Abstract
Genomic duplication-divergence processes are the primary source of new protein functions and thereby contribute to the evolutionary expansion of functional molecular networks. Yet, it is still unclear to what extent such duplication-divergence processes also restrict by construction the emerging properties of molecular networks, regardless of any specific cellular functions. We address this question, here, focusing on the evolution of protein–protein interaction (PPI) networks. We solve a general duplication-divergence model, based on the statistically necessary deletions of protein–protein interactions arising from stochastic duplications at various genomic scales, from single-gene to whole-genome duplications. Major evolutionary scenarios are shown to depend on two global parameters only: (i) a protein conservation index (M), which controls the evolutionary history of PPI networks, and (ii) a distinct topology index (M′) controlling their resulting structure. We then demonstrate that conserved, nondense networks, which are of prime biological relevance, are also necessarily scale-free by construction, irrespective of any evolutionary variations or fluctuations of the model parameters. It is shown to result from a fundamental linkage between individual protein conservation and network topology under general duplication-divergence evolution. By contrast, we find that conservation of network motifs with two or more proteins cannot be indefinitely preserved under general duplication-divergence evolution (independently from any network rewiring dynamics), in broad agreement with empirical evidence between phylogenetically distant species. All in all, these evolutionary constraints, inherent to duplication-divergence processes, appear to have largely controlled the overall topology and scale-dependent conservation of PPI networks, regardless of any specific biological function.
Keywords: evolutionary constraint, scale-free graph, functional motif, orthology, statistical model
The primary source of new protein functions is generally considered to originate from duplication of existing genes followed by functional divergence of their duplicate copies (1–3). In fact, duplication-divergence events have occurred and continue to occur at a wide range of genomic scales, from many independent duplications of individual genes† [10−3 fixed events per gene per million years (MY) (4)] to rare but evolutionary dramatic duplications of entire genomes [one fixed event per 100–200 MY (5)]. For instance, there have been between two and four consecutive whole-genome duplications in all major eukaryote kingdoms in the past 300–500 MY (5). This actually amounts to a more-or-less similar contribution of new genes from whole-genome duplication as from individual gene duplications [i.e., one fixed event per 100–200 MY ≃ 10−3 fixed events per gene per MY, assuming a 10% fixation rate after a whole-genome duplication with ≈10,000 genes (5)].
This succession of whole-genome duplications, together with the accumulation of individual gene duplications, must have greatly contributed to shaping the global structure of large biological networks, such as protein–protein interaction (PPI) networks, that control cellular activities. In fact, concordant empirical evidence reveals the evolutionary persistence of duplication-derived protein–protein interactions. For instance, there are clear enrichments of recent protein duplicates around common protein partners compared with randomly picked pairs of proteins (5, 6), although the fraction of proteins identified as having undergone a (recent) duplication (<200 MY) remains typically small in absolute terms, for example, 10% (4). Similarly, protein residues implicated in protein–protein interaction are generally the most conserved at the surface of proteins (7), revealing their duplication-derived origin,‡ with typically little more than one conserved binding interface per protein-binding domains.§
Ispolatov et al. (10) proposed an interesting local duplication-divergence model of PPI network evolution based on (i) the statistical deletion of individual, duplication-derived interactions and (ii) a time-linear increase in genome and PPI network sizes. Clearly, the deletion of redundant interactions arising from duplication is necessary to avoid the emergence of biologically irrelevant, densely connected PPI networks, lacking low-degree connectivities. Yet, we expect that independent local duplications and, a forteriori, partial- or whole-genome duplications all lead to exponential, not time-linear, evolutionary dynamics of PPI networks. In the long time limit, exponential dynamics should outweigh all time-linear processes that have been assumed in earlier PPI network evolution models (10–15). Models based on time-linear processes also assume that local evolutionary dynamics remain essentially frozen, as long as they are not directly affected by a local modification of the network. Yet, in reality, sequence mutations and environmental changes continue to affect the evolution of whole PPI networks, not just in the immediate surroundings of recently duplicated proteins.
In this article, we propose and asymptotically solve a general duplication-divergence model based on prevailing exponential dynamics¶ of PPI network evolution under local, partial, or global genome duplications. The only interaction changes that are considered are deletions of duplication-derived interactions. In particular, the rewiring dynamics of PPI networks by de novo creation of protein-binding interfaces (4) is neglected (10), as suggested by the empirical evidence mentioned earlier (see also Evolution of PPI network motifs). Indeed, our aim here is to establish a theoretical baseline from which other evolutionary processes beyond strict gene duplication and interaction loss events, such as shuffling of protein domains (5) or horizontal gene transfers, can then be considered.
