Abstract
Francisella tularensis is a highly virulent bacterium that causes tularemia, a disease that is often fatal if untreated. A live vaccine strain (LVS) of this bacterium is attenuated for virulence in humans but produces lethal disease in mice. F. tularensis has been classified as a Category A agent of bioterrorism. Despite this categorization, little is known about the components of the organism that are responsible for causing disease in its hosts. Here, we report the deletion of a well-characterized lipoprotein of F. tularensis, designated LpnA (also known as Tul4), in the LVS. An LpnA deletion mutant was comparable to the wild-type strain in its ability to grow intracellularly and cause lethal disease in mice. Additionally, mice inoculated with a sublethal dose of the mutant strain were afforded the same protection against a subsequent lethal challenge with the LVS as were mice initially administered a sublethal dose of the wild-type bacterium. The LpnA-deficient strain showed an equivalent ability to promote secretion of chemokines by human monocyte-derived macrophages as its wild-type counterpart. However, recombinant LpnA potently stimulated primary cultures of human macrophages in a Toll-like receptor 2-dependent manner. Although human endothelial cells also were activated by recombinant LpnA, their response was relatively modest. LpnA is clearly unnecessary for multiple functions of the LVS, but its inflammatory capacity implicates it and other Francisella lipoproteins as potentially important to the pathogenesis of tularemia.
Keywords: lipoprotein, inflammation, Francisella, virulence, mice
Introduction
Francisella tularensis is a small, non-motile, aerobic, Gram-negative coccobacillus, first isolated by G.W. McCoy in 1911 [1]. There are two main subspecies of F. tularensis that are pathogenic for humans. F. tularensis subsp. tularensis (also known as type A) is highly virulent in humans and causes the majority of tularemia cases in North America. F. tularensis subsp. holarctica (type B) causes a milder form of tularemia and is responsible for the bulk of tularemia cases reported throughout Eurasia [2]. A live vaccine strain (LVS) of F. tularensis was developed from a type B isolate [3]. Although this strain is attenuated for virulence in humans, it remains highly virulent in mice, making it a useful model for the study of the pathogenesis of tularemia [4].
Despite the high degree of virulence of F. tularensis, little is known about how this facultative intracellular pathogen causes lethal disease in its hosts; only recently have some of the factors involved in the pathogenesis of tularemia been identified. The ability of this organism to replicate to high numbers in murine macrophages, as well as to cause fatal disease in mice, is associated with IglC, a 23-kDa protein that is highly expressed during intramacrophage growth [5–8]. Mutation of the pdpA and pdpD genes of F. tularensis subsp. novicida also results in defects in virulence and growth within macrophages [9]. The pdpA and pdpD genes, along with iglC and several others, are thought to represent a Francisella pathogenicity island regulated by MglA, another protein that is required for virulence [9,10]. Type IV pili play a major role in the pathogenicity of many bacteria [11]. The genes required for the production of type IV pili exist in Francisella, and ultrastructural analysis of the LVS revealed the presence of long protrusions with the appearance of type IV pili [12]. Deletion of one of the putative pilin genes in a type B strain of F. tularensis results in attenuation of virulence [13], further implicating type IV pili in the pathogenesis of tularemia. Finally, deletion of the tolC or ftlC genes in the LVS decreases the resistance of the organism to anti-bacterial agents, and, in the case of the former, causes attenuation of virulence in mice [14].
The lipopolysaccharide (LPS) of F. tularensis possesses an atypical lipid A moiety and core structure [15]. Mice deficient in Toll-like receptor (TLR) 4, which binds LPS, do not gain increased resistance to F. tularensis, succumbing to disease as rapidly as do wild-type mice [16]. The purified LPS of F. tularensis LVS does not stimulate production of tumor necrosis factor-α, interferon-γ, interleukin (IL)-12, IL-10, or nitric oxide by murine macrophages [17,18]. Nonetheless, F. tularensis LVS induces inflammation in mice in vivo [19,20] and can elicit the production of proinflammatory mediators by cultured murine macrophages and dendritic cells [21,22]. Further, human cells of innate immunity respond to culture with the LVS by producing an array of chemokines, cytokines, and adhesion molecules [23,24]. Together, these data suggest that F. tularensis possesses components other than LPS that are capable of initiating inflammation and regulating virulence. Recent findings have implicated TLR2, a receptor for lipoproteins, as important in the host response to infection with Francisella [21,22,25,26]. Therefore, the lipoproteins of F. tularensis may be important in the pathogenesis of tularemia.
LpnA (also called Tul4) is an immunodominant 17-kDa lipoprotein [27] that is conserved throughout Francisella strains [28]. LpnA is associated with the outer membrane [29], stimulates T cells from F. tularensis-primed individuals [30,31], confers partial protective immunity to tularemia in mice when administered in recombinant form [32], and is predominantly expressed relative to other F. tularensis lipoproteins [27]. Herein, we set out to determine what role LpnA plays in the pathogenesis of tularemia. We show that absence of LpnA does not affect the intracellular replication of F. tularensis LVS or its virulence in mice, yet the recombinant lipoprotein induces production of proinflammatory mediators by primary human endothelial cells and macrophages. Our results clearly demonstrate that LpnA is not required for virulence of F. tularensis. However, its ability to stimulate cells of innate immunity implicates it and other F. tularensis lipoproteins as important mediators of the host response during tularemia.
Results
F. tularensis LVS lpnA was deleted by allelic exchange
To generate a strain of F. tularensis LVS with a deletion of lpnA, we utilized the conjugation method of Golovliov et al. [7], with changes as described in Materials and Methods. Briefly, Escherichia coli was transformed with pPV/ΔlpnA, a plasmid harboring a deleted version of lpnA, the sacB gene (which confers resistance to sucrose), and a chloramphenicol-resistance cassette. F. tularensis LVS was conjugated with this strain and plated on agar supplemented with chloramphenicol. A primary recombination event yielded an intermediate strain of the LVS with pPV/ΔlpnA incorporated into its genome. The intermediate strain then was plated on sucrose-containing agar, leading to a secondary recombination event that eliminated genomic lpnA along with the plasmid components of pPV.
The first and second recombination events were verified by polymerase chain reaction (PCR) (Fig. 1) and by growth on selective media. Primers specific to sacB generated the expected product of approximately 500 base pairs (bp) from the DNA of the intermediate colonies that had undergone a first recombination event, as well as from the pPV vector. Primers flanking lpnA (LpnA1; LpnA4) produced amplicons of approximately 3000 bp from the pPV vector, a selected intermediate colony, and the resulting final deletion strain, as well as a product of approximately 3500 bp from the wild-type LVS. The intermediate should have yielded both 3000- and 3500-bp products; however, we suspect that the primers preferentially amplified the smaller product. Verification of the deletion of lpnA was carried out using a primer specific to the deleted region of the gene and primer LpnA4. These primers generated amplicons from the LVS and the intermediate but not from the final deletion strain, indicating successful elimination of lpnA (Fig. 1). F. tularensis-specific DNA microarray analysis was performed to determine whether any transcriptional alterations occurred as a result of the lpnA deletion. Comparison of mRNA from wild-type and LpnA-deficient (ΔlpnA) F. tularensis LVS showed no change in transcript levels for any gene other than, as expected, lpnA itself (data not shown).
