Abstract
Although participation of PEROXIN11 (PEX11), FISSION1 (FISl), and DYNAMIN-RELATED PROTEIN (DRP) has been well established during induced peroxisome proliferation in response to external stimuli, their roles in cell cycle–associated constitutive replication/duplication have not been fully explored. Herein, bimolecular fluorescence complementation experiments with Arabidopsis thaliana suspension cells revealed homooligomerization of all five PEX11 isoforms (PEX11a-e) and heterooligomerizations of all five PEX11 isoforms with FIS1b, but not FIS1a nor DRP3A. Intracellular protein targeting experiments demonstrated that FIS1b, but not FIS1a nor DRP3A, targeted to peroxisomes only when coexpressed with PEX11d or PEX11e. Simultaneous silencing of PEX11c-e or individual silencing of DRP3A, but not FIS1a nor FIS1b, resulted in ∼40% reductions in peroxisome number. During G2 in synchronized cell cultures, peroxisomes sequentially enlarged, elongated, and then doubled in number, which correlated with peaks in PEX11, FIS1, and DRP3A expression. Overall, these data support a model for the replication of preexisting peroxisomes wherein PEX11c, PEX11d, and PEX11e act cooperatively during G2 to promote peroxisome elongation and recruitment of FIS1b to the peroxisome membrane, where DRP3A stimulates fission of elongated peroxisomes into daughter peroxisomes, which are then distributed between daughter cells.
INTRODUCTION
Peroxisomes are relatively small, pleomorphic, single-membrane-bound organelles that house a wide variety of critical metabolic pathways, including fatty acid β-oxidation, auxin and jasmonate metabolism, and photorespiration reactions (Zolman et al., 2000; Afitlhile et al., 2005; Mano and Nishimura, 2005; Nyathi and Baker, 2006; Reumann and Weber, 2006; Theodoulou et al., 2006). New peroxisomes can either be derived de novo from the endoplasmic reticulum (ER) (Tabak et al., 2003; Hoepfner et al., 2005; Kim et al., 2006) or from fission of preexisting peroxisomes (Schrader and Fahimi, 2006; Motley and Hettema, 2007). Preexisting peroxisomes can increase in number through at least two partially overlapping mechanisms: (1) proliferation, the substantive increase in peroxisome number in response to external stimuli; or (2) replication/duplication, the binary fission of peroxisomes in response to cues from the cell cycle machinery (Yan et al., 2005; Schrader and Fahimi, 2006). During both proliferation and replication, key molecules are required to stimulate fission of the organelle. The mechanics of these fission events have been the recent focus of an extensive body of research in plants, yeast, and mammals (Mano et al., 2004; Thoms and Erdmann, 2005; Yan et al., 2005; Lingard and Trelease, 2006; Schrader and Fahimi, 2006; Fagarasanu et al., 2007; Orth et al., 2007).
At least three classes of proteins, including PEROXIN11 (PEX11), DYNAMIN-RELATED PROTEINs (DRPs), and FISSION1 (FIS1) isoforms are required for peroxisome fission. The PEX11 family in mammals is composed of three members (PEX11α, PEX11β, and PEX11γ) (Abe et al., 1998; Schrader et al., 1998b; Li et al., 2002a, 2002b; Li and Gould, 2002) and yeasts (Pex11p, Pex25p, and Pex27p) (Marshall et al., 1995; Smith et al., 2002; Rottensteiner et al., 2003; Tam et al., 2003). By contrast, the PEX11 family in Arabidopsis thaliana is composed of five members (PEX11a, PEX11b, PEX11c, PEX11d, and PEX11e) (Lingard and Trelease, 2006), which can be separated into three groups based on sequence homology: PEX11a, PEX11b, and PEX11c-e (Lingard and Trelease, 2006).
Overexpression of PEX11 homologs in mammals, plants, trypanosomes, and fungi leads to profound increases in peroxisome number, suggestive of a role for PEX11 proteins in the promotion of peroxisome fission (Marshall et al., 1995; Abe et al., 1998; Lorenz et al., 1998; Schrader et al., 1998b; Li et al., 2002a, 2002b; Li and Gould, 2002; Smith et al., 2002; Rottensteiner et al., 2003; Tam et al., 2003; Lingard and Trelease, 2006; Orth et al., 2007). Further evidence for the involvement of these proteins in peroxisome fission comes from cells lacking PEX11 isoforms. Saccharomyces cerevisiae, Candidia boidinii, or Mus musculus cells lacking one or more PEX11 homolog exhibit reduced numbers of enlarged and/or elongated peroxisomes (Marshall et al., 1995; Sakai et al., 1995; Li et al., 2002a, 2002b; Li and Gould, 2002; Rottensteiner et al., 2003; Tam et al., 2003). In Arabidopsis plants, individual silencing of PEX11a and PEX11b, as well as simultaneous silencing of PEX11c, PEX11d, and PEX11e, leads to decreases of up to 75% in peroxisome number (Orth et al., 2007). Furthermore, simultaneous silencing of PEX11c, PEX11d, and PEX11e can also lead to dramatic increases in peroxisome size, and simultaneous silencing of PEX11a and PEX11b can lead to slight increases in peroxisome size (Nito et al., 2007).
DRPs are large GTPases that likely are the direct actuators of mitochondrion and peroxisome fission (Thoms and Erdmann, 2005). Mammalian, yeast, or plant cells lacking the appropriate DRP homologs (DLP1, Vps1p/Dnm1p, or DRP3A, respectively) have dramatically elongated peroxisomes, whereas overexpression of mammalian and yeast versions has no effect on peroxisome abundance (Hoepfner et al., 2001; Koch et al., 2003, 2004; Li and Gould, 2003; Kuravi et al., 2006). DRPs lack the pleckstrin homology domain, which is necessary for membrane binding in standard dynamins (Thoms and Erdmann, 2005); thus, DRP associations with organelle membranes are mediated through interactions with tether proteins.
The mitochondrial and peroxisomal membrane tether for mammalian DLP1 is FIS1 (Yoon et al., 2003; Koch et al., 2005; Yu et al., 2005; Schrader and Yoon, 2007). FIS1 homologs possess only a single membrane-spanning domain located near their C-terminal and thus are considered members of the tail-anchored (Nout-Cin) family of membrane proteins (Borgese et al., 2007). Human and S. cerevisiae FIS1 targets to both peroxisomes and mitochondria (Koch et al., 2005; Kuravi et al., 2006). Silencing or deletion of these proteins results in a decrease in the number of peroxisomes, whereas overexpression of human Fis1 promotes an increase in peroxisome number. In Arabidopsis, two genes, FIS1a and FIS1b, are homologous to yeast and mammalian FIS1. Although mutants with T-DNA insertions in FIS1a exhibit decreased numbers of mitochondria (Scott et al., 2006), it is not known whether FIS1b participates in mitochondrial division nor whether FIS1a or FIS1b is involved in the fission of plant peroxisomes.
The sequence of events leading to peroxisome proliferation is best characterized in mammalian cells and involves a well-defined sequence of morphological changes, including sequential elongation, constriction, and, ultimately, fission (Schrader et al., 1998b; Schrader and Fahimi, 2006). Interactive roles of mammalian PEX11, FIS1, and DRP during peroxisome proliferation are beginning to emerge. Overexpression of PEX11β or FIS1 in human fibroblasts lacking DLP1 promotes peroxisome elongation and constriction, but not multiplication, suggesting that both PEX11β and FIS1 act upstream of DLP1 (Li and Gould, 2003; Koch et al., 2004, 2005). Furthermore, immunoprecipitation experiments suggest that human FIS1 and DLP1, but not PEX11β and DLP1, directly interact with each other (Yoon et al., 2003; Koch et al., 2005). Likewise, in Chinese hamster ovary cells, PEX11β, FIS1, and DLP1 occur in a trimeric complex wherein the association of PEX11β with DLP1 is mediated through interaction with FIS1 (Kobayashi et al., 2007). By contrast, experimental evidence is virtually absent for any mechanistic relationships among these proteins during cell cycle–related peroxisome replication. Furthermore, it has not been established whether molecular mechanisms similar to those cited above for mammalian peroxisomes are engaged in either proliferation or cell cycle replication of plant peroxisomes.
We sought to ascertain the physical and temporal relationships among PEX11 isoforms a-e, FIS1 isoforms a and b, and DRP3A in preexisting peroxisomes undergoing replication in dividing Arabidopsis cells. We report that physical interactions exist among all five PEX11 isoforms and FIS1b, but not with FIS1a or DRP3A. We show also that PEX11c, PEX11d, PEX11e, FIS1b, and DRP3A play distinct yet overlapping roles during peroxisome replication wherein PEX11 isoforms promote peroxisome elongation and recruitment of FIS1b to the peroxisome membrane. FIS1b likely then recruits DRP3A to the peroxisome membrane, stimulating fission, the final step of peroxisome replication. Overall, our data not only corroborate current models that portray a fission process as being responsible for the replication of preexisting peroxisomes (Mullen and Trelease, 2006; Schrader and Fahimi, 2006; Fagarasanu et al., 2007), but additionally provide new mechanistic evidence for cooperative interactions that accomplish cell cycle–associated constitutive self-replication of preexisting peroxisomes. This semiautonomous peroxisome growth and division process is envisaged as the essential means for maintaining peroxisome homeostasis in actively growing and dividing plant cells.
RESULTS
Subcellular Localizations of FIS1a, FIS1b, and DRP3A
To examine the subcellular localization of FIS1a, FIS1b, and DRP3A, N-terminal myc epitope–tagged versions of each protein were transiently expressed individually or in combinations with PEX11 isoforms for ∼16 h within Arabidopsis suspension cells (Figure 1). We reported previously that all five PEX11 isoforms were peroxisomal membrane proteins (Lingard and Trelease, 2006). Because FIS1a and FIS1b are likely tail-anchored membrane proteins, which normally possess C-terminal membrane targeting signals (Borgese et al., 2007), the myc epitope was appended to their N termini to avoid interfering with putative targeting information. The myc epitope was also appended to the N terminus of DRP3A to mirror the N-terminal labeling with the green fluorescent protein (GFP-DRP3A) used for previous localization studies (Arimura and Tsutsumi, 2002; Mano et al., 2004).
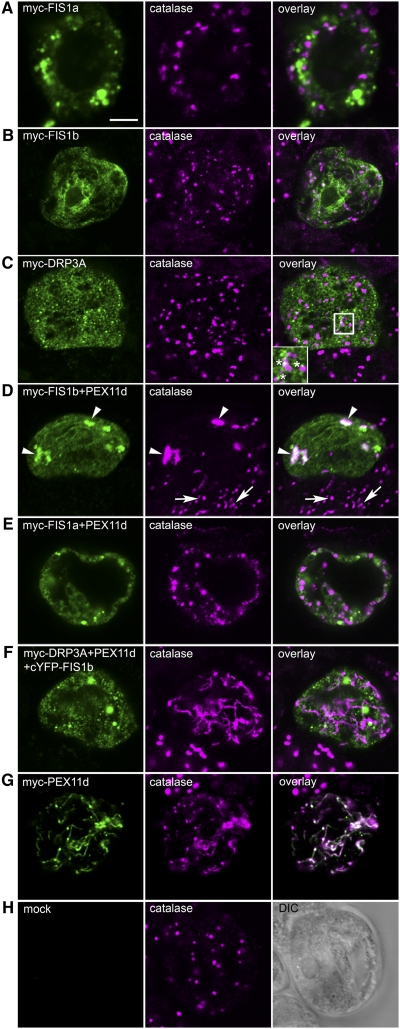
Figure 1.
