Abstract
Most diseases are the consequence of the breakdown of cellular processes, but the relationships among genetic/epigenetic defects, the molecular interaction networks underlying them, and the disease phenotypes remain poorly understood. To gain insights into such relationships, here we constructed a bipartite human disease association network in which nodes are diseases and two diseases are linked if mutated enzymes associated with them catalyze adjacent metabolic reactions. We find that connected disease pairs display higher correlated reaction flux rate, corresponding enzyme-encoding gene coexpression, and higher comorbidity than those that have no metabolic link between them. Furthermore, the more connected a disease is to other diseases, the higher is its prevalence and associated mortality rate. The network topology-based approach also helps to uncover potential mechanisms that contribute to their shared pathophysiology. Thus, the structure and modeled function of the human metabolic network can provide insights into disease comorbidity, with potentially important consequences for disease diagnosis and prevention.
An important challenge of contemporary biology and medicine is to establish the relationship between disease phenotypes and disruptions in the underlying cellular functions (1–8). In the past decades, huge efforts have been devoted to a gene-based approach, identifying the specific genetic defects that, together with single nucleotide polymorphisms and copy number variations, predispose an individual to disease. However, it is increasingly understood that such an approach, although enormously successful, is far from sufficient, especially because most cellular components exert their function through intricate networks of regulatory, metabolic, and protein interactions (9–14). Therefore, the impact of different (and often disease-causing) genetic and epigenetic variations are not restricted but may spread in the intracellular network, affecting the activity and/or function of gene products that otherwise carry no defects. Because of these complex interdependencies among a cell's molecular components, the possibility of deep functional and causal relationships between apparently distinct disease phenotypes is apparent. Indeed, certain diseases, such as diabetes and obesity, or Gaucher disease and Parkinson disease, often cooccur in the same individual, sometimes one being considered a significant risk factor for the other.
From this perspective, metabolism-related diseases are of special interest because high-quality molecular interaction maps exist for human cell metabolism (15, 16), providing strict flux-based dependencies between reactions processing the same metabolite (17), and earlier attempts to uncover disease dependencies based on shared genes have been shown to be inefficient in grouping metabolic diseases (18). Classical metabolic diseases are associated with mutations that cause a metabolic enzyme to be nonexpressed, inactive, or functionally compromised. The disease phenotypes themselves are usually the consequence of the cell's inability to break down a metabolic substrate that is toxic above a threshold concentration or to produce a substrate that is essential for the cell's normal function. Indeed, metabolic disorders are often classified by the phenotype or the particular type of building block that is affected, with broad categories including disorders of carbohydrate, amino acid, or fatty acid metabolism, organic acidemias, lysosomal storage diseases, and so on. However, the effect of these enzyme defects (especially in their more subtle forms) may not always be confined to the metabolic reactions they catalyze. Cellular metabolism represents the integrated interconversion of thousands of metabolic substrates through enzyme-catalyzed biochemical reactions (16, 19–21). Thus, sets of consecutive reactions are functionally interrelated, and their activities (i.e. their flux rates) to a certain degree synchronized. A consequence of this interdependency is that an enzyme defect that leads to a failure at one reaction may potentially affect the fluxes of one or several subsequent reactions (22). Importantly, such cascading effects may also couple the metabolic diseases that are associated with subsequent reactions, resulting in comorbidity effects (meaning that diverse disease phenotypes are coexpressed). Therefore, the association between diseases and specific biochemical reactions may lead to direct connections between diseases associated with the same pathway. See supporting information (SI) Fig. S1 for information on disease–reaction relationships.
Here, we aim to test to what degree the known metabolic network-based coupling between diseases associated with enzymes is amplified in the human population, emerging as detectable comorbidity effects between diseases. We construct a metabolic disease network in which two disorders are connected if they are linked to potentially correlated reactions. We then test the validity of the proposed metabolic links between diseases by examining mRNA level correlations among their enzymes. We also explore to what degree the predicted relationships between often distinct phenotypes result in detectable comorbidity patterns in patients. Our results demonstrate that the predicted links among diseases are frequently observed in patients and that the underlying disease network offers insights into the factors contributing to the mortality rate of diseases.
Results
Construction of the Cell Metabolism-Based Human Disease Network.
As a starting point of our analysis, we used the Kyoto Encyclopedia of Genes and Genomes (KEGG) Ligand database (15) and a database of biochemically, genetically and genomically structured genome-scale metabolic network reconstructions (BiGG) (16), each representing a manually curated list of metabolic reactions in a generic human cell and the enzymes catalyzing them. We used the list of disorder–gene association pairs available in the Online Mendelian Inheritance in Man (OMIM) database (23) to identify the disorders associated with each of the enzymes present in the human metabolic network (Fig. 1a), finding that in the KEGG (BiGG) database 737 (1,116) among the total of 1,493 (3,742) metabolic reactions are associated with at least one disease. Similarly, 337 (378) among the 1,437 distinct disorders identified in OMIM are related to at least one metabolic reaction in KEGG (BiGG).