A visual overview of the model is shown in Fig. 1 including its two main effective parameters, M and M′, that control, respectively, the evolutionary history or conservation (M) and resulting structure or topology (M′) of PPI networks under duplication-divergence evolution. In this article, we demonstrate a fundamental relation between protein conservation (M) and network topology (M′), that is, M ≤ M′, that is strictly independent from any evolutionary variations or fluctuations of the model parameters. We then discuss simple consequences in terms of evolutionary linkage between individual protein conservation and PPI network topology. The approach is also extended to outline the evolutionary statistics of small-network motifs including two or more proteins. In particular, we show that network motifs, unlike individual proteins, cannot be indefinitely conserved under general duplication-divergence evolution, regardless of any network-rewiring dynamics. Throughout the article, theoretical assumptions and results are commented on with brief discussions highlighting their biological relevance.
Fig. 1.
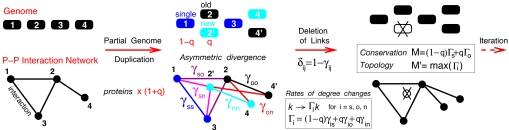
General duplication-divergence model for protein–protein interaction network evolution. Successive duplications of a fraction q of genes are followed by an asymmetric divergence of gene duplicates (e.g., 2 vs. 2′). New duplicates (n) are left essentially free to accumulate neutral mutations with the likely outcome of becoming nonfunctional and eventually deleted unless some new, duplication-derived interactions are selected; old duplicates (o), however, are more constrained to conserve old interactions already present before duplication. Interactions on the locally (q ≪ 1), partially (q < 1) or fully (q = 1) duplicated network are then preserved stochastically with different probabilities γij (0≤ γij ≤ 1, i, j = s, o, n) reflecting the recent history of each interacting partners, that are either singular, nonduplicated genes (s) or recently duplicated genes undergoing asymmetric divergence (o/n). Two effective parameters, M and M′, that depend on the rates of connectivity change, Γi, and underlying parameters q and γij, control the evolutionary history or conservation (M) and resulting structure or topology (M′) of PPI networks (see text).
Results
General Duplication-Divergence Model.
The general duplication-divergence (GDD) model is designed to capture PPI network properties caused by evolutionary constraints, inherent to duplication-divergence processes and independent of selective adaptation (3) or any specific biological function. Concretely, the GDD model analyzes the deletion statistics of protein–protein interactions that arise from stochastic duplications at various genomic scales, from single-gene to whole-genome duplications. This deletion statistics of duplication-derived interactions is indeed a necessary “background” dynamics of PPI network evolution to prevent the emergence of biologically irrelevant, densely connected PPI networks, lacking low-degree connectivities.
In practice, a fraction q of extant genes is randomly duplicated at each time step of the GDD model. The divergence of both duplicated and nonduplicated genes then leads to the stochastic deletion or conservation of their related interactions, before another round of duplication-divergence occurs (Fig. 1). In the following, we first solve the GDD model assuming that q is constant over evolutionary time scales. We then study more realistic scenarios combining, for instance, rare whole-genome duplications (q = 1) with more frequent local duplications of individual genes (q ≪ 1), and including also stochastic fluctuations in all microscopic parameters of the GDD model (see Fig. 1 and below). To analyze the deletion statistics of duplication-derived interactions, we assume that ancient and recently duplicated interactions are stochastically conserved with distinct probabilities γij's, depending only on the recently duplicated or nonduplicated state of each protein partners, as well as on the asymmetric divergence between “old” and “new” (or more “conserved” and more “divergent”) gene duplicates (5), see the Fig. 1 legend (“s” for “singular,” nonduplicated genes and “o”/“n” for old/new asymmetrically divergent duplicates). Here, we consider nonoriented PPI networks, that is, γij = γji, for i, j = s, o, n.
The first effective parameters derived from these microscopic evolutionary parameters are the average rates of connectivity change Γi (i.e., k → kΓi) for each type of node i = s, o, n, where Γi = (1 − q)γis + q(γio + γin) is independent from node connectivity k. In the following, we assume Γo ≥ Γn by definition of old and new duplicates caused by asymmetric divergence. Note that self-interacting proteins, corresponding to self-link loops, are not taken into account, for simplicity, in the main text, because they can be shown to have little effect on the asymptotic evolutionary regimes of the connectivity distribution (see SI Appendix, Fig. S3 and SI Text, for details).