Confocal Epifluorescence Images Illustrating Subcellular Localizations of Transiently Expressed FIS1a, FIS1b, and/or DRP3A in Biolistically Transformed Arabidopsis Suspension Cells.
Each panel shows a single optical section of a transformed and immunolabeled cell and is representative of ≥25 cells from at least two independent biolistic bombardments. After bombardment, cells were held for 16 h for gene expression and protein sorting, fixed in formaldehyde, then dual immunolabeled with anti-myc (green) and anticatalase (magenta) antibodies. Protein colocalizations, if present, are depicted in the far right overlay column as white images due to the overlay of the two fluorophores in the same cell. Bar = 10 μm.
(A) to (C) Single-transformed cells overexpressing myc-FIS1a (A), myc-FIS1b (B), or myc-DRP3A (C). The box in the overlay panel of (C) outlines the portion of the cell cytoplasm shown at higher magnification in the inset. Asterisks in (C) indicate examples of the close associations of punctate structures containing either expressed myc-DRP3A (green) or endogenous peroxisomal catalase (magenta).
(D) and (E) Dual-transformed cells overexpressing myc-FIS1b and PEX11d (D) or myc-FIS1a and PEX11d (E). Arrowheads in (D) point to aggregated peroxisomes in the transformed cell, whereas arrows point to normal peroxisomes in neighboring untransformed cells.
(F) Triple-transformed cell overexpressing myc-DRP3A, cYFP-FIS1b, and PEX11d.
(G) Single-transformed cell overexpressing myc-PEX11d.
(H) Mock cell transformed with empty vector (pRTL2-myc). DIC, differential interference contrast image.
The subcellular localization of each epitope-tagged protein was assessed in individual formaldehyde-fixed cells via confocal epifluorescence microscopy of antigen-bound anti-myc antibodies. Anticatalase antibodies were also used as markers for peroxisomes. Figures 1A and 1B show that myc-FIS1a and myc-FIS1b sorted mostly to punctate and perinuclear reticular structures, respectively, neither of which colocalized with endogenous catalase in peroxisomes (see merged image overlay). By contrast, Figure 1C illustrates that myc-DRP3A sorted to both the cytosol and small punctate structures that were dispersed throughout the cytoplasm. A subset of these myc-DRP3A-containing punctate structures was located immediately adjacent to individual peroxisomes (e.g., asterisks in Figure 1C overlay inset).
To further explore the subcellular localization(s) of myc-tagged FIS1a, FIS1b, and DRP3A, suspension cells were either cotransformed with the mitochondrial marker protein MITO-GFP (consisting of the F1-ATPase β-subunit N-terminal presequence linked to GFP) or labeled with fluorophore-conjugated Concanavalin A, an ER marker. Although myc-FIS1a did not colocalize with either the mitochondrial or ER marker, myc-FIS1b partially colocalized with the ER marker (see Supplemental Figure 1A online) but not with the mitochondrial marker. On the other hand, the small punctate structures containing myc-DRP3A frequently were located immediately adjacent to individual MITO-GFP-containing mitochondria (see Supplemental Figure 1B online). This juxtaposition of myc-DRP3A with mitochondria and peroxisomes (Figure 1C; see Supplemental Figure 1B online) is similar to the localizations reported previously for GFP-DRP3A in transiently transformed Arabidopsis roots (Mano et al., 2004) and tobacco (Nicotiana tabacum) suspension cells (Arimura and Tsutsumi, 2002), suggesting that the protein targets in a similar manner in all three cell types.
During the transient transformation experiments, we noted that cells expressing myc-FIS1b consistently possessed an increased number of spherical, often smaller peroxisomes (cf. catalase images in Figure 1B with 1A and 1H), consistent with a role for this protein in peroxisome fission as found in yeast and mammalian cells (Yoon et al., 2003; Koch et al., 2005; Yu et al., 2005). However, it is curious that myc-FIS1b is not associated with peroxisomes in our suspension cells, suggesting that the change in peroxisome number might reflect sorting to peroxisomes of an immunologically undetectable portion of the total expressed myc-FIS1b.
Co-overexpression experiments were conducted to test whether functional interactions occurred at the peroxisome membrane between PEX11d or PEX11e, which promote peroxisome elongation or an increase in number when expressed singly (Figure 1G; Lingard and Trelease, 2006), and FIS1a, FIS1b, or DRP3A. Figure 1D illustrates representative results of coexpressing myc-FIS1b with untagged PEX11d. In these cells, myc-FIS1b consistently colocalized with catalase within a few relatively large cytoplasmic structures (Figure 1D, arrowheads), distinctly different in size and number from the individual normal-appearing peroxisomes visible in the neighboring nontransformed cells (Figure 1D, arrows) or in mock-transformed cells (Figure 1H) or from the elongated peroxisomes observed in cells overexpressing myc-PEX11d alone (Figure 1G). The abnormally large structures in Figure 1D, possessing catalase, myc-FIS1b, and PEX11d likely resulted from peroxisome aggregation, as we previously observed in cells overexpressing peroxisome membrane proteins (Mullen et al., 2001; Lisenbee et al., 2003; Murphy et al., 2003; McCartney et al., 2005). In parallel experiments, coexpressions of myc-FIS1b with untagged PEX11e also resulted in the apparent recruitment of myc-FIS1b to aggregated peroxisomes. However, coexpressions of myc-FIS1a with PEX11d (Figure 1E) or with PEX11e did not result in the recruitment of myc-FIS1a to peroxisomes. Instead, myc-FIS1a sorted to unidentified reticulate and globular structures, the latter being morphologically similar to those observed when myc-FIS1a was expressed on its own (cf. myc-FIS1a images in Figures 1A and 1E).
Coexpression of individual untagged PEX11d or PEX11e isoforms with myc-DRP3A resulted in sorting of myc-DRP3A to the cytosol and small punctate structures similar to its localization shown in Figure 1C. Similarly, results of triple overexpression experiments with myc-DRP3A, cYFP-FIS1b, and untagged PEX11d (Figure 1F), or untagged PEX11e, revealed the same localization of myc-DRP3A to the cytosol and small punctate compartments. However, in contrast with results of the single or double transformations with myc-DRP3A, peroxisome morphology in triple transformed cells was profoundly altered (e.g., catalase image in Figure 1F); that is, all of the peroxisomes in these cells were elongated, remarkably similar to the elongated peroxisomes consistently observed in cells overexpressing PEX11d alone (Figure 1G).
To verify that the fluorescent signal observed in cells overexpressing myc-tagged proteins was attributable to expression of the epitope-tagged protein and not background or nonspecific labeling, mock-treated cells were transformed with the empty vector (Figure 1H). No fluorescence signal was detected in cells, shown in the differential interference image, labeled with anti-myc antibodies. In the same cell, normal-appearing round peroxisomes were labeled with anticatalase antibodies (Figure 1H).
Physical Interactions among the PEX11 Isoforms, the FIS1 Isoforms, and DRP3A
We employed bimolecular fluorescence complementation (BiFC) assays to determine whether in vivo physiological interactions occurred among the PEX11 isoforms, the FIS1 isoforms, and/or DRP3A (Table 1). Arabidopsis cells were transiently transformed with pairs of plasmids encoding a PEX11 isoform, a FIS1 isoform, and/or DRP3A. One protein of the pair was fused to the N-terminal half of the yellow fluorescent protein (YFP) and the other protein was fused to the C-terminal half of YFP (e.g., nYFP-PEX11c and cYFP-FIS1b). Because each half of the YFP is not intrinsically fluorescent, YFP fluorescence is observed only when intermolecular interactions occur between nYFP- and cYFP-tagged proteins (Citovsky et al., 2006). All cells were also cobombarded with a plasmid encoding a red fluorescent protein linked to the rice multifunctional protein (RFP-MFP), a well-characterized peroxisomal matrix enzyme (Chuong et al., 2002, 2005). Coexpressed RFP-MFP served as an internal standard for transformation efficiency as well as a convenient peroxisomal marker to assess colocalizations with BiFC signals and potential changes in peroxisome morphology.
Table 1.
BiFC Analyses of Interactions among PEX11 Isoforms, FIS1 Isoforms, and DRP3A
| PEX11a | PEX11b | PEX11c | PEX11d | PEX11e | FIS1a | FIS1b | DRP3A | |
|---|---|---|---|---|---|---|---|---|
| PEX11a | +a | +n | +n | +n,e | +n | −n,a | +n,a | −n,a |
| PEX11b | +n | +n | +n,e | +n,a | −n | +n | −n | |
| PEX11c | +e | +n,e | +n | −n | +n,a | −a | ||
| PEX11d | +e | +e | −n,e,a | +n,a | −a | |||
| PEX11e | +n | −n | +n | −n | ||||
| DRP3A | −n | −n | −n |
Arabidopsis suspension cells were cotransformed (via biolistic bombardments) with the PEX11 isoforms, FIS1 isoforms, and/or DRP3A fused to either the N- (rows) or C-terminal (columns) halves of YFP. Additionally, all cells were transformed with RFP-MFP, which served as a convenient means of identifying transformed cells and as a peroxisomal (morphology) marker protein. At 16 h after bombardment, cells were formaldehyde fixed and viewed by epifluorescence microscopy. Interactions were scored based on either the presence (+) or absence (−) of a BiFC (YFP) signal at the peroxisome membrane. All BiFC signals observed colocalized with (co)expressed peroxisomal RFP-MFP. For each pair of plasmids tested, ≥25 transformed cells were scored from at least two independent transformations. Peroxisome morphologies in transformed cells were scored as normal (n), elongated (e), and/or aggregated (a).
The results presented in Table 1 show that all five PEX11 isoforms interact in vivo as homooligomers and with each other as heterooligomers at the peroxisome membrane. Additional peroxisomal interactions were observed among all five PEX11 isoforms and FIS1b, but not FIS1a nor DRP3A. Furthermore, BiFC signal was not observed for coexpressed DRP3A, FIS1a, and FIS1b, suggesting that these proteins did not homo- or heterooligomerize. In control experiments, each of the PEX11-, FIS1-, and DRP3A-nYFP fusion constructs were expressed with a corresponding cYFP empty vector. The consistent lack of peroxisome fluorescence in these control cells provided convincing negative evidence that fluorescence observed in experimental BiFC assays resulted from specific protein–protein interactions.
Three categories of peroxisome morphologies were observed in transformed cells during BiFC assays (Table 1): (1) normal rounded or rod-shaped, as typically observed in untransformed suspension cells (Lingard and Trelease, 2006); (2) elongated (≥3 μm long; e.g., Figure 1F); and/or (3) aggregated (e.g., Figure 1D). Elongated peroxisomes were observed in the majority of cells coexpressing PEX11d, with the notable exception of cells coexpressing PEX11d with FIS1b or DRP3A. Elongated peroxisomes also were observed in cells expressing homooligomers of PEX11c; however, coexpression of PEX11c with any other PEX11 isoforms abrogated the peroxisome elongations that normally were observed during PEX11c overexpression (Lingard and Trelease, 2006).