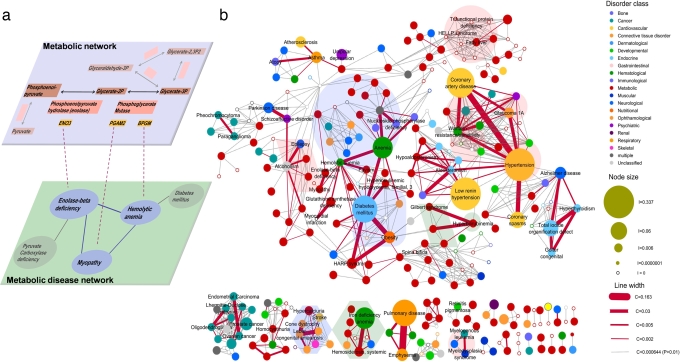
Fig. 1.
MDN. (a) Construction of the MDN. (Upper) A local region of the glycolysis, where the catalytic enzymes are shown with red background and their corresponding genes are shown with orange background. (Lower) A local neighborhood of the metabolic diseases (blue) associated with the shown reactions. The gene ENO3 encodes the enzyme catalyzing the conversion between phosphoenolpyruvate and glycerate-2P, and its mutation is involved in the development of enolase-β deficiency. The gene products of PGAM2 and BPGM, catalyzing the reaction involving glycerate-2P and glycerate-3P, are connected to myopathy and hemolytic anemia. Then the two diseases are not only connected with each other but also linked to enolase-β deficiency due to the adjacency of their associated reactions. (b) In the network representation, 308 nonisolated diseases (nodes) are connected by 878 metabolic links combining the potential links predicted by KEGG and OMIM reconstructions. The color of the nodes indicates the disease class (see SI Text and Dataset S1), and node size is proportional to the prevalence of each disease in the Medicare dataset. The width of the link between diseases is proportional to the comorbidity C of the two connected diseases. We show with red the links with significant (P < 0.01) comorbidity. Clusters of diseases associated with purine metabolism (blue shading), fatty acid metabolism (red shading), and porphyrin metabolism (green shading) are shown.
If the same substrate is shared between two metabolic reactions, the scarcity or abundance of that substrate may affect the fluxes of both reactions, potentially coupling their activity. For example, in Fig. 1a, if the phosphoglycerate mutase is not active, the production (or consumption) of glycerate-2P, and in turn of phosphoenolpyruvate, is expected to also be altered. In the following, we consider two metabolic reactions linked if they process a common metabolite, i.e. if they are adjacent to each other in a metabolic reaction map (see SI Text, Dataset S1, Dataset S2 and Dataset S3).
The altered activity of some metabolic enzymes is known to be associated with specific disorders. For example, mutations in the ENO3 gene (that encodes the enolase enzyme) are known to cause enolase-β deficiency, an autosomal recessive disorder characterized by muscle weakness and fatigability. Similarly, mutations in the BPGM gene (encoding one isoform of the phosphoglycerate mutase enzyme) can lead to hemolytic anemia. Our hypothesis is that, given that the two diseases can result from metabolic defects affecting coupled reactions, linked by glycerate-2P (Fig. 1a), their pathogenesis may also be related. That is, we hypothesize that the occurrence of one of the two diseases in a patient may enhance the likelihood of developing the other disease phenotype as well. The sum of all such cell metabolism-based links among disease phenotypes can be represented as a metabolism-based human disease network, hereafter referred to as metabolic disease network (MDN). In the MDN, each node corresponds to a disease and two diseases are connected if the metabolic reactions they are associated with are adjacent, suggesting that their fluxes may be coupled.
Characterizing the MDN.
The complete MDN is shown in Fig. 1b. The network has a large disease cluster, often called the giant component, in network theory (11, 24–26) and several smaller ones. The giant component includes 197 disorders of various disease classes, such as diabetes mellitus, obesity, Parkinson disease, asthma, unipolar depression, hypertension, and coronary artery diseases. The observed clustering of the MDN mirrors the existence of the distinct metabolic pathways. To illustrate this, in Fig. 1b, we highlighted with background colors the diseases associated with some of the better known pathways. For example, according to KEGG, human purine metabolism consists of 62 reactions associated with 33 diseases including congenital dyserythropoietic anemia and nucleoside phosphorylase deficiency. These diseases form a visually distinct cluster, highlighted with blue shading in Fig. 1b. Fatty acid metabolism, containing 34 reactions and 34 associated diseases, such as trifunctional protein deficiency and syndrome of hemolysis, elevated liver enzymes, and low platelet count (HELLP) appears again as a highly interlinked group (pink shading in Fig. 1b).