We study the GDD evolutionary dynamics of PPI networks in terms of ensemble averages 〈Q(n)〉 defined as the mean value of a feature Q over all realizations of the evolutionary dynamics after n successive duplications. This does not imply, of course, that all network realizations “coexist,” but only that a random selection of them is reasonably well characterized by the theoretical ensemble average. Although it is generally not the case for exponentially growing systems, here, we can show that ensemble averages over all evolutionary dynamics indeed reflect the properties of typical network realizations for biologically relevant regimes (see Statistical Properties of GDD Models in SI Appendix).
In the following, we focus on the number of proteins (or “nodes”) Nk of connectivity k in PPI networks, while postponing the analysis of GDD models for simple network motifs to the end of the article and the SI Appendix. The total number of nodes in the network is noted N = Σk≥0 Nk and the total number of interactions (or “links”) L = Σk≥0 kNk/2. The dynamics of the ensemble averages 〈Nk(n)〉 after n duplications is analyzed by using a generating function,
The evolutionary dynamics of F(n)(x) corresponds to the following recurrence deduced from the microscopic definition of the GDD model (see SI Appendix),
where we note for i = s, o, n,
where δij = 1 − γij are deletion probabilities (i, j = s, o, n) and A′i(1) = (1 − q)γis + q(γio + γin) = Γi, average rates of connectivity change for each type of nodes i = s, o, n (Fig. 1).
Network Expansion (Γ) and Protein Conservation (M).
The total number of nodes generated by the GDD model, F(n) (1), growths exponentially with the number of partial duplications, F(n) (1) = C·(1 + q)n, where C is the initial number of nodes, as a constant fraction of nodes q is duplicated at each time step. Yet, some nodes become completely disconnected from the rest of the graph during divergence and rejoin the disconnected component of size F(n) (0). From a biological point of view, these disconnected nodes represent genes that have presumably lost all biological functions and become pseudogenes before being simply eliminated from the genome. We neglect the possibility for nonfunctional genes to reconvert to functional genes again after suitable mutations, and remove them at each round of partial duplication‖ focusing solely on the connected part of the graph.
In particular, the link growth rate Γ = (1 − q)Γs + qΓo + qΓn obtained by taking the first derivative of Eq. 2 at x = 1, controls whether the connected part of the graph is exponentially growing (Γ > 1) or shrinking (Γ < 1).
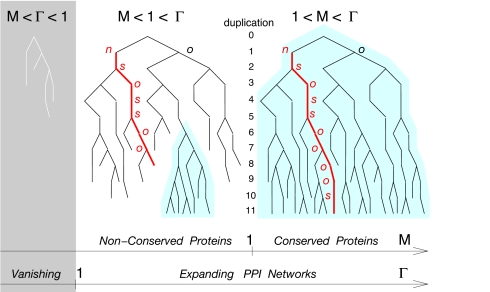
Let us now introduce another rate of prime biological interest, M = (1 − q)Γs + qΓo. It is the average rate of connectivity increase (M >1) of decrease (M < 1) for the most conserved duplicate lineage, which corresponds to a stochastic alternance between singular (s) and most conserved (o) duplicate descents. In particular, we have by construction, M < Γ = M + qΓn, independently from any evolutionary parameters, q and γij > 0. This implies three main evolutionary regimes from the perspective of network expansion (Γ) and protein conservation (M) (Fig. 2):
If M < Γ < 1. PPI networks are vanishing in this regime with seemingly little biological relevance.
If M < 1 < Γ. PPI networks are expanding, in this case, but their proteins are not conserved over long evolutionary time scales. This implies that the networks forget their evolutionary history exponentially fast, as most nodes eventually disappear and, with them, all traces of network evolution. These networks are not preserved over time, but instead are continuously renewed from duplication of the (few) most connected nodes (Fig. 2). Individual proteins of a given network realization are thus more similar to one another than to any protein of other network realizations, which can be seen, from a speciation perspective, as PPI networks of phylogenetically distant organisms. This is in sharp contrast to the widespread structural orthology observed across all extant life forms, even though functions of orthologs often differ (see Evolution of PPI Network Motifs).