Silencing of DRP3A, PEX11, and FIS1 Expression
To determine whether the PEX11 isoforms, FIS1 isoforms, and/or DRP3A were necessary for peroxisome replication, transient RNA interference (RNAi) was induced in suspension cell protoplasts transformed with gene-specific double-stranded RNA (dsRNA). RNAi specificity was achieved using dsRNA complementary to either the coding sequence (CDS) or the 3′ untranslated region (UTR) of each respective mRNA (see Supplemental Figure 2 online). Supplemental Table 1 online lists the cell names and corresponding dsRNA and DNA constructs used for preparing RNAi protoplasts.
In mock-transformed cells (Figure 2A) or in gfp-i cells transformed with irrelevant GFP dsRNA (Figure 2F), ACTIN2 (ACT2), PEX11c, PEX11d, and PEX11e transcripts were consistently detected using RT-PCR (with 25 amplification cycles) at 18 h after transformation. Conversely, the consistently low levels of PEX11b transcripts were detected in gfp-i control cells only after using additional PCR amplification cycles (Figure 2M). However, PEX11a transcripts were not detected even after additional amplification cycles (Figures 2L and 2M); hence, analysis of pex11a-i lines was not pursued.
Figure 2.
Gene-Specific Silencing of DRP3A, the Five PEX11 Isoforms, and the Two FIS1 Isoforms.
Arabidopsis suspension cell protoplasts were transformed with polyethylene glycol (PEG) with dsRNA complementary to GFP ([F], [K], and [M]), PEX11c/11d/11e (B), PEX11c (C), PEX11d (D), PEX11e ([E] and [G]), PEX11b (L), DRP3A (H), FIS1a (I), or FIS1b (J). Protoplasts for (G) were also cotransformed with plasmid DNA encoding the GFP-PEROX transgene. Mock transformations in (A) were done with buffer without RNA or DNA. After 18 h of expression, total RNA was extracted, and ACTIN2 (ACT2), FIS1a, FIS1b, DRP3A, PEX11a, PEX11b, PEX11c, PEX11d, and PEX11e mRNA were reverse transcribed and amplified via 25, 30, or 40 PCR cycles. In (M), ACT2 (reproduced from [L]), PEX11a, PEX11b, and PEX11e bands are from the same gel and were adjusted for brightness simultaneously before removing adjoining wells (gap in the image). All images are of fluorescence emitted from ethidium bromide–labeled PCR products (∼600 bp) separated in 1% ([A] to [G]) or 2% (w/v) ([H] to [M]) agarose gels.
In pex11c/11d/11e-i cells, PEX11c, PEX11d, and PEX11e mRNA levels were simultaneously reduced (Figure 2B) compared with mock or gfp-i controls (Figures 2A and 2F). In addition, in the pex11c-i, pex11d-i, or pex11e-i single mutants, PEX11c, PEX11d, or PEX11e mRNA levels, respectively, were individually reduced (Figures 2C to 2E). In pex11b-i cells, PEX11b, but not PEX11e, transcripts were effectively reduced compared with gfp-i cells (Figures 2L and 2M).
To verify that transformation of protoplasts with both dsRNA and plasmid DNA would not interfere with the RNAi mechanism, protoplasts were also cotransformed with GFP-PEROX plasmid DNA coding for GFP fused to a type 1 peroxisomal matrix targeting signal and with PEX11e dsRNA (pex11e-i), which resulted in specific silencing of PEX11e (Figure 2G).
We also observed silencing of DRP3A transcripts in drp3a-i cells (Figure 2H) compared with gfp-i cells (Figure 2K). However, obvious silencing was not observed for FIS1a or FIS1b transcripts in fis1a-i or fis1b-i cells, respectively (Figures 2I and 2J).
Kinetics of Transient RNAi during Protoplast Regeneration
To determine the kinetics of cell wall regeneration and cell cycle reinitiation following the formation of protoplasts, we quantified the number of protoplasts that underwent cell division from 1 to 6 d after transformation. Arabidopsis suspension cultures typically grow as microclusters of 20 to 30 cells (Figure 3A) that dissociate after incubation in protoplasting medium (Figure 3B). After protoplast transformation, incubation in growth medium stimulates cell wall resynthesis and cell cycle reinitiation. Figure 3C illustrates that after completion of cytokinesis, pairs of daughter cells are joined by a common cell wall (arrow). Subsequent cell divisions give rise to four or more daughter cells within nascent microclusters (Figure 3D). These common cell walls provide a simple and reliable means to identify protoplasts within a population of transformed cells that have divided one or more times. As shown in Figure 3E, the majority of mock-transformed protoplasts undergo at least one cell division 3 to 4 d after initial protoplasting (solid line with diamonds).
Figure 3.
Reinitiation of Cell Division and RNAi Persistence in Transiently Transformed Protoplasts.
(A) Typical microcluster of 20 to 30 Arabidopsis suspension cells with undigested cell walls (before protoplasting). All cell walls in (A) to (D) were stained with Fluorescence Brightener 28 (FB28).
(B) Single protoplast observed immediately after protoplasting (0 d), mock transformation, and staining of cell wall residues with FB28.
(C) Pair of daughter cells sharing an equatorial cell wall (arrow) (2 to 5 d after transformation).
(D) Cluster of four daughter cells with multiple internal cell walls (arrows) (3 to 6 d after transformation). For (A) to (D), bars = 10 μm.
(E) Percentage of protoplasts that underwent at least one cell division over a 6-d period (diamonds, left axis) and the number of peroxisomes (right axis) found in both daughter cells from pex11c/11d/11e-i (open triangles) or control gfp-i (closed triangles) cells. Protoplasts possessing at least one equatorial cell wall were defined as divided. Peroxisomes were either labeled with GFP-PEROX (pex11c/11d/11e-i) at 1 to 4 d after transformation or with anticatalase IgGs (gfp-i) at 2 to 4 d after transformation. The fragility of protoplasts precluded immunolabeling at 1 d after transformation. For cell division quantifications, values are averages of at least two independent transformations (n ≥ 300 per day). For peroxisome quantifications, values are averages ± se from at least 19 cells arising from two independent transformations.
(F) and (G) Quantification of PEX11 mRNA amounts in gfp-i (F) or pex11c/11d/11e-i (G) cells. Total RNA was isolated from protoplasts 1, 2, 3, and 4 d after transformation. Transcript levels (in fg) of PEX11a, PEX11b, PEX11c, PEX11d, and PEX11e in 4 ng total RNA were quantified via real-time PCR and normalized to ACT2 expression. Duplicate (PEX11a, -b, and ACT2) and triplicate (PEX11c, -d, and -e) technical replicates were acquired for each sample. Values are means ± sd.
We also quantified peroxisome number in gfp-i and pex11c/11d/11e-i cells to determine whether silencing of these three PEX11 isoforms impacted peroxisome number, and if so, when this effect occurred. Quantification of the number of peroxisomes in pairs of gfp-i daughter cells (Figure 3E, dashed line and closed triangles) revealed that peroxisome number gradually increased between 2 and 4 d after transformation. On the other hand, peroxisome number in pairs of pex11c/11d/11e-i daughter cells decreased dramatically between 2 and 3 d after transformation (Figure 3E, stippled line and open triangles).
To identify when gene silencing occurs and for how long it persisted after transformation, PEX11 mRNA was quantified (using real-time PCR) in gfp-i (Figure 3F) and pex11c/11d/11e-i (Figure 3G) cells from 1 to 4 d after transformation. Figure 3F shows that in gfp-i cells, PEX11a mRNA was relatively low and did not vary with time, whereas PEX11b expression increased gradually from 1 to 4 d after transformation. PEX11c mRNA exhibited a substantially higher expression level at all 4 d than the other PEX11 mRNA. PEX11d mRNA exhibited a steady increase from 1 d to a level almost five times the initial level at 4 d. PEX11e mRNA peaked at 2 d, with lower levels at 1, 3, and 4 d. Comparisons of data in Figures 3F with 3G show that in pex11c/11d/11e-i protoplasts, PEX11c, -d, and -e mRNA levels were substantially lower at all time points, whereas PEX11a and -b mRNA levels were the equivalent. Taken together, these results indicate that optimum time to observe the effects of transient RNAi on peroxisome number was 3 to 4 d after transformation.
Involvement of the PEX11 Isoforms in Peroxisome Replication
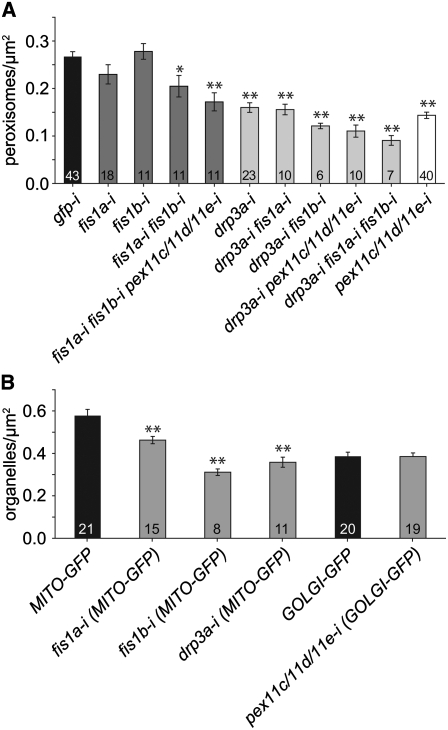
The data in Figures 2 and 3 established that our RNAi system effectively reduced PEX11 isoform mRNA levels in vivo and was thus suitable for studying peroxisome replication in living cells actively undergoing cell division. To determine whether the PEX11 isoforms were necessary for peroxisome replication, the average density of peroxisomes in the cytoplasm (peroxisomes/μm2) was determined for the progeny of RNAi cells. In each case, averages were derived from the number of peroxisomes in each of two daughter cells in a pair. The differences between each of the mean values presented in Figures 4A, 5A, 5B, and 6A were tested for significance with t tests (see Supplemental Tables 2 to 5 online).
Figure 4.
Cytoplasmic Peroxisome Density (Peroxisomes/μm2) in pex11-i Cells Pairs at 4 d after Transformation.
Protoplasts were transformed without RNA or DNA (mock), with GFP-PEROX, or cotransformed with various dsRNA (x axis) plus GFP-PEROX. Four days after transformation, protoplasts were fixed and stained with FB28. Bars for control cells are shaded black, and bars for single, double, and triple silencing are shaded in dark gray, light gray, and white, respectively.
(A) Peroxisome density in the cytoplasm of each of two transformed daughter cells. Values are means ± se of at least 26 daughter cell pairs from at least two independent transformations for all but pex11b-i (n = 12 from one transformation). Bars with asterisks are significantly different from gfp-i (* P ≤ 0.05; ** P ≤ 0.001).
(B) GFP-labeled peroxisomes in a control daughter cell pair transformed with GFP-PEROX only.
(C) Anticatalase/rhodamine redX-immunolabeled peroxisomes in a gfp-i daughter cell pair.
(D) GFP-labeled peroxisomes within a pex11d/11d/11e-i daughter cell pair cotransformed with GFP-PEROX. Images in (B) to (D) are representative of whole-cell projections acquired via nonconfocal epifluorescence microscopy. Bar = 10 μm.
Figure 5.
Cytoplasmic Peroxisome Density in fis1a-i, fis1b-i, and drp3a-i Daughter Cell Pairs at 4 d after Transformation.