Figure 1.
Verification of the lpnA deletion mutant. PCR amplification of the mutant strain (lane 3) with primers external to lpnA yielded a smaller-sized amplicon than that produced when the wild-type LVS (lane 2) was amplified with the same primers. Primers internal to lpnA did not generate an amplicon from the mutant strain or the mutagenesis vector pPVΔlpnA (lane 1) but did amplify the wild-type LVS and the intermediate strain (lane 4). Amplification of sacB occured in both the mutagenesis vector and the intermediate strain, but not the wild-type or mutant strain, indicating exclusion of the vector from the mutant strain. Lane 5: water control.
F. tularensis LVS ΔlpnA replicates to the same extent in murine macrophages as does wild-type LVS
F. tularensis LVS lacking lpnA grew in Mueller-Hinton (MH) broth at the same rate and to the same extent as the wild-type bacteria over a 48-h time period (data not shown). The ability of F. tularensis LVS to proliferate within murine macrophages has been well documented [5,24,33,34]. To determine whether a lack of LpnA affected intracellular replication, murine bone marrow-derived macrophages (muBMDM) were cultured with either wild-type or LpnA-deficient F. tularensis LVS for 2 h, after which the extracellular bacteria were killed with gentamicin. The F. tularensis strains then were allowed to multiply intracellularly for an additional 22 h. In three independent experiments, numbers of wild-type LVS increased 622 ± 66 fold in this span, and the mutant grew comparably, increasing 638 ± 370 fold over the same time period. Immunofluorescence microscopy demonstrated that similar percentages of muBMDM were infected with either wild-type or LpnA-deficient bacteria after 2 h and confirmed the extensive intracellular replication of these two strains over a subsequent 22-h period (Fig. 2).
Figure 2.
F. tularensis LVS lacking LpnA replicates to the same extent in muBMDM as does its wild-type counterpart. Cultured muBMDM were incubated with F. tularensis LVS or F. tularensis LVS ΔlpnA for 2 h at a MOI of ~50. Two-hour samples were washed and fixed immediately, whereas 24-h samples were washed, incubated for 1 h in the presence of 5 µg/ml of gentamicin, cultured in antibiotic-free medium for 21 h more, and fixed. F. tularensis LVS then was detected by indirect immunofluorescence. Samples were visualized by epifluorescence (columns 1 and 3) and phase microscopy. Columns 2 and 4 depict epifluorescent microscopic images superimposed on the phase microscopic images of the same field.
Deletion of lpnA from F. tularensis LVS does not alter its virulence in mice
Others have previously shown that when administered to C3H/HeN mice intradermally, F. tularensis LVS is lethal at doses of 106 and 107 bacteria but not when 105 or fewer organisms are given [4]. For this reason, we used infectious doses ranging from 105 to 107 bacteria to compare the abilities of wild-type and LpnA-deficient F. tularensis LVS to cause illness in mice. F. tularensis LVS ΔlpnA was no different than wild-type LVS in its ability to cause lethal disease (Table 1). At an infectious dose of 107 bacteria, one of 20 mice infected with the wild-type LVS survived, whereas three of 20 mice infected with the LpnA-deficient LVS survived infection. The median times to death for each strain were 4 and 5 days, respectively. Following infection with 106 total organisms, one of 15 mice survived from each of the wild-type- and mutant-infected groups, with a median time to death of 5 days for both. Mice infected with 105 organisms of F. tularensis LVS or F. tularensis LVS ΔlpnA were less susceptible to lethal disease, with seven of ten and eight of ten animals surviving, respectively (Table 1).Three days after inoculation with 106 organisms, mice harbored on average similar numbers of wild-type or mutant bacteria in the lung, liver, and spleen, although variability was greater among the animals that received the ΔlpnA strain (data not shown). To determine whether our above observations held true irrespective of the route of inoculation, C3H/HeN mice, in groups of five, were inoculated intranasally with a lethal dose of either wild-type F. tularensis LVS or the LpnA-deficient mutant strain. All mice from both groups died within 8 days of infection, with comparable times-to-death for each group (Table 1).
Table 1.
Virulence of wild-type and LpnA-deficient F. tularensis LVS in C3H/HeN mice
| Strain | Number of surviving/infected mice and median time-to-death (days) at an infectious dose of: | |||
|---|---|---|---|---|
| 107 CFU, i.d.a | 106 CFU, i.d.a | 105 CFU, i.d.a | 106 CFU, i.n.b | |
| F. tularensis LVS | 1/20; 4 | 1/15; 5 | 7/10; 6 | 0/5; 7 |
| F. tularensis LVS ΔlpnA | 3/20; 5 | 1/15; 5 | 8/10; 5 | 0/5; 7 |
Mice were infected intradermally (i.d.).
Mice were infected intranasally (i.n.).
LpnA is not necessary to confer immunity to F. tularensis LVS in mice
Mice surviving intradermal challenge with sublethal doses of F. tularensis LVS become immune to subsequent challenges with otherwise lethal inocula of the organism [4]. To determine whether LpnA is necessary to confer this immunity, C3H/HeN mice were administered a sublethal dose of wild-type LVS, LpnA-deficient LVS, or phosphate-buffered saline (PBS), followed by a challenge with an ordinarily lethal dose of F. tularensis LVS two months later. LpnA was not needed to confer immunity, as all ten mice from the group initially administered the ΔlpnA strain survived the secondary challenge. Similarly, eight of nine mice that were initially infected with wild-type LVS survived the challenge. All five mice that were initially given PBS, rather than bacteria, died within 6 days of the second inoculation (Fig. 3).
Figure 3.
LpnA is not required for immunoprotection of mice against F. tularensis LVS in mice. C3H/HeN mice were inoculated with a sublethal dose of F. tularensis LVS or F. tularensis LVS ΔlpnA, or with PBS. After 2 months, all mice were challenged with a normally lethal dose of F. tularensis LVS and monitored for survival for an additional 4 weeks.
Recombinant LpnA activates human macrophages and endothelial cells
Our laboratory previously demonstrated that human monocyte-derived macrophages (huMDM) and human umbilical vein endothelial cells (HUVEC) respond to stimulation with live or killed preparations of F. tularensis LVS by producing a wide array of proinflammatory mediators [23,24]. These mediators include the chemokines CXCL8 and CCL2 and the adhesion molecule E-selectin. E-selectin is expressed on the surface of activated endothelium and participates in the initial capture of circulating leukocytes [35]. Subsequently, chemokines induce the transendothelial migration of captured leukocytes and thus recruit these cells to extravascular sites of inflammation. CXCL8 and CCL2 are produced by both HUVEC and huMDM that have been exposed to the LVS and are attractants for neutrophils and mononuclear leukocytes, respectively [36].