The statistical characteristics of the MDN are shown in Fig. S2. We find that on average, a disease is connected to about five other diseases and that the degree distribution is much broader than that of a random network with the same number of nodes and links, indicating that there are considerable differences among the metabolism-based relatedness of various diseases. For example, some diseases, like hypertension, warfarin resistance/sensitivity, and hemolytic anemia, act as “hubs” (11, 24, 27), with links to 27, 19, and 17 other diseases, respectively. In contrast, the majority of diseases have links only to few other diseases (see Fig 1b, and Figs. S3 and S4). To a degree, this is expected because the studied disease phenotypes span a wide range of conditions, from simple Mendelian disorders, such as enolase-β deficiency (caused by deficiency of a single enzyme), to highly heterogeneous complex diseases, such as hypertension and diabetes (for which a fraction of the genetic contribution is in the form of susceptibility alleles that are neither necessary nor sufficient to cause the disease).
Gene Expression and Flux Coupling-Based Functional Relationships Among Disease Genes.
To examine the functional relevance of the MDN, next we explored to what degree the predicted links between metabolic diseases and the associated enzymes represent detectable functional relationships. By using published microarray data for gene expression in 36 normal human tissues (28), we computed the Pearson correlation coefficient (PCC) between the expression profiles of each pair of genes in the metabolic network. We find that the average coexpression of gene pairs connected by metabolic links is higher than the coexpression between genes for which no such metabolic link is known (29) (Fig. 2b and Fig. S5) with P < 10−8. For example, the genes ENO3 and PGAM2 (Fig. 1a) have a PCC = 0.66 with P < 10−5, a 7-fold increase over the average expectation.
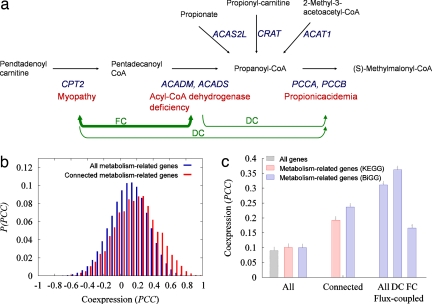
Fig. 2.
Flux coupling and coexpression of metabolic genes. (a) To illustrate the use of flux-coupling analysis, we show the reactions that display directional coupling (DC) with the reaction converting propanoyl–CoA to (S)-methylmalonyl–CoA. In blue, we indicate the genes encoding the corresponding enzymes, and in red, we indicate the associated diseases. The production (consumption) of pentadecanoyl–CoA is performed by a single reaction, catalyzed by CPT2 (ACADM, ACADS), and therefore the ratio of their fluxes should be a constant (full coupling; FC). On the contrary, propanoyl–CoA may be produced by four reactions and is consumed by only one reaction. Therefore a nonzero flux of any of those four reactions implies a nonzero flux of the reaction consuming propanoyl–CoA, but the opposite is not the case, which is DC. Because of the FC between the reactions producing and consuming pentadecanoyl–CoA, the reaction (CPT2) has DC also with the reaction (PCCA, PCCB). (b) Distribution of the PCC for all pairs of metabolism-related genes and for the pairs of genes connected by metabolic links based on the KEGG database. (c) Average PCC for all pairs of genes, all pairs of metabolism-related genes, genes connected by metabolic links, and genes associated with flux-coupled reactions displaying DC or FC. The coexpression is stronger for connected genes and significantly higher for flux-coupled genes.
The causal relationship among diseases may not be limited to those associated with adjacent reactions but could extend to disease pairs that are associated via reactions whose fluxes are coupled (22, 30, 31). By using the flux coupling finder methodology (22, 30–32), we identified two types of coupling between pairs of reactions i and j: (i) directional coupling (i → j), if a nonzero flux for i implies a nonzero flux for j but not necessarily the reverse; or (ii) full coupling (i ⇔ j), if a nonzero flux for i implies not only a nonzero but also a fixed flux for j and vice versa (31) (Fig. 2a). For the BiGG reconstruction, we identified 2,605 gene pairs catalyzing flux-coupled reactions. The average coexpression (PCC) of the flux coupled genes is 0.31, higher than 0.24 found for the genes catalyzing adjacent reactions and significantly higher than PCC = 0.10 characterizing all gene pairs (Fig. 2c). We also find that reactions connected by directional coupling show a significantly higher PCC (0.36) than those fully coupled (PCC = 0.17) (Fig. 2c). Taken together, these results confirm the existence of functional links between adjacent and flux-coupled reactions, suggesting the significance of these links for the coexistence of the related diseases in humans.