If 1 < M < Γ. By contrast, PPI networks remember their past evolution from the very beginning, in this case, as proteins statistically keep on increasing their connectivity once they have emerged from a duplication-divergence event. This implies that most proteins are conserved throughout the evolution process and preserve some interaction partners. This is indeed in broad agreement with empirical evidence, because traces of protein conservation are even observed within the core transcriptional and translational machineries across all three major living kingdoms (16).
Fig. 2.
Evolutionary growth (Γ) and protein conservation (M) of PPI networks. The constitutive constraint, M < Γ, defines three evolutionary regimes discussed in the text.
Evolution of PPI Network Degree Distribution.
We now turn to the evolution of the degree distribution and other topological properties of PPI networks, which correspond to the technical core of the GDD model. To this end, we rescale the exponentially growing connected graph by introducing a normalized generating function for the average degree distribution,
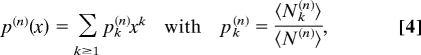 |
where 〈N(n)〉 = Σk≥1 〈Nk(n)〉, that is, after removing 〈N0(n)〉. F(n)(x) can be reconstructed from the shifted degree distribution, p̃(n)(x) = p(n)(x) − 1, as
which yields the following recurrence for p̃(n)(x),
where Δ(n) is the ratio between two consecutive graph sizes in terms of connected nodes, that is, Δ(n) = 〈N(n + 1)〉/〈N(n)〉,
Although Δ(n) is not known a priori and should, in general, be determined self-consistently with p̃(n)(x) itself, it is directly related to the evolution of the mean degree k̄(n) = Σk≥1 kpk(n) obtained by taking the first derivative of Eq. 6 at x = 1,
Hence, although connected networks grow exponentially both in terms of number of links (link growth rate Γ) and number of connected nodes (node growth rate Δ(n)), features normalized over these growing networks, such as node mean connectivity (Eq. 8) or distributions of node degree (or simple network motifs, see below), exhibit richer evolutionary dynamics in the asymptotic limit n → ∞, as we will now discuss.
Asymptotic Analysis of Node Degree Distribution (M′).
The node degree distribution can be shown (see SI Appendix) to converge toward a limit function p(x), with p̃(x) = p(x) − 1 solution of the functional Eq. 6.
where Δ = limn→∞Δ(n) with both Δ ≤ 1 + q, the maximum node growth rate, and Δ ≤ Γ, the link growth rate, because the number of connected nodes cannot increase faster than the number of links. Asymptotic regimes with Δ = Γ correspond to the same exponential growth of the network in terms of connected nodes and links, and will be referred to as linear regimes, hereafter, whereas Δ < Γ corresponds to nonlinear asymptotic regimes, which imply a diverging mean connectivity k̄(n) → ∞ in the asymptotic limit n → ∞ (Eq. 8).
To determine Δ and p(x) self-consistently, we first express successive derivatives of p(x) at x = 1 in terms of lower derivatives by using Eq. 9,
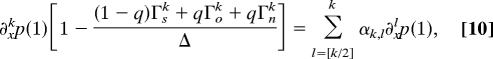 |
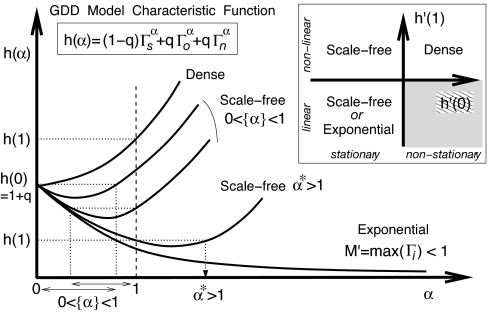
where αk,l are positive functions of the 1 + 6 parameters. Inspection of this expression readily defines two classes of asymptotic regimes, regular and singular regimes, depending on the value of a topology index M′ = maxi(Γi), for i = s, o, n. The detailed analysis relies on the “characteristic function” h(α) = (1 − q)Γsα + qΓoα +qΓnα, as outlined below and in Fig. 3(see SI Appendix, Asymptotic Methods, for proof details).
Fig. 3.
Asymptotic degree distribution for GDD models. Asymptotic regimes are deduced from the convex characteristic function h(α) and its derivatives h′(0) and h′(1) (see text).