(A) Cytoplasmic peroxisome density in single (fis1a-i, fis1b-i, and drp3a-i), double (fis1a-i fis1b-i, drp3a-i fis1a-i, drp3a-i fis1b-i, and drp3a-i pex11c/11d/11e-i), and triple (drp3a-i fis1a-i fis1b-i and fis1a-i fis1b-i pex11c/11d/11e-i) mutants. gfp-i and pex11c/11d/11e-i means are reproduced from Figure 4A. Bars corresponding to the basal level of peroxisomes in gfp-i and pex11c/11d/11e-i cells are shaded black and white, respectively, and bars reflecting analyses of fis1 and drp3a silencing are shown in dark and light gray, respectively.
(B) Cytoplasmic mitochondrial or Golgi body density in control (MITO-GFP and GOLGI-GFP) and RNAi (fis1a-i, fis1b-i, drp3a-i, and pex11c/11d/11e-i) transformed daughter cell pairs. Mitochondria and Golgi bodies were labeled in control and RNAi lines by (co)transforming cells with MITO-GFP or GOLGI-GFP plasmid DNA, respectively. Bars for controls are shaded black, and bars for RNAi lines are shaded gray.
Values in (A) and (B) are means ± se. n values for each sample are indicated within the histogram bars. Bars with asterisks are significantly different from gfp-i (A) or MITO-GFP (B) (* P ≤ 0.05; ** P ≤ 0.001).
Figure 6.
Necessity of PEX11c, PEX11d, FIS1b, and/or DRP3A for PEX11e-Induced Peroxisome Multiplication.
(A) Mean cytoplasmic peroxisome densities in pex11c-i pex11d-i, fis1b-i, and drp3a-i daughter cell pairs overexpressing GFP-PEX11e. Data for the means of gfp-i, drp3a-i, fis1b-i, pex11c/11d-i, and pex11c/11d/11e-i are reproduced from Figures 4A and 5A. All values are means ± se. n values for each sample are indicated within the histogram bars. Bars with asterisks are significantly different from GFP-PEX11e (* P ≤ 0.05; ** P ≤ 0.001). Bars for the control and corresponding GFP-PEX11e–overexpressing lines are shaded the same color.
(B) GFP-labeled peroxisomes in a pair of drp3a-i daughter cells overexpressing GFP-PEX11e.
(C) Anticatalase/rhodamine redX-immunolabeled peroxisomes in a pair of gfp-i daughter cells.
(D) GFP-PEX11e-labeled peroxisomes in a pair of daughter cells overexpressing GFP-PEX11e.
(E) GFP-PEROX–labeled peroxisomes in a pair of drp3a-i daughter cells. Images in (B) to (E) are representative whole-cell projections acquired via nonconfocal epifluorescence microscopy. Bar = 10 μm.
The histogram in Figure 4A shows that 4 d after transformation the average density of peroxisomes in the cytoplasm of daughter cell pairs that were either mock-transformed (without DNA or dsRNA), transformed with plasmid DNA alone (GFP-PEROX), or transformed with irrelevant dsRNA (gfp-i) was ∼0.25 peroxisomes/μm2. An ∼17% reduction in peroxisome number was observed in pex11c-i, pex11d-i, and pex11e-i single mutants, which was decreased further to an ∼30% reduction in the pex11d/11e-i, but not pex11c/11e-i or pex11d/11c-i, double mutants. The greatest reduction in peroxisome number (∼46%) was observed in pex11c/11d/11e-i triple mutants. These data indicate that peroxisome replication in actively dividing cells is dependent upon the cooperative function of PEX11c, PEX11d, and PEX11e.
Figures 4B and 4C show representative optical micrographs of peroxisomes in GFP-PEROX and gfp-i daughter cells. The relatively small, spherical, or rod-shaped peroxisomes in these control cells are characteristic of peroxisomes in untransformed cells. By contrast, peroxisomes in pex11c/11d/11e-i daughter cell pairs (Figure 4D) were fewer in number, larger than normal, and uniformly spherical. These results suggest that silencing of PEX11c, PEX11d, and PEX11e restricts only peroxisome elongation and division and that peroxisome growth is not regulated by PEX11 and can be uncoupled from peroxisome elongation and division.
Involvement of DRP3A, FIS1a, and FIS1b in Peroxisome Replication
We next silenced DRP3A, FIS1a, and FIS1b to determine whether these proteins were necessary for peroxisome replication. Although we were not able to detect an effect on transcript levels of the FIS1a and FIS1b targets (Figures 2I and 2J), the effects on peroxisome density and mitochondria shown below indicate that the dsRNA showed an effect, possibly at the translational level. Figure 5A shows that at 4 d after transformation, drp3a-i daughter cell pairs exhibited an ∼40% reduction in peroxisome number compared with gfp-i cells. However, the fis1a-i or fis1b-i single mutants showed no significant reductions in peroxisome number, in contrast with fis1a-i fis1b-i double mutants, which exhibited a significant (∼23%) reduction. Although no significantly enhanced reduction was observed in the drp3a-i fis1a-i double mutant compared with drp3a-i, the 40% reduction in drp3a-i peroxisome density was significantly enhanced to ∼55% in drp3a-i fis1b-i, but not drp3a-i fis1a-i, double mutants. The 46% reduction in peroxisome number in pex11c/11d/11e-i cells was enhanced to ∼59% in drp3a-i pex11c/11d/11e-i but was not enhanced in fis1a-i fis1b-i pex11c/11d/11e-i. The greatest reduction in peroxisome number of any mutant was observed in the drp3a-i fis1a-i fis1b-i triple mutant (66% reduction). Peroxisome number in the fisla-i fis1b-i pex11c/11d/11e-i mutant was not significantly different from peroxisome number in pex11c/11d/11e-i, fis1b-i pex11c/11d/11e-i, or the fis1a-i fis1b-i double mutant. Taken together, these data suggest that FIS1b is involved in peroxisome replication, specifically in collaboration with DRP3A, and that DRP3A also functions along with PEX11c, PEX11d, and PEX11e in peroxisome replication.
DRP3A and FIS1a were reported previously to be necessary for mitochondrial division (Mano et al., 2004; Scott et al., 2006). Figure 5B shows that the cytoplasmic densities of mitochondria in drp3a-i and fis1a-i daughter cell pairs were lower than in control cells expressing the mitochondrial marker protein, MITO-GFP, alone. Interestingly, the mitochondrial density in fis1b-i cells was substantially decreased relative to the density in control MITO-GFP cells, providing novel evidence that this protein is also involved in mitochondrial replication. Similar to these findings for fis1b-i, but not fis1a-i cells, Scott et al. (2006) also observed a reduction (∼43%) in the number of mitochondria in protoplasts isolated from two separate FIS1a T-DNA alleles. Notably, the reductions in mitochondrial number observed in drp3a-i, fis1a-i, and fis1b-i cells verified that DRP3A, FIS1a, and FIS1b were effectively silenced in our transient cell lines, even though this was not readily apparent in Figure 2 for fis1a-i and fis1b-i cells.
Figure 5B also shows that the cytoplasmic density of Golgi bodies in control GOLGI-GFP and pex11c/11d/11e-i (GOLGI-GFP) cells was not significantly different. These results suggest that silencing of PEX11c, PEX11d, and PEX11e and inhibition of peroxisome replication do not globally repress organelle replication in dividing cells.
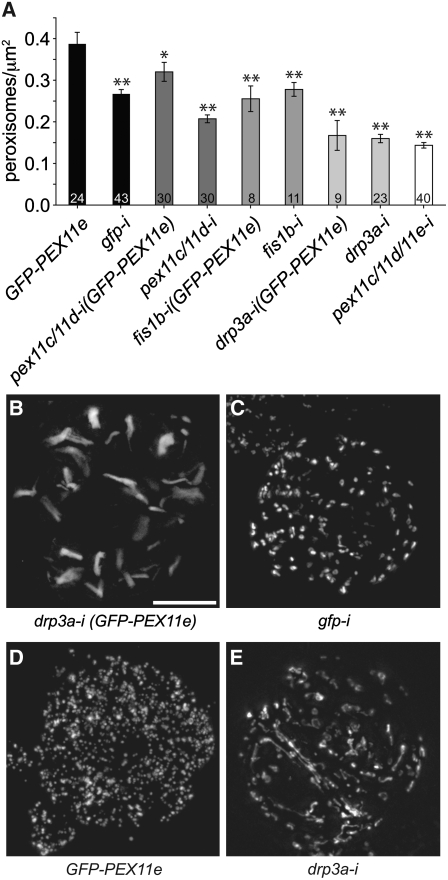
PEX11c, PEX11d, FIS1b, and DRP3A Are Necessary for PEX11e-Induced Peroxisome Multiplication
We reported previously that the overexpression of PEX11e results in a doubling of the number of peroxisomes per cell (Lingard and Trelease, 2006). To determine whether other members of the peroxisome fission machinery, including PEX11c, PEX11d, FIS1b, and/or DRP3A, were involved in this inducible multiplication, GFP-PEX11e was overexpressed in various RNAi backgrounds and peroxisomes were quantified in pairs of daughter cells. As expected, overexpression of GFP-PEX11e induced an ∼45% increase in peroxisome number compared with gfp-i control cells (Figure 6A). However, overexpression of GFP-PEX11e in a pex11c/11d-i double mutant background resulted in a significant inhibition of the PEX11e-induced peroxisome multiplication. When GFP-PEX11e was overexpressed in fis1b-i or drp3a-i backgrounds, peroxisome number was the same as in the normal fis1b-i and drp3a-i lines. These data suggest that PEX11c, PEX11d, PEX11e, FIS1b, and DRP3A are necessary for PEX11-induced peroxisome multiplication.
In addition to reductions in peroxisome number (Figure 6A), we noted that these transgenic cells exhibited dramatic alterations in peroxisome morphology. The fluorescence micrographs presented in Figures 6B to 6E illustrate that peroxisomes in drp3a-i (GFP-PEX11e) cells were substantially elongated (Figure 6B) compared with the uniformly spherical peroxisomes in gfp-i (Figure 6C) or the spherical but much more numerous peroxisomes in GFP-PEX11e cells (Figure 6D). Notably, the elongated drp3a-i (GFP-PEX11e) peroxisomes (Figure 6B) were thicker and shorter than peroxisomes in drp3a-i daughter cell pairs (Figure 6E), whose elongated/tubulated peroxisomes more closely resembled peroxisomes in cells overexpressing PEX11c or PEX11d (Lingard and Trelease, 2006).
Peroxisome Replication Occurs before M-Phase in Synchronized Suspension Cells and Is Correlated with Expression of DRP3A, PEX11, FIS1a, and FIS1b Expression
The relationships among (1) DRP3A, PEX11, FIS1a, and FIS1b expression, (2) peroxisome size and number, and (3) cell cycle stages were examined in synchronized suspension cell cultures. Cell cycle progression in nonprotoplasted suspension cells (e.g., Figure 3A) was synchronized using amphidicolin block and release (Menges and Murray, 2002), and synchronization efficiency was assessed by determining the percentage of cells in S- and M-phase at intervals from 0 to 32 h after amphidicolin release. To visualize and quantify the number of cells in S-phase, cells were incubated for 1 h in 5-bromo-2′-deoxyuridine (BrdU), which was incorporated into actively replicating DNA, prior to formaldehyde fixation and immunolabeling with anti-BrdU antibodies. Mitotic profiles (metaphase and anaphase chromosomes) typical of M-phase cells were identified by labeling nuclei with 4′,6-diamidino-2-phenylindole (DAPI).