Comorbidity Analysis.
Disease pathobiologies originate from a full or partial breakdown of physiological cellular processes together with subsequent (often compensatory) interactions among components of the genome, proteome, metabolome, and the environment. Therefore, the affected metabolic network activity is likely to contribute to disease progression and comorbidity on the cell, organ, and organismal level.
To examine whether the links in the MDN predict disease cooccurrences, we analyzed the Medicare records of 13,039,018 elderly patients in the United States who, over the period 1990–1993, had a total of 32,341,348 hospital visits. These records are highly complete and accurate and are frequently used for epidemiological and demographic research (33, 34). The present sample was abstracted from a complete set of all hospital visits of all elderly patients (aged 65–113) in the Medicare program, which is 96% of all elderly Americans. The sample of 13 million hospitalized patients has a mean age of 76.5 ± 7.5; 41.7% were male, and 90.1% were Caucasian (Fig. S6). Most patients were diagnosed with several diseases during the observation period, a cooccurrence that in some cases is accidental but is also often causal, i.e. one disease increases the likelihood of the development of other diseases(C.A. Hidalgo, N. Blumm, A.-L.B., and N.A.C., unpublished data; 36), perhaps in part because of causal effects rooted in the metabolic network-based links among the cellular components implicated in the particular disease.
To test whether the links of the MDN can be detected in the population as significant cooccurrences between metabolically linked diseases, for each pair of diseases X and Y, we computed the comorbidity index (CXY, SI Text), which captures to what degree the two diseases cooccur in the same group of patients. A positive comorbidity indicates that patients with disease X are likely to develop disease Y as well, whereas a negative comorbidity indicates a potential protective effect from a disease Y in a patient with disease X. We prepared a hand-curated mapping of the ICD-9-CM codes based on the genetic disorders in OMIM by using an expert coder and standard coding procedures implemented in hospitals for assigning ICD-9-CM codes to prose descriptions of diseases (e.g. converting “diabetes” to ICD-9-CM code 250), thus allowing us to compute the comorbidity of each pair of diseases CXY in the MDN, where X and Y are indices for the 337 diseases associated with KEGG and the 378 diseases associated with BiGG.
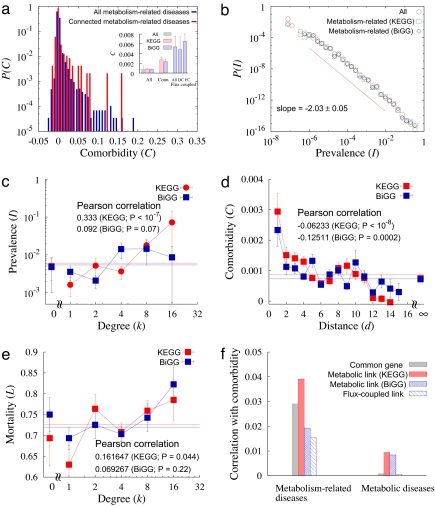
The overall tendency of the diseases to cooccur is supported by the right-skewed comorbidity distribution (Fig. 3a and Fig. S5), implying that in general, metabolically connected diseases show a higher than average comorbidity. The average comorbidity for all diseases is 0.0009 (0.0008) for the KEGG (BiGG) reconstruction, in contrast with metabolically connected pairs of diseases for which the average comorbidity is 0.0027 (0.0023), three times larger than the average for all diseases (P < 10−8). Furthermore, the average comorbidity of the diseases associated with the reactions whose fluxes are fully (directionally) coupled is 0.0062 (0.0041), ≈7 (5) times larger than the average for all diseases. In general, we find that 17% (16%) of all metabolic disease pairs for the KEGG (BiGG) reconstruction show significant (P < 0.01) comorbidity. This fraction is elevated to 31% (28%) for the disease pairs connected by a metabolic link and 28% for the flux-coupled diseases according to the KEGG (BiGG) reconstruction, a highly significant enhancement with P < 10−8.
Fig. 3.
Comorbidity and the human MDN. (a) Comorbidity distributions for all pairs of metabolism-related diseases and for connected diseases. (Inset) The average comorbidities. (b) Distribution of the prevalence of metabolism-related diseases, well approximated by a power–law with exponent −2.03 ± 0.05 (see red line). (c) Prevalence as a function of the degree of the disease in the MDN. The prevalence increases with the degree with the PCC 0.333 for KEGG database and 0.092 for BiGG database with P values <10−7 and ≈0.07, respectively. (d) Comorbidity as a function of the distance between diseases in the MDN, decreasing as the distance increases. The PCCs are −0.06233 and −0.12511 for the KEGG and BiGG databases, respectively, and the P values are <10−8 for KEGG and ≈0.0002 for BiGG database. (e) Mortality as a function of disease degree in the MDN. The mortality increases with the degree with the PCC 0.162 for KEGG database and 0.0693 for BiGG database with P values 0.044 and 0.22, respectively. (f) Correlation of potential disease comorbidity factors with disease comorbidity. PCCs between the presence of common associated genes, of metabolic links, and of flux-coupled links, with disease comorbidity are presented for metabolism-related diseases and classical metabolic diseases.
