998. PAUP*: Phylogenetic Analysis Using Parsimony (and Other Methods) 4.0 Beta. [Google Scholar]
- 33.Hillis DM, Huelsenbeck JP. Signal, noise, and reliability in molecular phylogenetic analyses. J Hered. 1992;83:189–195. doi: 10.1093/oxfordjournals.jhered.a111190. [DOI] [PubMed] [Google Scholar]
- 34.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 35.Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- 36.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 38.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 39.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 40.Farris JS, Källersjö M, Kluge AG, Bult C. Constructing a significance test for incongruence. Syst Biol. 1995;44:570–572. [Google Scholar]
- 41.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 42.Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, et al. ACT: the Artemis Comparison Tool. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of the L. biflexa, L. interrogans and L. borgpetersenii S10-spc-a genome sequences. Regions of greater than 80% sequence identity are shown as blue (between L. interrogans and L. borgpetersenii) and red (L. biflexa and L. interrogans). White regions indicate segments where sequence identity drops below 80%. Regions of similarity were determined using Blastn under default settings except the -m 8 output option was used. The display was generated using ACT. Note that the orientation of these sequences shown in the figure is consistent with the genomic sequence data in GenBank and are inverted relative to the direction of transcription. GenBank accession numbers for the genomes of L. interrogans, L. borgpetersenii and L. biflexa are AE016823, CP000348, CP000786, respectively.
(2.24 MB DOC)
Leptospira strains used for the S10-spc-α locus study.
(0.25 MB DOC)
All primers used for the S10-spc-α locus analysis.
(0.09 MB DOC)
Amplification through the S10-spc-α operon of Leptospira spp. Positive and negative PCR scores for amplification reactions along the locus from various strains.
(0.05 MB DOC)