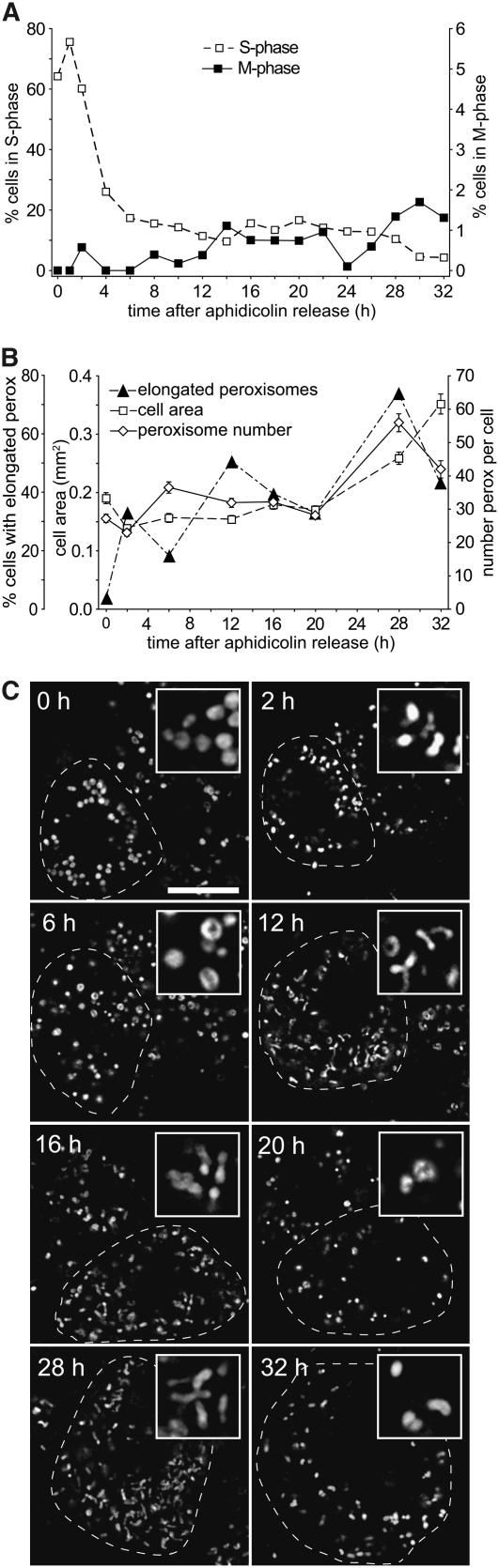
The dashed line in Figure 7A shows that 1 h after release of amphidicolin, ∼78% of the cells were in S-phase; thereafter, the percentage of cells decreased within 6 to 8 h to a background of ∼15%. These S-phase percentages resemble other published values for Arabidopsis suspension cell synchronization (Menges and Murray, 2002). The solid line in Figure 7A indicates that the percentage of cells in M-phase rose gradually from 6 to 14 h after release of the block and eventually reached a peak at 30 h of ∼2%. This maximum percentage was somewhat lower than that found by Menges and Murray (2002). However, Menges and Murray (2002) employed a novel fast-growing Arabidopsis suspension culture, which grew at least twice as fast as our cells and, after synchronization, transitioned from the peak of S-phase to the peak of M-phase in ∼10 h. By contrast, the transition from S-phase to M-phase lasted at least 24 h in our synchronized cultures (Figure 7), suggesting that the more dispersed (broader and shorter) M-phase peak in our synchronized culture was a product of the slower growth rate.
Figure 7.
Evidence for Synchronization of Suspension Cell Growth and Correlations with Distinct Changes in Peroxisome Morphologies.
(A) Percentage of cells in S-phase (open squares) or M-phase (closed squares) at varied time points in amphidicolin-synchronized cells. Aliquots from synchronized cell cultures at indicated time points (in hours) after amphidicolin release (x axis) were incubated in BrdU for 1 h and fixed. Actively replicating DNA in S-phase nuclei were labeled with anti-BrdU IgGs and secondary rhodamine redX antibodies. Mitotic chromosomes were identified with DAPI. At least 487 cells for each time point were examined.
(B) Percentage of cells bearing elongated peroxisomes (closed triangles), cell area (open squares), and the peroxisome number per cell (open diamonds) at varied time points in amphidicolin-synchronized cells Peroxisomes in the same cells in (A) were immunolabeled with anticatalase IgGs and secondary Cy2 antibodies. Values are averages ± se of at least 72 cells per time point, although sometimes the small differences in the error bars are not visible.
(C) Representative micrographs depicting peroxisomes (anticatalase and Cy2 secondary antibody labeling) in synchronized cells at the times after amphidicolin release indicated on each panel. In each panel, cell boundaries are outlined with a white dashed line, and examples of individual peroxisome morphologies are shown in ×3 magnification insets. Bar = 10 μm.
To determine whether cell cycle progression is synchronized with peroxisome replication, peroxisome number per cell, cell area, and peroxisome elongation were quantified in equatorial focal planes at several time points after release from the amphidicolin block (Figure 7B). The average cell area (Figure 7B, open squares) and the average number of peroxisomes per cell (Figure 7B, open diamonds) remained relatively constant throughout S-phase and most of G2 (20 h), then peroxisome number doubled near the onset of M-phase (28 h) and cell area doubled at the end of G2 near the onset of M-phase (32 h). These uniform increases in peroxisome number and cell area suggest that synchronization was maintained throughout the time course. The percentage of cells possessing at least one peroxisome longer than 3 μm (Figure 7B, closed triangles) exhibited distinct peaks at the end of S-phase (2 h), the middle of G2 (12 h), and at the end of G2 (28 h).
Representative optical micrographs presented in Figure 7C illustrate changes in peroxisome number per cell, morphology, and cell size that became apparent after amphidicolin release. Overall, cell size remained relatively constant during S-phase and G2 (0 to 20 h), then obviously increased at the onset and near the end of M-phase (28 to 32 h), corroborating the increases in cell area quantified in Figure 7A. Peroxisomes at 0 h were relatively small and spherical with virtually no elongated peroxisomal profiles. At 6 h, peroxisomes were still mostly spherical, although they also appeared somewhat enlarged and often donut shaped. Micrographs of single cells at 12, 16, and 28 h show the quantified changes (Figure 7C) in typical sizes and shapes for the elongated peroxisomes that were frequently observed among the cell populations. At 32 h, peroxisomes within a single enlarged cell were comparatively smaller and more spherical, similar to peroxisomes at 0 and 2 h, although peroxisomes at 32 h were more widely dispersed throughout the cell.
To assess whether endogenous expression patterns of DRP3A, the PEX11 isoforms, and/or the FIS1 isoforms were correlated with cell cycle events, mRNA levels were quantified via real-time PCR at various time points after amphidicolin release. HISTONE H4 expression, which is expressed predominately during S-phase (Chaubet et al., 1996), peaked sharply at 4 h and then remained relatively low throughout the remainder of the time course (Figure 8A). FIS1a, FIS1b, DRP3A, PEX11c, PEX11d, and PEX11e (Figures 8B and 8D) exhibited similar expression profiles at earlier time points with peak expression levels apparent at 2 and 12 h after amphidicolin release. FIS1a, DRP3A, PEX11c, and PEX11e each exhibited a third rise in expression level near the end of G2 (22 to 26 h after amphidicolin release) coincident with peak increases in both peroxisome elongation and number at 28 h (Figures 7B and 7C). Intriguingly, PEX11a and PEX11b, which under other conditions we have tested were either undetectable (PEX11a) or had very low expression levels (PEX11b), both underwent a single rise in expression during G2 (8 to 12 h after amphidicolin release) (Figure 8C).
Figure 8.
Quantification of HISTONE H4, DRP3A, FIS1, and PEX11 mRNA at Varied Time Points in Amphidicolin-Synchronized Cells.
Total RNA was isolated from synchronized cells at the indicated time points (in hours) after amphidicolin release (x axis) and reverse transcribed, and 4 ng total cDNA were quantified via real-time PCR for each data point. Values are means ± sd of at least two technical replicates.
(A) Absolute quantifications of HISTONE H4 mRNA.
(B) Absolute quantifications of FIS1a (diamonds), FIS1b (squares), and DRP3A (triangles) mRNA.
(C) Absolute quantifications of PEX11a (diamonds) and PEX11b (squares).
(D) Absolute quantification of PEX11c (triangles), PEX11d (x), and PEX11e (circles) mRNA.
DISCUSSION
Previously, we identified five functional Arabidopsis PEX11 homologs that, when overexpressed, sorted directly to the peroxisome membrane and stimulated peroxisome (1) doubling in number per cell (PEX11a and PEX11e), (2) aggregation or enlargement (PEX11b), and/or (3) elongation (PEX11a, PEX11c, and PEX11d) (Lingard and Trelease, 2006). In this study, we focus on the molecular mechanisms that are involved in maintaining a constitutive population of preexisting peroxisomes in actively dividing plant cells. In particular, we provide new evidence for the cooperative participation of PEX11c, PEX11d, PEX11e with FIS1b and DRP3A, in growth and subsequent equal division of preexisting peroxisomes during G2 phase just prior to mitosis. Hence, we provide evidence that the creation of new plant peroxisomes is not solely dependent upon direct origination from the ER, as portrayed in several recent models drawn from research with yeast and mammalian cells (Tabak et al., 2003; Kragt et al., 2005; Haan et al., 2006; Kim et al., 2006; Titorenko and Mullen, 2006). Our findings do not preclude origination of plant peroxisomes from the ER, although it is notable that ER-dependent peroxisome formation has not yet been described in plant cells (Trelease and Lingard, 2006). Instead, recent research in plant peroxisome biogenesis supports participation of the ER in the overall growth and division of preexisting plant peroxisomes through essential contributions of new membrane components (reviewed in Mullen and Trelease, 2006). A similar role for the ER was described recently in the replication of preexisting peroxisomes in S. cerevisiae (Motley and Hettema, 2007).
Interactions among PEX11 Isoforms, FIS1 Isoforms, and DRP3A
The homooligomerizations among PEX11 isoforms that we observed via BiFC experiments (Table 1) were consistent with previous published findings in mammals and yeast. For instance, immunoprecipitation experiments showed that mammalian PEX11α and PEX11β formed homodimers (Li and Gould, 2003), and yeast two-hybrid assays revealed that S. cerevisiae Pex11p, Pex25p, and Pex27p also formed homodimers (Rottensteiner et al., 2003). On the other hand, it is curious that we observed heterooligomerizations among all five of the Arabidopsis PEX11 isoforms (Table 1) because no heterooligomerizations were observed among mammalian PEX11 isoforms (Li and Gould, 2003), and only weak interactions were observed between S. cerevisiae Pex25p and Pex27p (Rottensteiner et al., 2003). This apparent difference may be due to the stabilization of short-lived intermolecular interactions that can occur after reconstitution of the YFP fluorophore during BiFC (Bracha-Drori et al., 2004; Walter et al., 2004).
An advantage of BiFC over yeast two-hybrid and immunoprecipitation experiments is that the subcellular localization of the interacting species can directly be observed via microscopy. For example, the reconstituted YFP fluorescence resulting from interactions between the PEX11 proteins and FIS1b revealed not only the interaction between these two molecules but also that FIS1b was recruited to the peroxisome membrane through its associations with the PEX11 isoforms, which was confirmed by the PEX11-dependent sorting of overexpressed myc-FIS1b to peroxisomes (Figures 1B and 1D).