We also identified the prevalence IX of each disease, defined as the fraction of the patients having disease X (Fig. 1b). The prevalence distribution is well approximated by a power–law with exponent −2.0 (Fig. 3b), indicating that although the vast majority of diseases are rare, a few affect a significant fraction of the examined patient population. Hypertension is one of the most prevalent diseases with prevalence 0.337 followed by coronary artery disease (0.246), diabetes mellitus (0.167), and pulmonary disease (0.147). Given this broad distribution of prevalence (Fig. 3b), it is plausible that the more links a disease has to other diseases in the MDN, the higher its prevalence is, given the increased likelihood that it will be induced by other diseases in the network. Therefore, we measured the correlation between the prevalence and the degree of connectivity of each disease in the MDN (Fig. 1b), finding that the average value of the disease prevalence (Dataset S4) increases with the degree (PCC is 0.333 for KEGG, P < 10−7, Fig. 3c). Thus, the more connected a disease is in the MDN, the higher the likelihood that it may contribute to the emergence of other diseases.
We next examined whether the comorbidity effects are limited to adjacent reactions or whether comorbidity relationships also can be discerned spreading over longer distances in the MDN (i.e. if disease X is linked to disease Y, which in turn is linked to disease Z, can one expect comorbidity between X and Z?). To address this question, we define the network distance between two diseases as the length (number of links) of the shortest reaction pathway connecting them within the MDN, a metric often used in network theory (10, 11, 24, 25). We find that the PCC between the network distance and comorbidity is −0.062 (−0.13) with P < 10−8 (P < 0.0002) for KEGG (BiGG), indicating that the comorbidity of two diseases decreases as their network distance in the MDN (Fig. 3d). This finding suggests that although the direct or local links are the most relevant for average comorbidity, measurable effects persist up to three links, leading to a potential clustering of diseases discerned in the comorbidity relationships. We also found that reactions associated with diseases are active in more than one tissue (Figs. S13 and S14). In particular, ≈27% (12%) of the reaction pairs associated with diseases displaying significant comorbidity are active in all tissues, from the KEGG (BiGG) database, suggesting that the reactions associated with diseases are located in the core of the metabolic network (37).
The widely different connectivities of various diseases (Fig. 1b) prompted us to ask whether the more connected diseases are associated with higher mortality rates (deaths) than the less connected ones. Therefore, we quantified the mortality rate associated with each disease, defined as the percentage of all elderly people who died in an 8-year period after the diagnosis with the particular disease. We find that the connectivity of a disease to other diseases in the MDN and its associated mortality rate display a PCC 0.16 (0.07) in the KEGG (BiGG) database (Fig. 3e). A potential explanation for this is that a patient diagnosed with a hub disease is very likely to also develop the diseases connected to it, whether they are diagnosed or not, and they together elevate the mortality of the hub disease.
Previous work has indicated that although most diseases can be grouped into a human disease network based on the genes the diseases share, metabolic diseases are the most disconnected class in this network (18). The main hypothesis behind the present work is that the potential relatedness of metabolic diseases is better predicted by the shared metabolites and correlated metabolic reactions than by shared genes. Therefore, we next tested whether metabolic links indeed offer a better measure of functional relatedness than shared genes by using multivariate analysis to quantify the contribution to comorbidity of the various potential links between diseases, distinguishing shared genes, metabolic links, or flux-coupled links. We find that when considering all diseases linked to metabolic enzymes (i.e., all nodes in Fig. 1b), the strongest comorbidity effects are predicted by the metabolic links in the KEGG database followed closely by shared genes (Fig. 3f). However, many diseases in Fig. 1b are not classical metabolic diseases but are related to metabolic diseases through multifunctional enzymes (6). To correct for those effects, we repeated the analysis for only diseases that are classified as metabolic diseases in the medical literature (shown as red symbols in Fig. 1b). For these, we find that the strongest predictors of comorbidity are the metabolic links, representing an equally strong effect in the KEGG and BiGG databases (Fig. 3f). In contrast, shared genes and, surprisingly, flux coupled enzymes offer a negligible predictive power. This result supports our initial hypothesis that for metabolic diseases, coupled metabolic reactions offer the best predictors for disease relatedness.
MDN-Predicted Significant Comorbidity Effects Between Diseases.