Purified LPS from the LVS activates cells of innate immunity poorly [37] (C. E. Noah and M. B. Furie, unpublished results), indicating that other components mediate the response elicited by live or killed bacteria. LpnA is potentially one such component. Because we have found that human macrophages have a far greater inflammatory response to F. tularensis LVS than do murine macrophages [24], we tested the capacity of LpnA to stimulate cells of human origin. There was no difference in the ability of the LpnA-deficient and wild-type strains of F. tularensis LVS to stimulate production of the chemokines CXCL8 and CCL2 by huMDM (Fig. 4). However, preparations of purified, recombinant LpnA ranging from 10 ng/ml to 1 µg/ml of protein induced huMDM to secrete both CXCL8 (Fig. 5A) and CCL2 (Fig. 5B), but not IL-1β (data not shown). LpnA was a potent stimulus, as even the lowest doses of LpnA tested elicited greater production of chemokines than did 10 ng/ml of E. coli LPS (Figs. 5A and 5B). All samples of LpnA were cleared of potential contaminating E. coli LPS prior to experimentation, as discussed in Materials and Methods. When TLR2 was blocked by antibody, a significant decrease in the production of both chemokines occurred, whereas no change was seen when the cells were pre-incubated with a control antibody of the same isotype (Fig. 6). As expected, the antibody to TLR2 inhibited secretion of CXCL8 by huMDM exposed to tripalmitoyl-Cys-Ser-(Lys)4, a known agonist for this receptor, but had no effect on cells stimulated by E. coli LPS (data not shown).
Figure 4.
Wild-type and LpnA-deficient F. tularensis LVS promote similar secretion of the chemokines CXCL8 and CCL2 by huMDM. HuMDM were incubated for 24 h at 37°C with medium alone (Unstim), a sham preparation of bacteria, live or killed F. tularensis LVS at a MOI of ~25, or live or killed F. tularensis LVS ΔlpnA at a MOI of ~25. Cell-free conditioned media were collected, and amounts of CXCL8 or CCL2 were measured by ELISA. Bars represent the means ± SD of three replicate samples. This experiment was repeated twice, yielding similar patterns of chemokine secretion.
Figure 5.
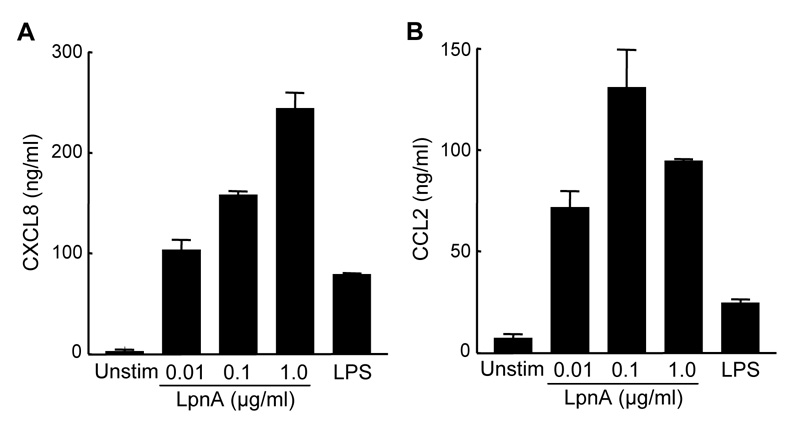
LpnA stimulates secretion of CXCL8 and CCL2 by huMDM. HuMDM were treated with medium alone (Unstim), recombinant LpnA ranging from 0.01 µg/ml to 1.0 µg/ml, or 0.01 µg/ml of E. coli LPS for 24 h at 37°C. All samples of LpnA caused significant increases in the production of CXCL8 (A) and CCL2 (B) relative to that of unstimulated macrophages as measured by ELISA of cell-free conditioned media (P < 0.001). Bars represent the means ± SD of three replicate samples. Although the magnitude of the response differed from donor to donor, LpnA at concentrations of 0.01 µg/ml or above significantly increased secretion of these chemokines in two additional experiments.
Figure 6.
Secretion of CXCL8 and CCL2 by huMDM in response to LpnA is TLR2-dependent. Addition of anti-human TLR2 antibody to huMDM subsequently stimulated with recombinant LpnA (0.01 µg/ml) resulted in the production of only basal levels of CXCL8 (A) or CCL2 (B), whereas macrophages incubated with an isotype-matched control antibody were unaffected. Bars represent the means ± SD of three replicate samples. *, Significantly different from samples stimulated with LpnA but not treated with anti-TLR2, P < 0.001. This experiment was repeated twice with similar results.
HUVEC responded to recombinant LpnA, but the amounts of lipoprotein required were 10- to 100-fold greater than those needed to stimulate macrophages. LpnA at 1 µg/ml induced secretion of modest levels of CXCL8 and CCL2 in five of eight independent experiments; stimulation was insignificant in the remaining three studies. Representative results are shown in Fig. 7, panels A and B. Upregulation of surface expression of the adhesion molecule E-selectin was observed in response to 100 ng/ml of LpnA (Fig. 7C). In contrast, stimulation of huMDM was reproducibly achieved with 10 ng/ml of LpnA (compare Fig. 5 and Fig. 7).
Figure 7.
HUVEC respond relatively poorly to LpnA. (A, B) HUVEC were cultured for 24 h with medium alone (Unstim), recombinant LpnA (0.01 µg/ml to 1.0 µg/ml), or E.coli LPS (0.5 ng/ml). Conditioned media then were collected and analyzed by ELISA. Significant increases in the levels of CXCL8 (A) and CCL2 (B) were observed in response to 1.0 µg/ml of LpnA. (C) HUVEC were incubated for 4 h with the same samples as described above. Whole-cell ELISA performed on living monolayers of endothelial cells showed significant upregulation of E-selectin in response to 0.1 µg/ml and 1.0 µg/ml of LpnA. Bars represent the means ± SD of three (A, B) or four (C) replicate samples. *, Significantly different from unstimulated controls, P < 0.001. These experiments were repeated once, yielding similar results.
Discussion
We regarded LpnA as potentially important in the pathogenesis of tularemia based on a number of previously published observations. These include the relatively high expression of LpnA in the outer membrane of F. tularensis [27], its high conservation among subspecies of Francisella [28], its immunogenicity [32], and its identity as a lipoprotein [27]. Bacterial lipoproteins can be predicted from genomic sequences by the presence of a lipobox, a consensus signal sequence at their N-terminus that is recognized and subsequently cleaved by the enzyme signal peptidase II [38]. The resultant lipid modification, N-acyl-S-diacylglyceryl cysteine, occurs at the N-terminus and is conserved throughout bacterial lipoproteins of many different species [39]. LpnA contains palmitic acid [27], which is the predominant amide-linked fatty acid in the murein lipoprotein of E. coli [38]. Moreover, all enzymes responsible for the N-acyl-S-diacylglyceryl modification, which include prolipoprotein diacylglyceryl transferase, signal peptidase II, and apolipoprotein N-acyltransferase [40], are encoded by the genome of the LVS (GenBank accession number AM23362). Nonetheless, we cannot be certain that the lipid modification of recombinant LpnA, expressed in E. coli, is identical to that of native LpnA.