Although the BiFC results clearly show that homo- and heterointeractions occur among the PEX11 isoforms, as well as FIS1b, they do not yield information about stoichiometric relationships within these complexes (e.g., whether the proteins exist within these complexes or have a more transitory interaction). Nevertheless, our BiFC results suggest that oligomerization of the PEX11 isoforms serves a regulatory function. For example, cells possessing PEX11d homo- and heterooligomers or PEX11c homooligomers also possessed unusually elongated peroxisomes (Table 1). Thus, heterooligomerization seemed to inhibit PEX11c-stimulated elongation but had no effect on PEX11d-stimulated elongation. Hetero-interactions among PEX11c, PEX11d, and PEX11e also seem to be required for PEX11e-induced peroxisome fission, as peroxisome multiplication was inhibited in pex11c/11d-i (GFP-PEX11e) cells (Figure 6A). Yeast and mammalian models also support a role for PEX11 oligomerization in the regulation of peroxisome fission. Inhibiting mammalian PEX11 homodimerization precludes peroxisome proliferation (Kobayashi et al., 2007), whereas inhibition of S. cerevisiae Pex11p homodimerization promotes peroxisome proliferation (Marshall et al., 1996). Given the widespread oligomerizations among the Arabidopsis PEX11 isoforms (Table 1), and the ability for some of these oligomers to promote peroxisome elongation, it seems likely that at least some types of oligomers are active.
Our data suggest that FIS1a, unlike FIS1b, does not participate in peroxisome replication. No interactions were observed between FIS1a and DRP3A, or between FIS1a and any PEX11 isoform (Table 1), and overexpressed FIS1a did not sort to peroxisomes (Figures 1A and 1E). Also, the number of peroxisomes in fis1a-i drp3a-i cells is not significantly different from drp3a-i cells, whereas the number of peroxisomes in fis1b-i drp3a-i cells is significantly lower (Figure 5A), suggesting that FIS1b, but not FIS1a, cooperates with DRP3A during peroxisome replication.
Although our targeting (Figure 1), BiFC (Table 1), and silencing data (Figure 5A) do not support a direct role for FIS1a in peroxisome division, this tail-anchored protein appears to play a role in mitochondrial biogenesis consistent with the results from a previous study (Scott et al., 2006). For instance, we showed in Figure 5B that the number of mitochondria in fis1a-i daughter cell pairs was lower than in control cells. It is surprising, however, that myc-FIS1a does not localize to mitochondria in our suspension cells, suggesting that the targeting mechanism for FIS1a, as in mammalian cells (Schrader and Yoon, 2007), is complex. Experiments are currently underway to determine whether FIS1a, similar to other tail-anchored membrane proteins (e.g., Bcl-2 and BAX) (Leber et al., 2007), may only target to the mitochondria from the cytosol in response to a specific developmental and/or environmental signal.
Although interactions between FIS1b and DRP3A were not observed in our BiFC experiments (Table 1), evidence for their cooperation can be inferred from the dual-silencing experiments. Because mRNA silencing only knocks down protein expression levels, double or triple silencing mutants within the same oligomeric complex, or acting within the same pathway, are expected to have enhanced phenotypes compared with the single mutants. Accordingly, the enhanced reduction in peroxisome number observed in drp3a-i fis1b-i, but not drp3a-i fis1a-i, cells (Figure 5A) suggests that these two proteins are jointly involved in peroxisome fission. It may be that the physical interaction between FIS1b and DRP3A is mediated through the action of an unidentified adaptor or bridge protein, as is observed during mitochondrial fission in yeast (reviewed in Schrader and Yoon, 2007). The existence of such a bridge protein in Arabidopsis may explain why our BiFC experiments revealed no direct contact between FIS1b and DRP3A (Table 1).
Other Roles for the PEX11 Isoforms during Peroxisome Fission
We reported previously that overexpression of PEX11e promotes peroxisome fission without discernable elongation (Lingard and Trelease, 2006). In this work, overexpression of PEX11e in cells with silenced DRP3A resulted in peroxisome elongation without fission (Figure 6B), indicating that in plants, as in other organisms, PEX11 isoforms are not directly responsible for peroxisome fission, but likely modify or tubulate the peroxisome membrane in preparation for fission (Schrader and Fahimi, 2006). In plants and mammals, overexpression of certain PEX11 isoforms leads to peroxisomal elongation, which often precedes membrane fission (Li and Gould, 2003; Koch et al., 2004; Lingard and Trelease, 2006). This elongation may be accomplished through direct interactions of a PEX11 isoform with phospholipids in the peroxisome membrane. For example, the sequence and predicted secondary structure of S. cerevisiae Pex11p is similar to the ligand binding domain of the peroxisome proliferator-activated receptor subfamily of nuclear hormone receptors (Barnett et al., 2000), which bind to fatty acids, suggesting that the PEX11 proteins may directly bind to phospholipids on the exterior surface of the peroxisome membrane (Fagarasanu et al., 2007).
The silencing experiments presented in Figures 3 to 5 demonstrate that peroxisomal growth and tubulation are separable events and that the latter is dependent upon PEX11 expression. Specifically, even though drp3a-i and pex11c/11d/11e-i cells had similar numbers of peroxisomes (Figure 5A), the peroxisomes in pex11c/11d/11e-i were rounded and enlarged (Figure 4D), whereas those in drp3a-i were elongated but not enlarged (Figure 6E). These results confirm that DRP3A is necessary for peroxisome fission and suggest that peroxisome elongation occurs constitutively in plant cells, as observed in mammals (Li and Gould, 2003; Koch et al., 2004). It is significant that peroxisomes in cells with reduced levels of PEX11c, PEX11d, and PEX11e enlarge but do not elongate, suggesting that PEX11 isoforms are required for elongation but not for growth. Notably, in synchronized cell cultures, the coordinated increases in peroxisome elongation did not always coincide with increases in peroxisome number (Figure 7, 2 and 12 h), but always coincided with increases in PEX11 expression (Figures 8C and 8D, 2 and 12 h), supporting a role for the PEX11 proteins in peroxisome elongation but not fission. The constitutive high levels of PEX11c, PEX11d, and PEX11e expression (Figures 2 and 3F) and the dramatic decreases in peroxisome number in pex11c/11d/11e-i, but not pex11b-i, pex11c-i, pex11d-i, or pex11e-i, cells (Figure 4), both suggest that PEX11c, PEX11d, and PEX11e, but not PEX11a or PEX11b, play distinct, but overlapping, roles in peroxisome replication.
Previously, we found that cells overexpressing PEX11b have enlarged peroxisomes (Lingard and Trelease, 2006), suggesting either that PEX11b acts as a negative regulator of peroxisome fission or that overexpression of PEX11b promotes peroxisome aggregation, as has been observed for other overexpressed peroxisome membrane proteins (Mullen et al., 2001). However, pex11b-i cells have normal numbers of peroxisomes (Figure 4A), which is not suggestive of a role for PEX11b in peroxisome replication. Orth et al. (2007) recently reported that overexpression of PEX11b in Arabidopsis leaf mesophyll cells resulted in peroxisome elongation, which was quite different from our previous findings that overexpression of PEX11b in Arabidopsis suspension-cultured cells led to peroxisome aggregation (Lingard and Trelease, 2006). In addition, Orth et al. (2007) reported that silencing of PEX11b led to an ∼75% decrease in peroxisome number per leaf mesophyll cell, whereas we found that silencing of PEX11b had no apparent effect on peroxisome abundance in suspension cells (Figure 4A). Although these results seem contradictory, recent results (Desai and Hu, 2008) offer a plausible explanation that they are not contradictory. They reported that Arabidopsis peroxisomes first elongate and then proliferate in number when etiolated seedlings were transferred to the light, which coincided with an upregulation of PEX11b expression. Furthermore, they found that silencing of PEX11b precluded this light-induced peroxisome proliferation and that PEX11b was a direct target of the photomorphogenic transcription factor HYH. Thus, it seems that the primary role for PEX11b is induction of peroxisome proliferation in response to light signals, not maintenance of peroxisome homeostasis within dividing cells. Because peroxisomes in our suspension cells do not increase in number per cell in response to an overexpression of PEX11b (Lingard and Trelease, 2006), nor do peroxisomes exhibit defects in the cell division process within pex11b-i cells (Figure 4), it seems likely that PEX11c, PEX11d, and PEX11e, but not PEX11b, are the dominant PEX11 isoforms in dark-grown, rapidly dividing cells. In concert with this assertion are the constitutive low level of PEX11b expression (Figures 2 and 3A) and the increases in PEX11b expression during protoplast cell wall regeneration and cell cycle reinitiation (Figure 3F). These collective results suggest that PEX11b fulfills a minor role that partially overlaps with those of PEX11c, PEX11d, and PEX11e in the maintenance of peroxisome homeostasis in dividing cells.
Peroxisome Division during Cell Cycle Transitions
We undertook a detailed morphological analysis of the changes in Arabidopsis peroxisomes that occur during cell cycle transitions. Numerous studies with mammalian and yeast cells describe the morphological changes associated with induced peroxisome proliferation (Marshall et al., 1995; Schrader et al., 1998a, 1998b, 1999; Koch et al., 2005) but do not correlate peroxisome morphological changes with cell cycle progression. Our data show that cell division, peroxisome elongation, and peroxisome replication are intimately connected (Figure 7) and that these events are coordinated with distinct peaks in PEX11, FIS1b, and DRP3A expression (Figure 8).
The strikingly similar expression profiles for the PEX11 isoforms, the FIS1 isoforms, and DRP3A in the synchronized culture (Figure 8) are highly suggestive for a role for these genes in cell cycle progression. In addition, it is possible to infer from results of our silencing experiments that at least part of the function of these proteins is the replication of peroxisomes. Whether these proteins play roles in cell cycle progression in addition to organelle replication is unknown. A potentially analogous scenario has been identified in rat kidney cells where some, but not all, proteins that are necessary for Golgi body fission are also necessary for entrance into mitosis (Sutterlin et al., 2002), implying that organelle homeostasis is intimately linked with cell division. Interestingly, Golgi division can also be controlled through action of some cyclin-dependent kinases (Lowe et al., 1998, 2000; Draviam et al., 2001), a class of proteins classically involved in the regulation of cell cycle transitions. In plants, CDT1a and CDT1b (for CDC10-dependent transcript 1a and b, which encode proteins necessary for initiation of DNA replication in yeast) are necessary for both DNA synthesis and plastid division (Raynaud et al., 2005), supporting the hypothesis that organelle replication and cell division are interrelated.
To fully explore the links between plant organelle replication and cell division, it would be necessary to generate transgenic cell lines or plants defective in key genes, such as the PEX11 isoforms, the FIS1 isoforms, or DRP3A. Although growth defects were not observed in FIS1a or PEX11 mutants (Scott et al., 2006; Orth et al., 2007), plants defective in DRP3A exhibited severe growth defects (Mano et al., 2004). Further experiments will be needed to determine whether normal peroxisome replication is necessary for cell cycle progression and, if so, whether the PEX11 isoforms, the FIS1 isoforms, and DRP3A are also involved in other aspects of cell division.