The MDN-based methodology allowed us to uncover 193 pairs of diseases that are metabolically linked according to either the KEGG or the BiGG dataset and also show significant comorbidity. The full list is provided in Dataset S5), and the subset of diseases connected in both datasets and showing the highest level of comorbidity is shown in Table S1. Among the pairs of diseases having high gene coexpression and high comorbidity are diabetes and obesity, a well known comorbidity relationship (38), but less obvious pairs, such as glutathione synthetase deficiency and myocardial infarction, are also apparent.
We also find that a detailed analysis of individual disease pairs can help to understand the way by which disturbance in the underlying metabolic network may contribute to shared pathophysiology and suggest other potential disease-modifying factors. For example, diabetes mellitus and hemolytic anemia show higher than expected comorbidity (Table S1); in our database, we find 1,656 patients that are diagnosed with both diseases, in contrast with the expected 1,215 if the two diseases are to occur independently (P < 10−8). Inspecting the relationship between the genes associated with the two diseases, we find that some of the mutated genes associated with them encode enzymes catalyzing adjacent metabolic reactions (Fig. S7). Indeed, NADPH deficiency due to glucose-6-phosphate dehydrogenase deficiency causes a reduction in the levels of glutathione that is a main factor in protecting against oxidative damage. In turn, impaired glucose uptake due to glucokinase mutation may not only alter the threshold of insulin release in pancreatic β-cells but may also increase their sensitivity to oxidative damage by reducing substrate flow toward the pentose phosphate pathway (that produces NADPH). Thus, single nucleotide polymorphisms (SNPs) in the coding region of enzymes directly or indirectly affecting the redox capacity of cells (39, 40) are expected to be among the different factors that affect the phenotype and penetrance of either or both diseases (Fig. S7).
Finally, similar disease cooccurrence associations, linking the metabolic dependency and the MDN to comorbidity, can be found for many other disease pairs, such as hypertension and coronary spasm (Fig. S8), glutathione synthetase deficiency and myocardial infarction (Fig. S9), alcoholism and epilepsy (Fig. S10), and asthma and atherosclerosis (Fig. S11), together indicating the MDN based approach's utility in discovering comorbidity effects and highlighting their potential mechanisms.
Discussion
A fundamental question in biology and medicine is to what degree the topological connectivity of cellular networks is related to the manifestation of human diseases, possibly leading to phenotypic interdependencies. For example, metabolic fluxes are tightly correlated because of the stringent constraints of flux balance. These correlations are bound to have an effect on disease cooccurrences, a hypothesis that we aimed to test in the present work. We find that (i) connected diseases show higher comorbidity than those that have no metabolic link between them; (ii) the more connected a disease is in the MDN, the higher is its prevalence in the population, and the higher is the chance that it will result in death; (iii) comorbidity effects are not only pairwise but can spread in the MDN, exhibiting associations in high-order network links; and (iv) for classical metabolic diseases, the strongest predictor of comorbidity are the metabolic links and not shared genes. Taken together, these results offer strong support for the functional importance of the MDN and its relevance to human health.
We note that the completeness of the MDN is limited by our expanding knowledge of disease–gene associations. The associations used here are based on the OMIM database, which represents the best currently available depository of stringent disease–gene associations and continues to expand rapidly thanks to new technologies for identifying disease–gene associations. Such growth will likely uncover the metabolic origin of new diseases, expanding the MDN. Significant improvements are expected from more accurate reconstructions of the human metabolic network, from the appearance of cell type-specific metabolic network reconstructions, and from improvements in flux balance-based modeling approaches (17, 41). Although these improvements are essential, we do not expect to significantly alter the basic results of this study, i.e. the relevance of shared metabolic pathways to the cooccurrence of human diseases.
The finding that 31% disease pairs in the MDN show a statistically significant tendency to cooccur in the population raises an important question. Why do we not observe a significant comorbidity for the rest of disease pairs connected in the MDN? One explanation may have to do with the presence of multifunctional or moonlighting enzymes, which are enzymes with additional nonenzymatic functional activities, linking to the metabolic network diseases that do not have a metabolic origin. For example, Fig. 1b reveals that only 46% of the diseases listed in the MDN are in fact classical metabolic diseases; the rest of them may be associated with metabolism through such multifunctional enzymes. In addition to the effects induced by these moonlighting enzymes, a number of other effects, from the tissue specificity of metabolic activity (and gene expression) to the role of cofactors, may also alter the impact of the induced correlations, leading to diminished comorbidity effects. Note also that the development of most of the diseases depends on the contribution of a number of environmental (diet, drug exposure) and patholophysiological factors, and thus an impaired flux activity will often not be sufficient to initiate the disease phenotype.