Lipoproteins are important factors in the pathogenesis of bacterial diseases. For example, lipoproteins from Borrelia burgdorferi, the causative agent of Lyme disease, provoke a proinflammatory response in endothelial cells [41] and leukocytes [42] and play an essential role in transmission of the infection [43]. The proinflammatory nature of lipoproteins has been attributed to their attached lipid, and not the protein itself. Lipoproteins from Borrelia burgdorferi and Brucella abortus are able to induce inflammation in host cells only after they have been processed and modified with lipid [41,42,44]. However, lipoproteins display significant functional diversity that likely can be attributed to the variability in their associated protein moieties. They can serve structural purposes in the bacterial membrane, function as transporters across membranes, or work as toxins in some bacterial species [39]. Thus, it can be expected that lipoproteins of all bacteria display both unique and common functions.
Our observation that LpnA-deficient F. tularensis LVS could induce secretion of chemokines by human macrophages to the same extent as the wild-type LVS was not entirely surprising. A search of the annotated F. tularensis subsp. tularensis Schu S4 genome [45] reveals 38 potential lipoproteins, all of which are also present in the genome of the LVS. If LpnA were involved in inflammation, it would likely be due to its lipid attachment, which should be the same in all F. tularensis lipoproteins. Ablation of only one of these putative lipoproteins, even if predominantly expressed, may be insufficient to affect the inflammatory capacity of the organism. Our finding that recombinant LpnA elicits production of proinflammatory factors by host cells strongly supports this possibility. Consequently, it is plausible that the proinflammatory capacity of LpnA is shared by all Francisella lipoproteins. Compared to huMDM, HUVEC were relatively resistant to activation by LpnA. Recently, we observed that mice infected with the LVS contain substantial numbers of extracellular organisms in the blood throughout the course of their illness [46]. Perhaps a poor response of endothelium to LpnA permits these bacteria to persist in the circulation without provoking overwhelming, systemic inflammation.
We did anticipate a role for LpnA in the pathogenesis of tularemia. F. tularensis is known for its ability to proliferate within the macrophages of mice and humans. In fact, this is one of the few established properties that contribute to the virulence of the organism, as mutants lacking a gene required for intra-macrophage growth are avirulent in mice [7,8,10]. Others have shown that an LpnA-deficient strain of F. tularensis subspecies novicida replicates 10-fold less in the murine macrophage-like J774 cell line than does its parental strain [47]. As a consequence, one might expect this decreased growth rate to correspond to a decrease in virulence of the mutant strain. Although this assumption was not tested with the LpnA-deficient strain of F. tularensis subspecies novicida, we show here that it does not hold true for the LVS lacking LpnA; LpnA-deficient F. tularensis LVS did not display any changes in virulence in mice when administered via either the intradermal or intranasal route. Moreover, we did not observe any deficiency in growth of the mutant strain within muBMDM relative to the wild-type LVS. It is possible that LpnA is more essential to F. tularensis subsp. novicida than it is to the LVS, leading to the observed decrease in intracellular replication of the LpnA-deficient strain of the former [47]. In F. tularensis LVS, however, LpnA clearly makes no indispensable contribution to virulence, despite its predominant expression compared to other F. tularensis lipoproteins [27] and its ability to elicit an immune reaction [30,32,48].
Others have shown previously that immunity to F. tularensis can be attained after exposure to sublethal doses of the organism [4]. In addition, mice can be immunized against F. tularensis LVS by administration of its purified LPS alone [49,50]. The observations that LpnA stimulates T cells of individuals previously exposed to F. tularensis [31] and that it can confer partial resistance to challenge with the LVS when given to mice in recombinant form [32,48] suggested a potentially important role for the lipoprotein in acquired immunity to F. tularensis. Despite these previous findings, we determined that mice immunized with either wild-type or LpnA-deficient F. tularensis LVS are equally resistant to subsequent challenge with a typically lethal dose of the bacterium. Thus, LpnA is not required for full immunoprotection of mice against infection with F. tularensis LVS.
TLRs are a group of pattern-recognition receptors that play important roles in host responses to invading pathogens. Bacterial lipoproteins and atypical LPS of certain bacteria signal via TLR2, whereas LPS from most bacteria signals through TLR4 [51]. The weak proinflammatory activity of LPS from F. tularensis has been well described [17,18], and mice deficient in TLR4 are as susceptible to lethal tularemia as are naïve mice of the same background [16,52]. In contrast, recent reports have indicated an important role for TLR2 in the pathogenesis of tularemia [21,22,25,26]. The fact that recombinant LpnA induced inflammation in human macrophages in a manner that was dependent on TLR2 implicates it and other F. tularensis lipoproteins as important mediators of the host response in tularemia. Lipoproteins from several different bacterial species are released during growth of the bacteria and upon killing of the organisms with antibiotics, and they subsequently stimulate the production of proinflammatory mediators in vitro [53]. These data support the notion that biogenesis of lipoproteins can be an important factor in microbial pathogenesis. Our findings clearly rule out LpnA as an essential component in the pathogenesis of tularemia in the murine model. However, the strong proinflammatory capacity of LpnA indicates that it and other lipoproteins of F. tularensis may play a key role in the host response to infection.
Materials and Methods
Culture of bacteria
F. tularensis LVS (American Type Culture Collection No. 29684) was maintained and cultured as previously described [23]. For each experiment, a frozen stock of bacteria was thawed and streaked on solid medium composed of MH II agar supplemented with 1% bovine hemoglobin and 1% IsoVitaleX Enrichment (all from BD Biosciences, Lincoln Park, NJ). The bacteria were grown for 2 to 3 days at 37°C in a 5% CO2 /95% air environment to allow for formation of colonies. A single colony was inoculated into supplemented MH broth [23] and grown to late-log phase for 16 to 18 h at 37°C with shaking at 100 rpm in a 5% CO2 atmosphere. Aliquots of bacterial culture were centrifuged, and the pellets were resuspended in culture medium appropriate to the experiment being performed. E. coli strains S17-1 and DH5α were grown on Luria-Bertani (LB) plates or in LB broth at 37°C. Stocks of these two strains were maintained at −80°C in LB broth containing 10% glycerol.
Generation of LpnA-deficient F. tularensis LVS
The oligonucleotides 5’-ATTTGCGGCCGCGTCGACTTGTTCTGGTGTTTCAGCC-3’ (LpnA1), 5’-AGGTCTGCAGTCATCAGAGCCACCTAACCC-3’ (LpnA2), 5’GCAACTGCAGAACTTCTGTAGCGGTAAGTGGAC-3’ (LpnA3), and 5’ATTTGCGGCCGCGTCGACATCAATTTCATGCGTCGTAG-3’ (LpnA4) were used to amplify the upstream (LpnA1 and LpnA2) or downstream (LpnA3 and LpnA4) flanking regions of lpnA in F. tularensis LVS by PCR. Primers LpnA1 and LpnA4 incorporated both NotI and SalI restriction sites into their products (shown in bold). Primers LpnA2 and LpnA3 generated PstI restriction sites in their products. The amplification size for each set of primers was approximately 1500 bp, and the resulting products incorporated the first 98 bp and the last 87 bp of lpnA. These products were ligated directly into the pGEM-T Easy vector (Promega, Madison, WI) and electroporated into DH5α. Transformants were selected on LB agar supplemented with 100 µg/ml of ampicillin. Plasmid DNA then was isolated from the transformants using a Spin Miniprep Kit (Qiagen, Valencia, CA) and digested with NotI and PstI. The 1500-bp fragments corresponding to the upstream and downstream regions of lpnA were recovered with the Qiaquick Gel Extraction Kit (Qiagen) and ligated simultaneously into pPCR-Script (Stratagene, La Jolla, CA) that had been previously digested with the same two enzymes. The resultant plasmid, pPCR-ScriptΔlpnA, was recovered as described above. This protocol culminated in the joining of the upstream and downstream regions of lpnA, with the central region of the gene being absent. This insert then was digested out of pPCR-Script with SalI and ligated into SalI-digested pPV [7], which was provided by Anders Sjöstedt (Umeå University, Umeå, Sweden). The resultant mutagenesis plasmid was designated pPVΔlpnA. This plasmid then was used to generate an LpnA-deficient strain of F. tularensis, designated F. tularensis LVS ΔlpnA, by following a previously described conjugation protocol [7], with some modifications [14].