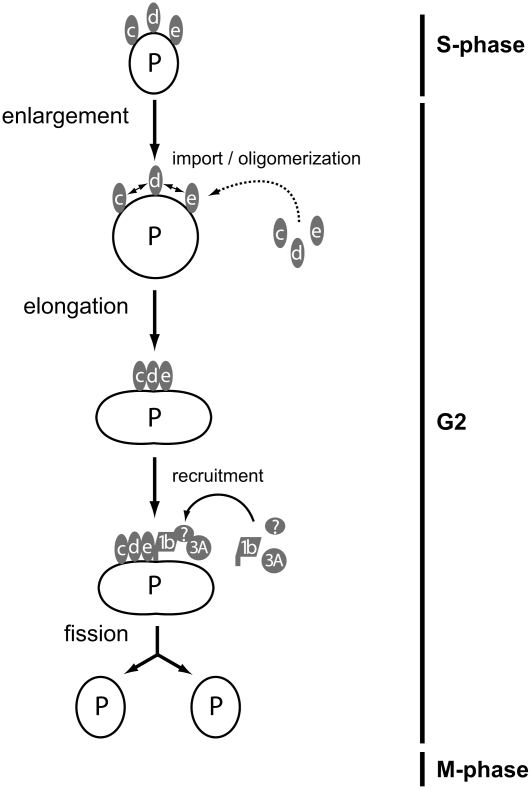
Our data provide insights for updating and modifying the Mullen and Trelease (2006) model for the biogenesis of preexisting plant peroxisomes. Figure 9 presents a working model for replication of preexisting peroxisomes in actively dividing Arabidopsis cells. After entry from S-phase into the second growth phase (G2) of the cell cycle, preexisting peroxisomes undergo a defined series of morphological changes. First, peroxisome enlargement, but not elongation, occurs independent of any PEX11 isoform. Then, inactive forms of membrane-bound PEX11c, PEX11d, and/or PEX11e undergo homo/heterooligomerization events within the membrane to promote peroxisome elongation. Alternatively/concomitantly, newly synthesized PEX11c, PEX11d, and/or PEX11e are inserted into the peroxisome membrane where they promote peroxisome elongation (dashed arrows). PEX11 oligomers next recruit from the cytosol FIS1b and DRP3A, the latter indirectly, possibly through an intermediary bridge protein(s). Recruited DRP3A then stimulates fission of the elongated peroxisome as the cell prepares to enter M-phase. Notably, this model does not include PEX11a, PEX11b, or FIS1a, as our data do not reveal a role for these proteins in basal peroxisome replication. The final result is the growth and division of a preexisting peroxisome, giving rise to daughter peroxisomes that segregate into daughter somatic cells and thereby maintain an ongoing population of functional peroxisomes.
Figure 9.
Model for Peroxisome Replication in Dividing Arabidopsis Cells.
S, S-phase; G2, second growth phase; M, M-phase; P, preexisting peroxisome; c, PEX11c; d, PEX11d; e, PEX11e; 1b, FIS1b; 3A, DRP3A; ?, putative (unknown) intermediary bridge protein(s). Note: PEX11a, PEX11b, and FIS1a are not shown since our data do not suggest a role for these proteins during replication of preexisting peroxisomes.
METHODS
Chemicals, Reagents, and Plasmids
Enzymes and reagents used for DNA and/or RNA isolations and manipulations were purchased from Eppendorf, Fermentas, New England Biolabs, Promega, and Qiagen. Custom oligonucleotide primers were purchased from Genetech Biosciences, Integrated DNA Technologies, and Invitrogen. DAPI was purchased from Invitrogen. Macerozyme R10 and Cellulase Y-C were purchased from Karlan Research Products. All other chemicals were purchased from Sigma-Aldrich.
Plant expression plasmids containing FIS1a, FIS1b, and DRP3A fused to the myc epitope tag were constructed as follows. Sequences corresponding to the entire open reading frame (ORF) of Arabidopsis thaliana FIS1a, FIS1b, or DRP3A were amplified by PCR using the appropriate plasmid (template) DNA and forward and reverse mutagenic primers. Sequences of all primers used in this study are shown in Supplemental Table 6 online. PCR products encoding FIS1A and FIS1B were digested with BamHI and XbaI, ligated into the corresponding sites of the multiple cloning region of pRTL2/myc-BX, a modified version of the plant expression vector pRTL2ΔN/S (Lee et al., 1997) that includes the 35S cauliflower mosaic virus promoter and sequences encoding an initiator Met, Gly linkers, and the myc epitope tag (underlined: MGEQKLISEEDLG-) (Fritze and Anderson, 2000), followed by in-frame BamHI and XbaI sites (Shockey et al., 2006). The resulting plasmids were referred to as pRTL2/myc FIS1A and pRTL2/myc FIS1B. Similarly, PCR products encoding DRP3A were digested with XmaI and subcloned in frame with a 5′ myc epitope tag into XmaI-digested pRTL2/myc-MCS, yielding pRTL2/myc DRP3A. pRTL2/myc-MCS was constructed by inserting sequence encoding restriction enzymes including XmaI immediately downstream of the XbaI site in PRTL2/myc-X.
GFP-PEX11e and untagged PEX11d were prepared previously (Lingard and Trelease, 2006). GOLGI-GFP (GmMan1-GFP) was provided by Andreas Nebenführ (University of Tennessee, Knoxville, TN) and Andrew Staehelin (University of Colorado, Boulder, CO) (Nebenführ et al., 1999). GFP-PEROX encodes GFP fused to an amino acid sequence containing a type 1 peroxisomal targeting signal -SRY (U. Schumann and R.T. Mullen, unpublished data). GFP-MFP and RFP-MFP encode GFP and RFP, respectively, linked to the rice peroxisomal matrix multifunctional protein (Chuong et al., 2005). MITO-GFP encodes the 60–amino acid N-terminal presequence of the β-subunit of F1-ATPase (Chaumont et al., 1994) linked to GFP (R. Di Leo and R.T. Mullen, unpublished data).
Arabidopsis Cell Culture
Arabidopsis thaliana var Landsberg erecta suspension cell cultures (a gift from Steven Neill, University of West England, Bristol, UK) were propagated as described (Lingard and Trelease, 2006) except that the Murashige and Skoog (MS) salt and vitamin mixture (Invitrogen; no longer available) was replaced with a custom-made MS basal medium with vitamins (PhytoTechnology Laboratories) modified to match the Invitrogen MS mixture by reducing nicotinic acid and pyridoxine HCl concentrations from 0.5 mg/L to 0.05 mg/L and substituting 27.8 mg/L ferric sulfate for 27.8 mg/L ferrous sulfate. Over a 20-d period, the same growth pattern was documented for cells grown in both the custom PhytoTechnology and the standard Invitrogen media. Alternatives were to prepare media with either PhytoTechnology MS basal salt mixture without vitamins or Sigma-Aldrich MS Basal salt mixture, both supplemented with myo-inositol (0.1 g/L), nicotinic acid (0.05 mg/L), pyroxidine HCl (0.05 mg/L), and thiamine (0.1 mg/L). In both media, cells exhibited normal growth, even though a fine white precipitate was routinely found in both types of media.
Protoplast Preparation and Transformation
Solutions and procedures used for protoplast preparation and transformation were described by others (Doelling and Pikaard, 1993; Lisenbee et al., 2005). Our protocol was modified as follows. Suspension cells were pelleted at room temperature in sterile 50-mL conical tubes for 2.5 min at setting 3 in an IEC HN-SII centrifuge (Thermo Electron). For washing steps, cells were pelleted, the supernatants aspirated, and the cell pellets resuspended in media/solution and then incubated with rocking inversion for 5 min at room temperature.
To prepare protoplasts, 3- to 4-d-old cells were pelleted (5- to 6-mL packed cell volume) and resuspended in 40 mL of unbuffered 0.4 M d-mannitol. After 5 min of incubation at room temperature, cells in a 15-mL aliquot of this cell suspension were pelleted (2- to 3-mL packed cell volume). This cell pellet was resuspended in 12.5 mL of filter-sterilized (0.2 μm filter; Nalgene) enzyme solution (Arabidopsis cell growth medium plus 0.4 M mannitol, 0.1% [w/v] Macerozyme R10, and 1% [w/v] Cellulase Y-C). Cells were digested at 30°C for 3 h in the dark with slow rocking inversion. Following cell wall digestions, the protoplast suspension was poured onto a 40-μm nylon filter (BD Falcon) to remove undigested cell aggregates. The protoplasts that passed through the filter were washed first in 30 mL and then in 15 mL of W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 5 mM glucose, and 0.03% [w/v] MES, pH 5.8). An aliquot of the 15-mL protoplast suspension was diluted (one part to nine parts) in MaMg solution (15 mM MgCl2, 0.1% [w/v] MES, and 0.4 M mannitol, pH 5.6), and the number of protoplasts per mL was counted using a hemacytometer.
Protoplasts were chemically transformed essentially as described by J. Sheen (http://genetics.mgh.harvard.edu/sheenweb/) with the following modifications. All centrifugations to pellet-transformed protoplasts were done in a MSE GT-2 tabletop centrifuge (setting 7) for 1 min. Prior to transformation, protoplasts in 15 mL of W5 solution were pelleted and resuspended in 15 mL of MaMg solution. For (immuno)fluorescence experiments, 1.0 × 105 protoplasts in 250 μL MaMg solution were mixed with dsRNA and/or DNA in sterile 2-mL microfuge tubes. For RT-PCR, 4.0 × 105 protoplasts in 1 mL of MaMg solution were mixed with dsRNA and/or DNA in sterile, round-bottomed Pyrex culture tubes. One volume [275 or 1100 μL for (immuno)fluorescence or PCR experiments, respectively] of freshly prepared, filter-sterilized PEG solution [40% (w/v) PEG 4000, 0.4 M mannitol, and 0.25 M Ca(NO3)2] was added to each tube. Each tube was then inverted six to eight times by hand to mix dsRNA, DNA, protoplasts, and the PEG solution. The reaction mixture was then incubated at room temperature in the dark for 30 min without mixing. Transformed protoplasts were diluted in 2 volumes (1100 μL or 4400 μL, respectively) W5 solution, pelleted, and washed twice with 1 or 5 mL of recovery solution (Arabidopsis growth media plus 0.4 M mannitol), respectively. For cell cycle initiation (Mathur et al., 1995), protoplasts were resuspended in 250 or 1000 μL recovery solution, respectively. Protoplasts were incubated in 2-mL microfuge tubes [(immuno)fluorescence experiments] or in flat-bottomed six-well culture dishes (one reaction per well) (RT-PCR). Protoplasts were incubated up to 6 d in darkness, at room temperature, without agitation.
dsRNA Preparation
dsRNA complementary to the coding sequence or 3′ UTRs of the PEX11 isoforms, the FIS1 isoforms, and DRP3A were synthesized using the T7 RiboMAX Express RNAi system (Promega). Primers and templates for dsRNA synthesis are listed in Supplemental Tables 1 and 6 online, respectively. A T7 RNA polymerase promoter (see Supplemental Table 6 online, bolded bases) was added to 5′ ends of each gene-specific primer, resulting in PCR products with T7 RNA polymerase promoter sites at 5′ sense and 5′ antisense ends of each UTR or CDS. pex11b-i, pex11c/11d/11e-i, gfp-i, fis1a-i, and fis1b-i PCR products were ∼600 bp and corresponded to the entire CDS for each mRNA (see Supplemental Table 1 online). The drp3a-i PCR product was 102 bp and corresponded to a unique sequence near the 5′ end of the CDS. pex11c-i, pex11d-i, pex11e-i, and fis1b-i PCR products were 200 to 300 bp and corresponded to the entire UTR for each mRNA. RNA synthesis, annealing, and cleanup were performed according to the T7 Ribomax Express protocol. After DNase and RNase digestions, dsRNA integrity and concentration were estimated via agarose gel electrophoresis. For (immuno)fluorescence experiments, protoplasts were transformed with 25 μg of individual dsRNA and/or with 25 μg plasmid DNA (e.g., pex11c-i), 20 μg each of dsRNA pairs (e.g., pex11c-i pex11d-i), or 15 μg each of three dsRNA (e.g., pex11c-i pex11d-i fis1a-i). Amounts of dsRNA and/or DNA were quadrupled for RT-PCR experiments.