Finally, our definition of diseases as individual nodes, although appropriate for monogenic disorders, leaves room for improvement for complex diseases. To check the validity of our results under more stringent conditions, we have repeated the measurements of Fig. 3 c and e for monogenic diseases only, finding that the trends reported in this paper survive for this disease class (Fig. S12). We realize, however, that many diseases are rather heterogeneous in nature. For example, diabetes can result from many abnormalities including immune dysfunction, insulin resistance, or pancreatic failure, arguing for the need to distinguish disease subphenotypes as well. The current datasets do not allow such systematic study at this moment, however. Similarly, as a proof of principle, in this paper, we focused only on the impact of metabolic links on comorbidity. In reality, a comprehensive disease network should incorporate genetic, transcriptional, regulatory, coexpression, and potential environmental links as well, offering a higher level of detail and stronger predictive power.
Despite these methodological and biological limitations, our results offer a comprehensive picture, demonstrating the importance of the large-scale structure of metabolic networks in understanding human diseases. Indeed, most functions of the human cell are carried by complex networks of genes, proteins, and metabolites interacting via biochemical and physical interactions. Thus, the impact of disease-causing mutations is often not limited to the products of the mutated gene but may spread and affect the activity of other cellular components, causing apparently unrelated disease phenotypes and resulting in comorbidity. Given the unknown environmental, lifestyle, and treatment-related factors that all contribute to comorbidity, it is not a priori evident if the metabolic network-based dependencies are strong enough to manifest themselves at the individual and population level. Our results indicate that, in fact, such metabolic dependencies lead to detectable disease comorbidity. Most important, we find that the systematic mapping of such metabolism-based links between diseases helps us uncover some critical disease comorbidities and helps to explain their pathophysiology and metabolic origin. These findings suggest that the network-based, top-down methodologies with the extant systematic large-scale databases offer an increasingly potent tool to explore and understand the interplay between cellular networks and human diseases (9, 35).
Supplementary Material
Acknowledgments.
We thank C. Hidalgo for help in compiling the Medicare patient data and Laurie Meneades for assistance with data preparation. Research at the Northeastern University and at the University of Pittsburgh was supported by National Institutes of Health (NIH) Grant A1070499-01. Research at the Northeastern University was supported by NIH Grants HG4233 (Centers of Excellence in Genomic Science) and CA113004 and at Harvard University by NIH Grant AG17548-01.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 9849.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802208105/DCSupplemental.
References
- 1.Giallourakis C, Henson C, Reich M, Xie X, Mootha VK. Disease gene discovery through integrative genomics. Annu Rev Genomics Hum Genet. 2005;6:381–406. doi: 10.1146/annurev.genom.6.080604.162234. [DOI] [PubMed] [Google Scholar]
- 2.Argmann CA, Chambon P, Auwerx J. Mouse phenogenomics: The fast track to “systems metabolism.”. Cell Metab. 2005;2:349–360. doi: 10.1016/j.cmet.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Loscalzo J, Kohane I, Barabási A-L. Human disease classification in the postgenomic era: A complex systems approach to human pathobiology. Mol Syst Biol. 2007;3:124. doi: 10.1038/msb4100163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lamb J, et al. The connectivity map: Using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 5.Lage K, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol. 2007;25:309–316. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- 6.Kann MG. Protein interactions and disease: Computational approaches to uncover the etiology of diseases. Brief Bioinform. 2007;8:333–346. doi: 10.1093/bib/bbm031. [DOI] [PubMed] [Google Scholar]
- 7.Schadt EE, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oti M, Huynen MA, Brunner HG. Phenome connections. Trends Genet. 2008;24:103–106. doi: 10.1016/j.tig.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 9.Friedman A, Perrimon N. Genetic screening for signal transduction in the era of network biology. Cell. 2007;128:225–231. doi: 10.1016/j.cell.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 10.Barabási A-L, Oltvai ZN. Network biology: Understanding the cell's functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 11.Albert R. Scale-free networks in cell biology. J Cell Sci. 2005;118:4947–4957. doi: 10.1242/jcs.02714. [DOI] [PubMed] [Google Scholar]