F. tularensis DNA microarray
RNA was prepared from overnight cultures of LpnA-deficient or wild-type F. tularensis LVS using an RNeasy Midi Kit (Qiagen). All subsequent steps were performed following protocols provided by The Institute for Genomic Research (TIGR) [54]. F. tularensis-specific microarray chips were obtained through the National Institute of Allergy and Infectious Diseases’ Pathogen Functional Genomics Research Center, managed and funded by the Division of Microbiology and Infectious Diseases, NIAID, NIH, DHHS and operated by TIGR.
Preparation of murine bone marrow-derived macrophages
Murine BMDM were obtained from the femurs of female C3H/HeN mice [55]. Bone marrow was collected from femurs flushed with Dulbecco’s Modified Eagle Medium (DMEM, Life Technologies Inc., Gaithersburg, MD) containing 5% heat-inactivated (HI) (56°C for 30 min) fetal bovine serum (FBS; HyClone, Logan UT) and washed with the same medium. After the final wash, the cells obtained were resuspended in DMEM containing 2 mM L-glutamine, 1 mM sodium pyruvate (both from Life Technologies), 20% HIFBS, and 30% medium previously conditioned by L929 cells. The conditioned medium was obtained by plating 2 × 105 L929 cells in 75-cm2 culture flasks in Minimal Essential Medium (Life Technologies) containing 2 mM L-glutamine, 1 mM sodium pyruvate, 1 mM non-essential amino acids (all three from Life Technologies), and 10% FBS and collecting the medium after 10 days. The bone marrow cells were then seeded in 100 mm-diameter dishes (Nunc, Naperville, IL) at a concentration of 4 × 106 cells per plate. After 5 days at 37°C, muBMDM were detached with ice-cold PBS, resuspended in culture medium, and seeded in 24-well plates at a concentration of 1.5 × 105 cells per well. The muBMDM were used for experiments the following day.
Infection of muBMDM
Murine BMDM grown in 24-well plates were exposed to F. tularensis LVS at a multiplicity of infection (MOI) of approximately 50 bacteria per cell. After 2 h of co-culture at 37°C, leukocytes were washed extensively and incubated with 5 µg/ml of gentamicin for 1 h to kill extracellular bacteria [23]. To measure intracellular content of viable F. tularensis LVS, some samples were lysed with 1.0 ml of water for 10 min. Serial dilutions were plated to determine the numbers of intracellular bacteria. Other infected cultures were incubated for an additional 21 h in antibiotic-free medium before lysis.
Immunofluorescent staining of intracellular bacteria
F. tularensis LVS organisms that had been internalized by murine leukocytes were visualized by indirect immunofluorescence. Murine BMDM were plated on 12 mm-diameter glass coverslips in 24-well dishes and incubated with F. tularensis LVS at a MOI of approximately 50 for 2 h. Some samples were washed and immediately fixed. Others were incubated for 1 h in medium containing 5 µg/ml of gentamicin, followed by an additional 21-h incubation in antibiotic-free medium before fixation. The cells were fixed with 2.5% paraformaldehyde (Electron Microscopy Sciences, Ft. Washington, PA) in PBS, permeabilized with 0.5% Triton X-100 (Sigma-Aldrich), and incubated with a polyclonal rabbit anti-serum to F. tularensis (BD Biosciences) for 30 min. Cells then were washed with PBS and incubated with fluorescein isothiocyanate-labeled goat anti-rabbit Ig (BD Biosciences) for 30 min. Coverslips were inverted onto slides containing Slowfade® mounting medium (Molecular Probes, Eugene, OR) and viewed by epifluorescence and phase contrast microscopy with a Zeiss Axioplan2 microscope equipped with a 40X objective. Images were captured using a Spot camera (Diagnostic Instruments, Inc., Sterling Heights, MI) and processed using Adobe Photoshop 6.0.
Infection of mice with F. tularensis
All studies with mice were approved by the Institutional Animal Care and Use Committee of Stony Brook University. Murine infections used 6 to 8 week-old C3H/HeN mice (Taconic Laboratories, Germantown, NY) infected via the intradermal or intranasal route. Mice infected intradermally were inoculated in groups of five with infectious doses of F. tularensis LVS or F. tularensis LVS ΔlpnA ranging from 105 to 107 total bacteria in a volume of 100 µl. An inoculum of 106 bacteria was used to compare dissemination of bacteria in mutant and wild-type mice. After 3 days, the liver, spleen, and one lung were removed from each mouse. Organs were weighed and homogenized in a Stomacher 80 Biomaster laboratory blender (Seward Ltd., Worthing, West Sussex, UK), and dilutions of the homogenates were plated on MH II agar to determine the number of bacteria per gram of tissue.
Mice inoculated intranasally were anaesthetized with a mixture of 100 mg/ml of Ketamine and 20 mg/ml of Xylazine (0.01 ml of mixture/gram of body weight, both from Henry Schein, Melville, NY), placed into groups of five, and infected with F. tularensis LVS or F. tularensis LVS ΔlpnA. Inocula of 106 wild-type or mutant F. tularensis organisms per 20 µl of PBS (Life Technologies) were generated, and 10 µl was applied to each nare for each mouse.
Immunoprotection of mice by wild-type and mutant F. tularensis
Groups of ten C3H/HeN mice were infected intradermally with a sublethal inoculum [4] consisting of 104 F. tularensis LVS or F. tularensis LVS ΔlpnA bacteria. As a control, five mice were administered a solution of sterile PBS. After 2 months, all mice received a second intradermal challenge with a normally lethal dose of 107 F. tularensis LVS organisms. Animals were monitored for an additional 4 weeks.