Regular regimes, if M′ = maxi(Γi) < 1, for i = s, o, n. In this case, the only possible solution is Δ = h(1) (i.e., linear regime). Hence, since M′ < 1, h(1) > h(k), and successive derivatives ∂xkp(1) are thus finite and positive for all k ≥ 1. This corresponds to an exponential decrease of the node degree distribution for k ≫ 1, pk ∝ e−μk with a power law prefactor. The limit average connectivity (Eq. 8) is finite in this case, k̄ < ∞.
Singular regimes, if M′ = maxi(Γi) > 1, for i = s, o, n. In this case, Eq. 10 suggests that there exists an integer r ≥ 1 for which the rth derivative is negative, ∂xrp(1) < 0, which is impossible by definition. This simply means that neither this derivative nor any higher ones exist (for k ≥ r). We thus look for self-consistent solutions of the “characteristic equation” h(α) = Δ (with r − 1 < α ≤ r) corresponding to a singularity of p(x) at x = 1 and a power law tail of pk, for k ≫ 1 (17),
where the singular term (1 − x)α is replaced by (1 − x)r ln(1 − x) for α = r exactly. Several asymptotic behaviors are predicted from the convex shape of h(α) (∂α2h ≥ 0), depending on the signs of its derivatives h′(0) and h′(1) (Fig. 3 Inset).
If h′(0) < 0 and h′(1) < 0. There exists an α★ > 1 so that h(α★) = h(1) and the condition Δ ≤ h(1) implies that α★ ≥ α ≥ 1. The solution α = 1 requires h′(1) = 0 and should be rejected in this case. Hence, because k̄ < ∞ for α > 1, we must have Δ = h(1) (linear regime) and a scale-free limit degree distribution with a unique α = α★ > 1, pk ∝ k−α★−1 for k ≫ 1.
If h′(0) < 0 and h′(1) = 0. α = 1, Δ = h(1), and pk ∝ k−2 for k ≫ 1 (k̄(n) → ∞ as n → ∞).
If h′(0) < 0 and h′(1) > 0. The general condition Δ ≤ min(h(0), h(1)) leads a priori to a whole range of possible α ∈]0, 1] corresponding to stationary scale-free degree distributions with diverging mean degrees k̄(n) → ∞. Yet, numerical simulations suggest that there might still be a unique asymptotic node growth rate Δ regardless of initial conditions or evolutionary trajectories, although convergence is extremely slow (see SI Appendix, Numerical simulations).
If h′(0) ≥ 0 and h′(1) > 0. Δ = h(0) = 1 + q, implying that all duplicated nodes are selected in this case. No suitable α exists as the node degree distribution is exponentially shifted toward higher and higher connectivities. This is a dense, nonstationary regime with seemingly little relevance to biological networks.
Finally, note that the characteristic equation Δ = h(α) can be recovered directly from the average change of connectivity k → kΓi and the following continuous approximation (by using N(n) = Σk Nk(n) ≃ ∫u Nu(n) du and 〈Nk(n)〉 ∝ k−α−1),
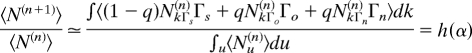 |
Local (q ≪ 1) and Global (q = 1) Duplication Limits.
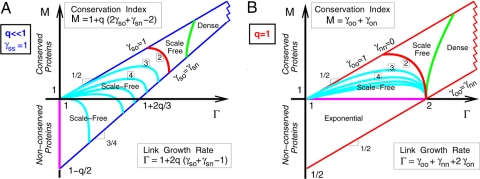
The asymptotic degree distribution of the GDD model can be conveniently mapped into the (Γ, M) plane for two limit regimes of prime biological relevance: (i) for local duplication events (q ≪ 1 and γss = 1; Fig. 4A) and (ii) for whole-genome duplication events (q = 1; Fig. 4B). See SI Appendix for details.
Fig. 4.
Asymptotic phase diagram of PPI networks under the GDD model. (A) Local duplication-divergence limit (q ≪ 1 and γss = 1). (B) Whole-genome duplication-divergence limit (q = 1). Boxed figures are power law exponents (α + 1) of scale-free regimes (Eq. 11).
The local duplication-divergence limit leads to scale-free limit degree distributions for both conserved and nonconserved networks, with power law exponents 1 < α + 1 ≤ 3 if γso ≃ 1 (i.e., which ensures that most previous interactions are conserved in at least one copy after duplication). By contrast, the whole-genome duplication-divergence limit leads to a wide range of asymptotic behaviors from nonconserved, exponential regimes to conserved, scale-free regimes with arbitrary power law exponents. Conserved, nondense networks require, however, an asymmetric divergence between old and new duplicates (γoo ≠ γnn) (5) and lead to scale-free limit degree distributions with the same range of exponents 1 < α + 1 ≤ 3 for maximum divergence asymmetry (γoo ≃ 1 and γnn ≃ 0).