- 12.Almaas E. Biological impacts and context of network theory. J Exp Biol. 2007;210:1548–1558. doi: 10.1242/jeb.003731. [DOI] [PubMed] [Google Scholar]
- 13.Cusick ME, Klitgord N, Valle D, Hill D. Interactome: Gateway into systems biology. Hum Mol Genet. 2005;14:R171–R181. doi: 10.1093/hmg/ddi335. [DOI] [PubMed] [Google Scholar]
- 14.Zhu X, Gerstein M, Snyder M. Getting connected: Analysis and principles of biological networks. Genes Dev. 2007;21:1010–1024. doi: 10.1101/gad.1528707. [DOI] [PubMed] [Google Scholar]
- 15.Kanehisa M, et al. From genomics to chemical genomics. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duarte ND, et al. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci USA. 2007;104:1777–1782. doi: 10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price ND, Reed JL, Palsson BO. Genome-scale models of microbial cells: Evaluating the consequences of constraints. Nat Rev Microbiol. 2004;2:886–897. doi: 10.1038/nrmicro1023. [DOI] [PubMed] [Google Scholar]
- 18.Goh K-I, et al. The human disease network. Proc Natl Acad Sci USA. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Almaas E. Optimal flux patterns in cellular metabolic networks. Chaos. 2007;17 doi: 10.1063/1.2737828. 026107. [DOI] [PubMed] [Google Scholar]
- 20.Jeong H, Tombor B, Albert R, Oltvai ZN, Barabási A-L. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 21.Wagner A, Fell D. The small world inside large metabolic networks. R Soc London Ser B. 2001;268:1803–1810. doi: 10.1098/rspb.2001.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jamshidi N, Palsson BO. Systems biology of SNPs. Mol Syst Biol. 2006;2:38. doi: 10.1038/msb4100077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caldarelli G. Scale-Free Networks: Complex Webs in Nature and Technology. New York: Oxford Univ Press; 2007. [Google Scholar]
- 25.Caldarelli G, Vespignani A. Large Scale Structure and Dynamics of Complex Networks: From Information Technology to Finance and Natural Science. Singapore: World Scientific Publishing; 2007. [Google Scholar]
- 26.Dorogovtsev SN, Mendes JFF. Evolution of Networks. New York: Oxford Univ Press; 2003. [Google Scholar]
- 27.Barabási A-L, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- 28.Ge X, et al. Interpreting expression profiles of cancers by genome-wide survey of breadth of expression in normal tissues. Genomics. 2005;86:127–141. doi: 10.1016/j.ygeno.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 29.Kharchenko P, Church GM, Vitkup D. Expression dynamics of a cellular metabolic network. Mol Syst Biol. 2005;1 doi: 10.1038/msb4100023. 2005.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papin JA, Reed JL, Palsson BO. Hierarchical thinking in network biology: The unbiased modularization of biochemical networks. Trends Biochem Sci. 2004;29:641–647. doi: 10.1016/j.tibs.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 31.Burgard AP, Nikolaev EV, Schilling CH, Maranas CD. Flux coupling analysis of genome-scale metabolic network reconstructions. Genome Res. 2004;14:301–312. doi: 10.1101/gr.1926504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nikolaev EV, Burgard AP, Maranas CD. Elucidation and structural analysis of conserved pools for genome-scale metabolic reconstructions. Biophys J. 2005;88:37–49. doi: 10.1529/biophysj.104.043489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lauderdale D, Furner SE, Miles TP, Goldberg J. Epidemiological uses of Medicare data. Am J Epidemiol. 1993;15:319–327. doi: 10.1093/oxfordjournals.epirev.a036123. [DOI] [PubMed] [Google Scholar]
- 34.Mitchell JB, et al. Using Medicare claims for outcomes research. Med Care. 1994;32:JS38–JS51. [PubMed] [Google Scholar]
- 35.Barabási A-L. Network medicine—From obesity to the “diseasome.”. New Eng J Med. 2007;357:404–407. doi: 10.1056/NEJMe078114. [DOI] [PubMed] [Google Scholar]
- 36.Rzhetsky A, Wajngurt D, Park N, Zheng T. Probing genetic overlap among complex human phenotypes. Proc Natl Acad Sci USA. 2007;104:11694–11699. doi: 10.1073/pnas.0704820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Almaas E, Oltvai ZN, Barabási A-L. The activity reaction core and plasticity of metabolic networks. PLoS Comput Biol. 2005;1 doi: 10.1371/journal.pcbi.0010068. 0557–0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nath D, Heemels M-T, Anson L. Nature insight: Obesity and diabetes. Nature. 2006;444:839–888. [Google Scholar]
- 39.Goth L, Eaton JW. Hereditary catalase deficiencies and increased risk of diabetes. Lancet. 2000;356:1820–1821. doi: 10.1016/S0140-6736(00)03238-4. [DOI] [PubMed] [Google Scholar]
- 40.Wijk V, Solinge V. The energy-less red blood cell is lost: Erythrocyte enzyme abnormalities of glycolysis. Blood. 2005;106:4034–4042. doi: 10.1182/blood-2005-04-1622. [DOI] [PubMed] [Google Scholar]
- 41.Beg QK, et al. Intracellular crowding defines the mode and sequence of substrate uptake by Escherichia coli and constrains its metabolic activity. Proc Natl Acad Sci USA. 2007;104:12663–12668. doi: 10.1073/pnas.0609845104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.