Preparation of recombinant LpnA
LpnA was amplified from the genome of F. tularensis LVS using the oligonucleotides 5’GACTGTCGACCGTAATGTTAGCTGTATCATC 3’ (LpnA5) and 5’GACTGCGGCCGCTTAGTGATGGTGATGGTGATGAATATTTATTGAATCAGAAGCGATTACTTC 3’ (LpnA6). LpnA5 incorporated an N-terminal SalI restriction site (in bold), and LpnA6 incorporated a C-terminal NotI restriction site (in bold) and a 6X-histidine tag (underlined). Amplified products were ligated into the pGEM-T Easy vector and transformed into E. coli DH5α, followed by purification of the LpnA-containing plasmid as described above. Next, the region corresponding to LpnA was excised using SalI and NotI and ligated into the pET30a vector (Novagen, Madison, WI). Ligation products were expressed in DH5α followed by purification of the recombinant vector, designated pET30a-LpnA. This vector then was sent to the Northeast Biodefense Center Protein Expression Core (Wadsworth Center, Albany, NY) for purification of LpnA by Triton X-114 phase partitioning [56]. This method has been previously employed to isolate functional, acylated proteins [44,56]. Potential contaminating E. coli LPS was removed from the LpnA preparations using Endotoxin Removal Affinity Resin (Cape Cod Associates, East Falmouth, MA). The QCL-1000 Chromogenic Limulus Amebocyte Lysate Endpoint Assay (Cambrex, East Rutherford, NJ) confirmed that only a trace amount of LPS (4 pg per µg of LpnA) remained. This amount of LPS was too small to stimulate HUVEC or macrophages (data not shown). Nonetheless, 20 µg/ml of the LPS inhibitor polymyxin B (Sigma-Aldrich, St. Louis, MO) was included in all samples containing recombinant LpnA in all experiments.
Preparation of human monocyte-derived macrophages
Monocytes were obtained from the venous blood of healthy adult volunteer donors, using 0.12% disodium EDTA as an anticoagulant. Leukocytes were separated by density gradient centrifugation using Accu-Prep™ Lymphocytes (Accurate Chemical Co., Westbury, NY). Monocytes were negatively selected by immunomagnetic sorting using the Monocyte Isolation Kit II (Miltenyi Biotec, Auburn, CA) according to the instructions of the manufacturer. Isolated monocytes were resuspended in RPMI medium (Life Technologies) containing 10% HIFBS, added to 24-well plates at a concentration of 2.0 × 105 cells per well, and cultured for 5 to 7 days in the presence of 10 ng/ml of recombinant human macrophage colony-stimulating factor (Sigma-Aldrich) to induce formation of huMDM.
Culture of endothelial cells
Endothelial cells were isolated from human umbilical veins via collagenase perfusion and grown to confluence in 60-mm dishes in a 37°C, 5% CO2 /95% air environment in Medium 199 (M199; Life Technologies) supplemented with 20% HIFBS, 100 U/ml of penicillin, 100 µg/ml of streptomycin, and 2 µg/ml of amphotericin B [57]. Cells were cultured for 3 to 5 days, trypsinized, and passaged onto 48- or 96-well plates (both from BD Biosciences) at a concentration of 2.3 × 105 cells/ml.
Quantitation of cytokines
To assess production of cytokines by huMDM or HUVEC, cells were treated for 24 h with preparations of live or gentamicin-killed (50 µg/ml for 1 h) F. tularensis, recombinant LpnA, E. coli LPS (serotype 0111:B4, Sigma-Aldrich), a sham preparation of bacteria consisting of uninoculated MH broth processed in the same fashion as the bacterial culture, or control medium. Conditioned media were collected and clarified by centrifugation. Amounts of CXCL8, CCL2, or IL-1β in the supernatants were quantitated using commercial enzyme-linked immunosorbent assay (ELISA) kits (Antigenix America, Inc., Franklin Square, NY).
Quantitation of E-selectin
The ability of recombinant LpnA to induce expression of E-selectin on HUVEC was evaluated by whole-cell ELISA. HUVEC were incubated with 100 µl of control medium (M199 with 20% HIFBS), LpnA, or E. coli LPS at 37°C for 4 h. ELISAs to detect surface expression of E-selectin on living endothelial monolayers then were performed as described previously [58].
Blocking of TLR2
To assess whether LpnA stimulates cells via TLR2, a murine neutralizing monoclonal antibody to human TLR2 (clone TLR2.1; eBioscience, San Diego, CA) was used in some experiments. Human MDM were cultured with medium alone, 20 µg/ml of antibody, or 20 µg/ml of an isotype-matched control antibody (mouse IgG2a, κ; eBiosciences) for 30 min. An equal volume of medium containing stimuli then was added, so that 10 µg/ml of antibody remained throughout the experiment. Measurement of the production of E-selectin, CXCL8, or CCL2 was performed as described above. A similar protocol was used to determine the ability of the antibody to inhibit stimulation of huMDM by 1 ng/ml of tripalmitoyl-Cys-Ser-(Lys)4 (EMC Microcollections, Tübingen, Germany), a known agonist for TLR2, or 10 ng/ml of E. coli LPS, which signals through TLR4 [51].
Statistics
To determine statistical significance, data were subjected to an unpaired analysis of variance and the Tukey-Kramer multiple-comparison test using Instat version 3.01 (GraphPad Software, San Diego, CA).
Acknowledgements
This work was supported by National Institutes of Health Grant P01 AI055621.
We thank Chandra Shekhar Bakshi, Vitaly Pavlov, and Timothy Sellati for their assistance with the techniques utilized to create a defined gene deletion in F. tularensis. We acknowledge assistance with protein purification from the Northeast Biodefense Center Protein Expression Core at the Wadsworth Center (Northeast Biodefense Center U54-AI057158-Lipkin). We thank Gloria Monsalve and Patricio Mena for their assistance in the handling of mice. We thank Anders Sjöstedt for kindly providing us with the pPV plasmid.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tärnvik A, Berglund L. Tularaemia. Eur Respir J. 2003;21:361–373. doi: 10.1183/09031936.03.00088903. [DOI] [PubMed] [Google Scholar]
- 2.Ellis J, Oyston PCF, Green M, Titball RW. Tularemia. Clin Microbiol Rev. 2002;15:631–646. doi: 10.1128/CMR.15.4.631-646.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tigertt WD. Soviet viable Pasteurella tularensis vaccines. A review of selected articles. Bacteriol Rev. 1962;26:354–373. doi: 10.1128/br.26.3.354-373.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fortier AH, Slayter MV, Ziemba R, Meltzer MS, Nacy CA. Live vaccine strain of Francisella tularensis: infection and immunity in mice. Infect Immun. 1991;59:2922–2928. doi: 10.1128/iai.59.9.2922-2928.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Golovliov I, Ericsson M, Sandström G, Tärnvik A, Sjöstedt A. Identification of proteins of Francisella tularensis induced during growth in macrophages and cloning of the gene encoding a prominently induced 23-kilodalton protein. Infect Immun. 1997;65:2183–2189. doi: 10.1128/iai.65.6.2183-2189.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gray CG, Cowley SC, Cheung KK, Nano FE. The identification of five genetic loci of Francisella novicida associated with intracellular growth. FEMS Microbiol Lett. 2002;215:53–56. doi: 10.1111/j.1574-6968.2002.tb11369.x. [DOI] [PubMed] [Google Scholar]