Evolutionary Variations of Model Parameters.
The previous analysis with fixed parameters {q, γij} can be readily extended to combine local and global PPI network duplications (Fig. 4 A and B) or even include any evolutionary variations and stochastic fluctuations of the GDD model parameters with arbitrary series {q(n),γij(n)}R (see SI Appendix). Protein conservation is then found to be controlled by the cumulated product of connectivity growth/decrease rates following the most conserved, old duplicate lineage,
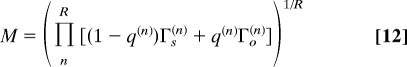 |
with conserved (resp. nonconserved) protein evolutionary regimes corresponding to M > 1 (resp. M < 1).
A similar geometric average also controls the nature of the asymptotic degree distribution as the network topology index now reads,
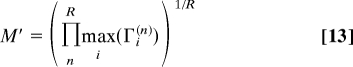 |
with M′ < 1 corresponding to exponential networks and M′ > 1 to scale-free (or dense) networks with an effective node degree exponent α and effective node growth rate Δ that are self-consistent solutions of the generalized characteristic equation,
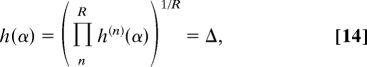 |
where h(n)(α) = (1 − q(n))Γs(n)α + q(n)Γo(n)α + q(n)Γn(n)α, as before. This leads to exactly the same discussion for singular regimes as with constant q and Γi (Fig. 3) because of the convexity of the generalized function h(α) (∂α2h(α) ≥ 0; see SI Appendix for details and discussion on the R → ∞ limit).
In particular, because (1 − q(n))Γs(n) + q(n)Γo(n) ≤ maxi(Γi(n)) for all q(n) and Γi(n) (i = s, o, n), we always have M ≤ M′. This relation implies a fundamental linkage between protein conservation and network topology under general duplication-divergence evolution, regardless of all possible evolutionary variations of the model parameters, q(n) and Γi(n). We expect, in particular, that all conserved networks are necessarily scale-free (or dense) (1 < M ≤ M′), whereas exponential networks can never be conserved (M ≤ M′ < 1), under general duplication-divergence evolution.
Evolution of PPI Network Motifs.
The generating function approach, introduced for the one-node degree distribution pk(n) (Eqs. 1–6), can be generalized to analyze the evolutionary statistics of multinode correlation functions and related clustering coefficient, distribution of first-neighbor average connectivity gk (18) (see Fig. 6) and small-network motifs. Yet, although M′ also controls transitions between major evolutionary regimes for multinode correlation functions, their analysis remains technically involved (SI Appendix).
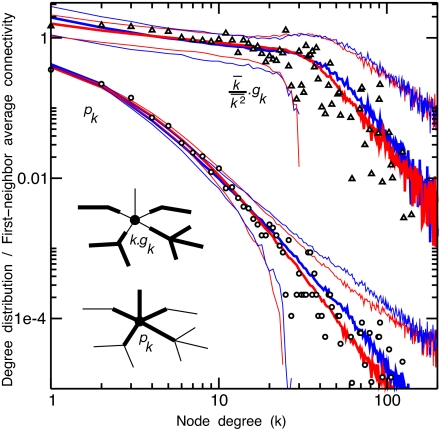
Fig. 6.
Comparison between empirical PPI network data (20) and finite-size, duplication-divergence simulations. Protein physical interaction data for yeast are taken from the Biomolecular Interaction Network Database (20) (4,576 proteins, 9,133 physical interactions, k̄ = 3.99, k̄2 = 106). Both connectivity distribution pk (open circles) and first-neighbor average connectivity gk (18) (open triangles) are shown. Best one-parameter fit of the data (blue curves) (±2σ for expected deviations) for the most asymmetric genome duplication-divergence model with γon = 0.26 (and q = 1, γoo = 1, γnn = 0) (5). Best two-parameter fit of the data (red curves) (±2σ for expected deviations) with the same most asymmetric whole-genome duplication limit but at the level of protein domains instead of entire proteins, with γon = 0.1 and λ = 0.3, which corresponds to 1/(1 − λ) = 1.5 average number of binding domains per proteins. See SI Appendix, Fig. S6 and ref. 5 for details about this model including domain shuffling.
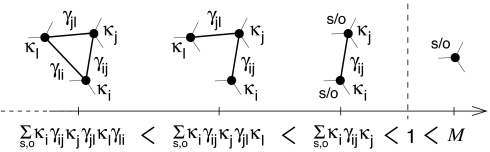
By contrast, the conservation property of network motifs under general duplication-divergence evolution turns out to be remarkably simple, as outlined in Fig. 5. We derive conservation indices for specific network motifs by summing over all possible combinations of s nodes (with probability κs = 1 − q) or o nodes (with probability κo = q) and the corresponding γij (i, j = s, o) (Fig. 5). Clearly, network motifs with a larger number of interactions, p ≥ 1, have lower conservation indices, Mp ≃ O(γijp) (Fig. 5). Moreover, because the probability to conserve a specific interaction γij cannot be exactly 1, because of deleterious mutations (i.e., γij < 1), motif conservation indices Mp must all be <1, regardless of any parameter variations, q(n) and γij(n).
Fig. 5.
Motif conservation indices. Although individual proteins are typically conserved (if M > 1), network motifs including two or more proteins cannot be indefinitely conserved under general duplication-divergence evolution (κo = 1 − κs = q; see text).
Hence, network motifs cannot be indefinitely conserved under duplication-divergence evolution, even though their individual proteins are typically conserved in the network (if M > 1) (Fig. 2). This implies that structural orthology between individual proteins from phylogenetically distant species cannot indefinitely coincide with functional orthology at the level of protein interactions and complexes, in broad agreement with empirical evidences (19). The resulting turnover toward more and more divergent interaction partners is a simple evolutionary consequence of the GDD model, regardless of any network-rewiring dynamics (that have been neglected here). In particular, even the most conserved orthologous proteins (s/o descents) must eventually perform different functions, but conserved ancestral functions are inevitably passed down to less conserved protein complexes (s/o/n descents) in phylogenetically distant species. This inherent evolutionary constraint of the GDD model sets conceptual and practical limitations on functional annotation transfer between genomes (19).
Comparison with Empirical PPI Network Data.
Although the GDD model can, in principle, accommodate 1q + 6γij fitting parameters, and any evolutionary variations or fluctuations q(n) and γij(n), we have restricted all quantitative comparisons with the empirical data (20) to biologically relevant regimes, limited to one or two effective fitting parameters, only.
The results, corresponding to 103 to 104 protein nodes and low network growth rates in the conserved, scale-free regime (1< M ≤ M′), are compared with Yeast PPI network data (20) (Fig. 6), both for the connectivity distribution pk and the first-neighbor average connectivity gk (18) (see Fig. 6 legend and SI Appendix for model and fitting details).
Extending these numerical simulations to larger PPI network sizes (>105 nodes) shows little changes in these distributions for small k ≤ 20 (SI Appendix, Figs. S5–S7). This suggests that the available empirical PPI network for yeast (20) has essentially reached its asymptotic degree distribution, for k ≤ 20. By contrast, global convergence of the GDD model is typically quite slow for large growth rate regimes with little biological relevance, as shown in SI Appendix, Numerical simulations, Figs. S5–S7.
Conclusions
Although conservation and network topology have a priori no reason to be related features of emerging PPI networks in the course of evolution, we demonstrate in this article that they are, in fact, linked properties under general duplication-divergence evolution, because of a fundamental linkage between protein conservation and network topology indices, that is, M ≤ M′, regardless of any variations or fluctuations of the model parameters.
By contrast, we also showed that conservation of network motifs cannot be indefinitely preserved under general duplication-divergence evolution (independently from any network rewiring dynamics). This underlies intrinsic limitations on functional annotation transfer across phylogenetically distant genomes (19).
All in all, these evolutionary constraints, inherent to duplication-divergence processes and independent from selective adaptation (3), appear to have largely controlled the overall topology and scale-dependent conservation of PPI networks, regardless of any specific biological function.
Supplementary Material
Acknowledgments.
We thank U. Alon, R. Bruinsma, M. Cosentino-Lagomarsino, T. Fink, R. Monasson, and M. Vergassola for discussion. This work was supported by the Ministry of Higher Education and Research, Centre National de la Recherche Scientifique, and Institut Curie.
Footnotes
The authors declare no conflict of interest.