- 7.Golovliov I, Sjöstedt A, Mokrievich A, Pavlov V. A method for allelic replacement in Francisella tularensis. FEMS Microbiol Lett. 2003;222:273–280. doi: 10.1016/S0378-1097(03)00313-6. [DOI] [PubMed] [Google Scholar]
- 8.Twine S, Byström M, Chen W, Forsman M, Golovliov I, Johansson A, et al. A mutant of Francisella tularensis strain SCHU S4 lacking the ability to express a 58-kilodalton protein is attenuated for virulence and is an effective live vaccine. Infect Immun. 2005;73:8345–8352. doi: 10.1128/IAI.73.12.8345-8352.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nano FE, Zhang N, Cowley SC, Klose KE, Cheung KK, Roberts MJ, et al. A Francisella tularensis pathogenicity island required for intramacrophage growth. J Bacteriol. 2004;186:6430–6436. doi: 10.1128/JB.186.19.6430-6436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lauriano CM, Barker JR, Yoon SS, Nano FE, Arulanandam BP, Hassett DJ, et al. MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoebae and intramacrophage survival. Proc Natl Acad Sci U S A. 2004;101:4246–4249. doi: 10.1073/pnas.0307690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pizarro-Cerdá J, Cossart P. Bacterial adhesion and entry into host cells. Cell. 2006;124:715–727. doi: 10.1016/j.cell.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 12.Gil H, Benach JL, Thanassi DG. Presence of pili on the surface of Francisella tularensis. Infect Immun. 2004;72:3042–3047. doi: 10.1128/IAI.72.5.3042-3047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forslund AL, Kuoppa K, Svensson K, Salomonsson E, Johansson A, Byström M, et al. Direct repeat-mediated deletion of a type IV pilin gene results in major virulence attenuation of Francisella tularensis. Mol Microbiol. 2006;59:1818–1830. doi: 10.1111/j.1365-2958.2006.05061.x. [DOI] [PubMed] [Google Scholar]
- 14.Gil H, Platz GJ, Forestal CA, Monfett M, Bakshi CS, Sellati TJ, et al. Deletion of TolC orthologs in Francisella tularensis identifies roles in multidrug resistance and virulence. Proc Natl Acad Sci U S A. 2006;103:12897–12902. doi: 10.1073/pnas.0602582103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vinogradov E, Perry MB, Conlan JW. Structural analysis of Francisella tularensis lipopolysaccharide. Eur J Biochem. 2002;269:6112–6118. doi: 10.1046/j.1432-1033.2002.03321.x. [DOI] [PubMed] [Google Scholar]
- 16.Chen W, KuoLee R, Shen H, Bùsa M, Conlan JW. Toll-like receptor 4 (TLR4) does not confer a resistance advantage on mice against low-dose aerosol infection with virulent type A Francisella tularensis. Microb Pathog. 2004;37:185–191. doi: 10.1016/j.micpath.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 17.Ancuta P, Pedron T, Girard R, Sandström G, Chaby R. Inability of the Francisella tularensis lipopolysaccharide to mimic or to antagonize the induction of cell activation by endotoxins. Infect Immun. 1996;64:2041–2046. doi: 10.1128/iai.64.6.2041-2046.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kieffer TL, Cowley S, Nano FE, Elkins KL. Francisella novicida LPS has greater immunobiological activity in mice than F. tularensis LPS, and contributes to F. novicida murine pathogenesis. Microbes Infect. 2003;5:397–403. doi: 10.1016/s1286-4579(03)00052-2. [DOI] [PubMed] [Google Scholar]
- 19.Golovliov I, Sandström G, Ericsson M, Sjöstedt A, Tärnvik A. Cytokine expression in the liver during the early phase of murine tularemia. Infect Immun. 1995;63:534–538. doi: 10.1128/iai.63.2.534-538.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cole LE, Elkins KL, Michalek SM, Qureshi N, Eaton LJ, Rallabhandi P, et al. Immunologic consequences of Francisella tularensis live vaccine strain infection: role of the innate immune response in infection and immunity. J Immunol. 2006;176:6888–6899. doi: 10.4049/jimmunol.176.11.6888. [DOI] [PubMed] [Google Scholar]
- 21.Katz J, Zhang P, Martin M, Vogel SN, Michalek SM. Toll-like receptor 2 is required for inflammatory responses to Francisella tularensis LVS. Infect Immun. 2006;74:2809–2816. doi: 10.1128/IAI.74.5.2809-2816.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cole LE, Shirey KA, Barry E, Santiago A, Rallabhandi P, Elkins KL, et al. Toll-like receptor 2-mediated signaling requirements for Francisella tularensis live vaccine strain infection of murine macrophages. Infect Immun. 2007;75:4127–4137. doi: 10.1128/IAI.01868-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forestal CA, Benach JL, Carbonara C, Italo JK, Lisinski TJ, Furie MB. Francisella tularensis selectively induces proinflammatory changes in endothelial cells. J Immunol. 2003;171:2563–2570. doi: 10.4049/jimmunol.171.5.2563. [DOI] [PubMed] [Google Scholar]
- 24.Bolger CE, Forestal CA, Italo JK, Benach JL, Furie MB. The live vaccine strain of Francisella tularensis replicates in human and murine macrophages but induces only the human cells to secrete proinflammatory cytokines. J Leukoc Biol. 2005;77:893–897. doi: 10.1189/jlb.1104637. [DOI] [PubMed] [Google Scholar]
- 25.Li H, Nookala S, Bina XR, Bina JE, Re F. Innate immune response to Francisella tularensis is mediated by TLR2 and caspase-1 activation. J Leukoc Biol. 2006;80:766–773. doi: 10.1189/jlb.0406294. [DOI] [PubMed] [Google Scholar]
- 26.Malik M, Bakshi CS, Sahay B, Shah A, Lotz SA, Sellati TJ. Toll-like receptor 2 is required for control of pulmonary infection with Francisella tularensis. Infect Immun. 2006;74:3657–3662. doi: 10.1128/IAI.02030-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sjöstedt A, Tärnvik A, Sandström G. The T-cell-stimulating 17-kilodalton protein of Francisella tularensis LVS is a lipoprotein. Infect Immun. 1991;59:3163–3168. doi: 10.1128/iai.59.9.3163-3168.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sjöstedt A, Kuoppa K, Johansson T, Sandström G. The 17 kDa lipoprotein and encoding gene of Francisella tularensis LVS are conserved in strains of Francisella tularensis. Microb Pathog. 1992;13:243–249. doi: 10.1016/0882-4010(92)90025-j. [DOI] [PubMed] [Google Scholar]
- 29.Huntley JF, Conley PG, Hagman KE, Norgard MV. Characterization of Francisella tularensis outer membrane proteins. J Bacteriol. 2007;189:561–574. doi: 10.1128/JB.01505-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sjöstedt A, Sandström G, Tärnvik A, Jaurin B. Molecular cloning and expression of a T-cell stimulating membrane protein of Francisella tularensis. Microb Pathog. 1989;6:403–414. doi: 10.1016/0882-4010(89)90082-x. [DOI] [PubMed] [Google Scholar]
- 31.Sjöstedt A, Sandström G, Tärnvik A, Jaurin B. Nucleotide sequence and T cell epitopes of a membrane protein of Francisella tularensis. J Immunol. 1990;145:311–317. [PubMed] [Google Scholar]
- 32.Sjöstedt A, Sandström G, Tärnvik A. Humoral and cell-mediated immunity in mice to a 17-kilodalton lipoprotein of Francisella tularensis expressed by Salmonella typhimurium. Infect Immun. 1992;60:2855–2862. doi: 10.1128/iai.60.7.2855-2862.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anthony LSD, Burke RD, Nano FE. Growth of Francisella spp. in rodent macrophages. Infect Immun. 1991;59:3291–3296. doi: 10.1128/iai.59.9.3291-3296.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lai XH, Golovliov I, Sjöstedt A. Francisella tularensis induces cytopathogenicity and apoptosis in murine macrophages via a mechanism that requires intracellular bacterial multiplication. Infect Immun. 2001;69:4691–4694. doi: 10.1128/IAI.69.7.4691-4694.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sperandio M. Selectins and glycosyltransferases in leukocyte rolling in vivo. FEBS J. 2006;273:4377–4389. doi: 10.1111/j.1742-4658.2006.05437.x. [DOI] [PubMed] [Google Scholar]
- 36.Luster AD. Chemokines - chemotactic cytokines that mediate inflammation. New Engl J Med. 1998;338:436–445. doi: 10.1056/NEJM199802123380706. [DOI] [PubMed] [Google Scholar]
- 37.Duenas AI, Aceves M, Orduna A, Diaz R, Sanchez CM. Garcia-Rodriguez C. Francisella tularensis LPS induces the production of cytokines in human monocytes and signals via Toll-like receptor 4 with much lower potency than E. coli LPS. Int Immunol. 2006;18:785–795. doi: 10.1093/intimm/dxl015. [DOI] [PubMed] [Google Scholar]
- 38.Hayashi S, Wu HC. Lipoproteins in bacteria. J Bioenerg Biomembr. 1990;22:451–471. doi: 10.1007/BF00763177. [DOI] [PubMed] [Google Scholar]
- 39.Madan Babu M, Sankaran K. DOLOP--database of bacterial lipoproteins. Bioinformatics. 2002;18:641–643. doi: 10.1093/bioinformatics/18.4.641. [DOI] [PubMed] [Google Scholar]
- 40.Juncker AS, Willenbrock H, Von Heijne G, Brunak S, Nielsen H, Krogh A. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 2003;12:1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sellati TJ, Abrescia LD, Radolf JD, Furie MB. Outer surface lipoproteins of Borrelia burgdorferi activate vascular endothelium in vitro. Infect Immun. 1996;64:3180–3187. doi: 10.1128/iai.64.8.3180-3187.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weis JJ, Ma Y, Erdile LF. Biological activities of native and recombinant Borrelia burgdorferi outer surface protein A: dependence on lipid modification. Infection and Immunity. 1994;62:4632–4636. doi: 10.1128/iai.62.10.4632-4636.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Templeton TJ. Borrelia outer membrane surface proteins and transmission through the tick. J Exp Med. 2004;199:603–606. doi: 10.1084/jem.20040033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giambartolomei GH, Zwerdling A, Cassataro J, Bruno L, Fossati CA, Philipp MT. Lipoproteins, not lipopolysaccharide, are the key mediators of the proinflammatory response elicited by heat-killed Brucella abortus. J Immunol. 2004;173:4635–4642. doi: 10.4049/jimmunol.173.7.4635. [DOI] [PubMed] [Google Scholar]
- 45.Larsson P, Oyston PC, Chain P, Chu MC, Duffield M, Fuxelius HH, et al. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat Genet. 2005;37:153–159. doi: 10.1038/ng1499. [DOI] [PubMed] [Google Scholar]
- 46.Forestal CA, Malik M, Catlett SV, Savitt AG, Benach JL, Sellati TJ, et al. Francisella tularensis has a significant extracellular phase in infected mice. J Infect Dis. 2007;196:134–137. doi: 10.1086/518611. [DOI] [PubMed] [Google Scholar]
- 47.Lauriano CM, Barker JR, Nano FE, Arulanandam BP, Klose KE. Allelic exchange in Francisella tularensis using PCR products. FEMS Microbiol Lett. 2003;229:195–202. doi: 10.1016/S0378-1097(03)00820-6. [DOI] [PubMed] [Google Scholar]
- 48.Sjöstedt A, Sandström G, Tärnvik A. Immunization of mice with an attenuated Salmonella typhimurium strain expressing a membrane protein of Francisella tularensis. A model for identification of bacterial determinants relevant to the host defence against tularemia. Res Microbiol. 1990;141:887–891. doi: 10.1016/0923-2508(90)90126-b. [DOI] [PubMed] [Google Scholar]
- 49.Dreisbach VC, Cowley S, Elkins KL. Purified lipopolysaccharide from Francisella tularensis live vaccine strain (LVS) induces protective immunity against LVS infection that requires B cells and gamma interferon. Infect Immun. 2000;68:1988–1996. doi: 10.1128/iai.68.4.1988-1996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fulop M, Manchee R, Titball R. Role of lipopolysaccharide and a major outer membrane protein from Francisella tularensis in the induction of immunity against tularemia. Vaccine. 1995;13:1220–1225. doi: 10.1016/0264-410x(95)00062-6. [DOI] [PubMed] [Google Scholar]
- 51.Akira S, Sato S. Toll-like receptors and their signaling mechanisms. Scand J Infect Dis. 2003;35:555–562. doi: 10.1080/00365540310015683. [DOI] [PubMed] [Google Scholar]
- 52.Chen W, KuoLee R, Shen H, Bùsa M, Conlan JW. Toll-like receptor 4 (TLR4) plays a relatively minor role in murine defense against primary intradermal infection with Francisella tularensis LVS. Immunol Lett. 2005;97:151–154. doi: 10.1016/j.imlet.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 53.Zhang H, Niesel DW, Peterson JW, Klimpel GR. Lipoprotein release by bacteria: potential factor in bacterial pathogenesis. Infect Immun. 1998;66:5196–5201. doi: 10.1128/iai.66.11.5196-5201.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hegde P, Qi R, Abernathy K, Gay C, Dharap S, Gaspard R, et al. A concise guide to cDNA microarray analysis. Biotechniques. 2000;29:548–556. doi: 10.2144/00293bi01. [DOI] [PubMed] [Google Scholar]
- 55.Celada A, Gray PW, Rinderknecht E, Schreiber RD. Evidence for a gamma-interferon receptor that regulates macrophage tumoricidal activity. J Exp Med. 1984;160:55–74. doi: 10.1084/jem.160.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Radolf JD, Chamberlain NR, Clausell A, Norgard MV. Identification and localization of integral membrane proteins of virulent Treponema pallidum subsp. pallidum by phase partitioning with the nonionic detergent triton X-114. Infect Immun. 1988;56:490–498. doi: 10.1128/iai.56.2.490-498.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang AJ, Furie MB, Nicholson SC, Fischbarg J, Liebovitch LS, Silverstein SC. Effects of human neutrophil chemotaxis across endothelial cell monolayers on the permeability of these monolayers to ions and macromolecules. J Cell Physiol. 1988;135:355–366. doi: 10.1002/jcp.1041350302. [DOI] [PubMed] [Google Scholar]
- 58.Sellati TJ, Burns MJ, Ficazzola MA, Furie MB. Borrelia burgdorferi upregulates expression of adhesion molecules on endothelial cells and promotes transendothelial migration of neutrophils in vitro. Infect Immun. 1995;63:4439–4447. doi: 10.1128/iai.63.11.4439-4447.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]