Duplicated protein domains or subdomains are also quite common even within ancestral proteins, such as the ubiquitous aquaporin membrane proteins in eubacteria, archaea, and eukaryotes, or the TATA-binding protein from archaea and eukaryotes.
Except for a few interesting cases of protein-binding mimicry, typically found in virus–host protein–protein interactions (8).
Except for domains that self-assemble into homo-oligomers, which must have at least two binding interfaces, see table 2 in ref. 9.
Results from the time-linear duplication-divergence model (10) are recovered as a special limit, see supporting information (SI) Appendix.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804119105/DCSupplemental.
Note, however, that pseudogenes may still have a critical role in evolution by providing functional domains that can be fused to adjacent genes. This supports a view of PPI network evolution in terms of protein domains instead of entire proteins (SI Appendix, Fig. S6B, and ref. 5). Yet, we showed in ref. 5 that extensive domain shuffling does not change the resulting network topology from duplication-divergence models.
References
- 1.Ohno S. Evolution by Gene Duplication. New York: Springer; 1970. [Google Scholar]
- 2.Li WH. Molecular Evolution. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- 3.Lynch M. The Origins of Genome Architecture. Sunderland, MA: Sinauer; 2007. [Google Scholar]
- 4.Berg J, Lässig M, Wagner A Structure and evolution of protein interaction networks: a statistical model for link dynamics and gene duplications. BMC Evol Biol. 2004;4:51. doi: 10.1186/1471-2148-4-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evlampiev K, Isambert H. Modeling protein network evolution under genome duplication and domain shuffling. BMC Syst Biol. 2007;1:49. doi: 10.1186/1752-0509-1-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maslov S, Sneppen K, Eriksen KA, Yan KK. Upstream plasticity and downstream robustness in evolution of molecular networks. BMC Evol Biol. 2004;4:9. doi: 10.1186/1471-2148-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones S, Thornton JM. Principles of protein-protein interactions. Proc Natl Acad Sci USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henschel A, Kim WK, Schroeder M. Equivalent binding sites reveal convergently evolved interaction motifs. Bioinformatics. 2006;22:550–555. doi: 10.1093/bioinformatics/bti782. [DOI] [PubMed] [Google Scholar]
- 9.Kim WK, Henschel A, Winter C, Schroeder M. The many faces of protein-protein interactions: A compendium of interface geometry. PLoS Comput Biol. 2006;2:e124. doi: 10.1371/journal.pcbi.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ispolatov I, Krapivsky PL, Yuryev A Duplication-divergence model of protein interaction network. Phys Rev E Stat Nonlin Soft Matter Phys. 2005;71 doi: 10.1103/PhysRevE.71.061911. 061911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Albert R, Barabási A-L. Statistical mechanisms of complex networks. Rev Mod Phys. 2002;74:47–97. [Google Scholar]
- 12.Barabási A-L, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 13.Raval A. Some asymptotic properties of duplication graphs. Phys Rev E Stat Nonlin Soft Matter Phys. 2003;68 doi: 10.1103/PhysRevE.68.066119. 066119. [DOI] [PubMed] [Google Scholar]
- 14.Vázquez A, Flammini A, Maritan A, Vespignani A. Modeling of protein interaction networks. ComPlexUs. 2003;1:38–44. doi: 10.1038/nbt825. [DOI] [PubMed] [Google Scholar]
- 15.Ispolatov I, Krapivsky PL, Mazo I, Yuryev A. Cliques and duplication-divergence network growth. N J Phys. 2005;7:145. doi: 10.1088/1367-2630/7/1/000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartman H, Favaretto P, Smith TF. The archaeal origins of the eukaryotic translational system. Archaea. 2006;2(1):1–9. doi: 10.1155/2006/431618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flajolet P, Sedgewick R. [Accessed March 22, 2007];Analytic Combinatorics. 2006 Available at http://algo.inria.fr/flajolet/Publications/books.html.
- 18.Maslov S, Sneppen K. Specificity and stability in topology of protein networks. Science. 2002;296:910. doi: 10.1126/science.1065103. [DOI] [PubMed] [Google Scholar]
- 19.Yu H, et al. Annotation transfer between genomes: Protein-protein interologs and protein-DNA regulogs. Genome Res. 2004;14:1107–1118. doi: 10.1101/gr.1774904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alfarano C, et al. The Biomolecular Interaction Network Database and related tools 2005 update. Nucleic Acids Res. 2005;33:D418–D424. doi: 10.1093/nar/gki051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.