RT-PCR
Total RNA was extracted from protoplasts and synchronized suspension cells with the RNeasy mini kit (Qiagen). Contaminating genomic DNA was removed with the Qiagen RNAe-Free DNase Set. For RT- PCR in Figure 2, PCR products were amplified from 10 ng (Figures 2A to 2G) or 45 ng (Figures 2H to 2M) cDNA (prepared from total RNA). Primers used for these reactions are listed in Supplemental Table 6 online. The following thermal profile was used for all RT-PCR amplifications: 94°C for 5 min; 25, 30, or 40 cycles of 94°C for 30 s, 57.5°C for 30 s, and 72°C for 1 min; and 72°C for 7 min. PCR products were analyzed via agarose gel electrophoresis as described by Lingard and Trelease (2006).
Quantitative PCR (qPCR) was performed in optical 384-well plates using an ABI Prism 7900 HT sequence detection system (Applied Biosystems). Each 10-μL qPCR consisted of qPCR MasterMix for SYBR Green (Eurogentec), 4 ng cDNA, and 0.1 μM each of isoform-specific primer pairs (see Supplemental Table 6 online). The following thermal profile was used for qPCR: 95°C for 10 min; 40 cycles of 95°C for 15 s and 60°C for 1 min; and a melt curve analysis of 95°C for 15 s, 60°C for 15 s, and 95°C for 15 s. Because of the high nuclei acid sequence identity among the PEX11c, PEX11d, and PEX11e coding sequences, qPCR primers were designed that corresponded to their unique 3′ UTRs (see Supplemental Table 6 online). Absolute quantities of PEX11 mRNA were determined from standard curves prepared from serial dilutions of known quantities of PEX11a, PEX11b, PEX11c, PEX11d, PEX11e, ACT2, FIS1a, FIS1b, DRP3A, and HISTONE H4 plasmid DNA. Mean absolute quantities and sd were calculated by the Applied Biosystems SDS 2.1 software. Melt curve analyses were used to verify that a single product was produced from each qPCR.
BiFC
Plasmids used in BiFC experiments were constructed as follows. Sequences corresponding to the entire ORF of Arabidopsis PEX11a, PEX11b, PEX11c, PEX11d, PEX11e, FIS1a, FIS1b, or DRP3A were amplified by PCR using the appropriate plasmid (template) DNA and forward and reverse mutagenic primers (see Supplemental Table 6 online). PCR products encoding either the PEX11 isoforms, FIS1a, or FIS1b were digested with BamHI and ligated into the BamHI site of the multiple cloning region of pSAT4-nEYFP-C1 and/or pSAT4-cEYFP-C1(B), plant expression vectors that contain a tandem 35S cauliflower mosaic virus promoter, tobacco etch virus translation leader sequence, and encode the N-terminal (amino acid residues 1 to 174, nEYFP) and C-terminal (amino acid residues 175 to 239, cEYFP) halves of the enhanced yellow fluorescent protein (EYFP), respectively (kindly provided by Stanton Gelvin, Purdue University) (Citovsky et al., 2006). A similar procedure was used to construct pSAT4-nEYFP-C1 and pSAT4-cEYFP-C1(B) containing the DRP3A ORF [i.e., PCR products encoding the DRP3A ORF were digested with XhoI and ligated into XhoI-digested pSAT4-nEYFP-C1 and pSAT4-cEYFP-C1(B)]. Plasmids used as positive controls for BiFC included pE3012, encoding the Arabidopsis importin α protein (At IMPα4) fused to the N terminus of the C-terminal half of EYFP, and pE2946, encoding At IMPα4 fused to the N terminus of the N-terminal half of EYFP (Citovsky et al., 2006). pRTL2/RFP-MFP encodes the monomeric RFP fused to the full-length ORF of the rice peroxisomal MFP. pRTL2/RFP-MFP was constructed by digesting pRTL2/GFP-MFP (Chuong et al., 2005) with XbaI and ligating the resulting fragment into XbaI-digested pRTL2/RFP-MCS, a modified version of the plant expression vector pRTL2ΔN/S (Lee et al., 1997) that contains the ORF for a monomeric version of RFP (obtained from pmRFPC1; Clontech) followed by a multiple cloning site (MCS).
Biolistic bombardment of cells for BiFC experiments was performed essentially as described by Lingard and Trelease (2006) with the exception that tungsten particles were coated with 100 ng each of pSAT-nEYFP-C1 and pSAT-cEYFP-C1(B) plasmids containing the appropriate combinations of PEX11 isoforms, FIS1 isoforms, and DRP3A. Tungsten particles were coated also with 200 ng of pRTL2/RFP-MFP so that the expressed RFP-MFP fusion protein could serve as an internal reference marker to identify cells that were transformed and could be examined for the BiFC signal. Approximately 16 h after bombardment, cells were processed and examined by fluorescence microscopy (see below). The 16-h time point and the amounts of plasmid DNA employed in BiFC assays were chosen based on preliminary optimization experiments aimed at eliminating the possibility of any nonspecific (false positive) interactions that may occur due to protein overexpression (reviewed in Bhat et al., 2006). Overall, the conditions used here for BiFC were based on those in which nEYFP- or cEYFP-tagged versions of the PEX11 isoforms, FIS1 isoforms, or DRP3A coexpressed with empty vectors containing cEYFP or nEYFP alone yielded no significant YFP fluorescence. Also, transformations with MFP-RFP alone confirmed that there was no cross-bleed of the red fluorescence signal into the yellow channel.
Biolistic Cell Transformation and (Immuno)Fluorescence Microscopy
Transient transformations of Arabidopsis suspension cells and immunostaining were performed as previously described by Lingard and Trelease (2006). Briefly, transient transformations of unprotoplasted suspension cells (Figure 1) were performed using 5 μg of plasmid DNA (or 2.5 μg of each plasmid for cotransformations) with a biolistic particle delivery system (1000/HE; Bio-Rad Laboratories). Bombarded cells were incubated for 16 h to allow for expression and sorting of the introduced gene product(s), fixed in formaldehyde, incubated with 0.03% (w/v) cellulysin (Calbiochem) and 0.1% (w/v) pectinase (Sigma-Aldrich), and permeabilized with 0.3% Triton X-100 (v/v) (Sigma-Aldrich). Primary and secondary antibody sources and concentrations were as follows: mouse anti-myc monoclonal antibody 9E10 (1:250) (Santa Cruz Biotechnology) and affinity-purified (Protein A-Sepharose) rabbit anti-cottonseed catalase IgGs (1:500) (Kunce et al., 1988); goat anti-mouse Alexa Fluor 488 IgGs (1:1000) and goat anti-rabbit rhodamine red-X IgGs (1:500; Jackson ImmunoResearch Laboratories).
Confocal laser scanning microscopic images of Arabidopsis cells were acquired using a Leica DM RBE microscope with a Leica ×63 Plan Apochromat oil-immersion objective, a Leica TCS SP2 scanning head, and the Leica TCS NT software package (version 2.61). Fluorophore emissions were collected sequentially in double-labeling experiments. All single-labeling experiments showed no detectable crossover at the settings used for data collection. All images were acquired as single optical sections (∼0.4 μm) and were saved as 512 × 512-pixel digital images.
To prepare transformed protoplasts for (immuno)fluorescence observations, 250 μL of 0.8% (w/v) formaldehyde (prepared from paraformaldehyde) was added directly to 250 μL protoplasts, which were then mixed and incubated at room temperature in darkness for 45 min. After fixation, 1 mL of PBS (140 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, and 4.4 mM KH2PO4, pH 7.4) was added to protoplasts. GFP-transformed protoplasts were pelleted, washed in 1.5 mL PBS, and all but 250 μL of the supernatant was removed. To stain cell walls, the cells were incubated in 2.4 × 10−4% (w/v) FB28 in PBS for 7 min and then washed in 1 mL of PBS. Cells were adhered for at least 10 min to poly-l-lysine–coated microscope slides, covered with 1 volume N-propyl gallate (0.5 mg/mL in 45% [v/v] glycerol in PBS), and cover slipped.
Immunolabeling of protoplasts was done as described previously (Lisenbee et al., 2003; Flynn et al., 2005) with the following modifications. Briefly, 2 to 4 d after transformation, formaldehyde-fixed protoplasts (see above) were adhered to poly-l-lysine–coated slides for 30 min, incubated in 0.33% (v/v) Triton X-100 in PBS for 15 min, and washed twice in PBS. No cell wall digestion step was necessary for suitable immunolabeling of protoplasts. Cells were incubated for 45 min in primary antibodies (diluted in PBS and 5% [w/v] BSA), washed twice, and incubated 45 min in dye-conjugated secondary antibodies (diluted in PBS). Following the removal of secondary antibodies, the cell walls were stained, and then cells were treated with N-propyl gallate and cover slipped (see above). Cy2-conjugated secondary antibodies were purchased from Jackson ImmunoResearch Laboratories and diluted in PBS (1:250). Synchronized and bombarded suspension cells prepared in this study were fixed and immunolabeled as described previously (Lingard and Trelease, 2006).
Microscope observations were performed using either a Zeiss Axiovert 100 inverted or a Zeiss Axioskop 2 MOT microscope (Carl Zeiss). For the former, Cy2 and GFP fluorescence were observed using a modified filter set 09 (Carl Zeiss) in which the emitter was replaced with D540/40 (Chroma Technology). FB28 and DAPI fluorescence were observed using filter set 02 (Carl Zeiss). GFP and RFP fluorescence were observed with the Axioskop 2 MOT microscope using filter sets (Chroma Technology) with emitters HQ535/50 and HQ610/75, respectively.
For experiments that involved counting organelles in protoplasts, images of all (immuno)labeled organelles in overlapping z-sections (∼25 images per cell) were acquired with a Cool Snap ES CCD digital camera (Roper Scientific). For counting involving (unprotoplasted) synchronized cells, all organelles from an approximately equatorial plane were analyzed. Images were analyzed with MetaMorph software (Molecular Devices). Out-of-focus signal in three-dimensional reconstructions of cells was removed with the flatten background function, cell boundaries were outlined, and the number of organelles and flattened (two-dimensional) cell areas were recorded. Descriptive statistics and t tests were derived in Excel or SPSS 14.0. Images of cells for BiFC experiments were acquired with a Retiga 1300 CCD camera (Qimaging) and OpenLab software (version 5.0; Improvision). All image levels were adjusted and figures were compiled using Adobe Photoshop CS.
Synchronization of Arabidopsis Suspension Cells
Synchronization of cells was accomplished essentially as described (Menges and Murray, 2002). Briefly, 20 mL of a 10-d-old cell suspension were transferred into 40 mL of fresh growth media followed by the addition of 10.4 μg/mL amphidicolin (Sigma-Aldrich). After 36 h of incubation in amphidicolin, the cells were washed three times each in 100 mL growth media to remove the drug. At this point, staining with the vital dye Evans Blue revealed that virtually all of the cells were viable. Cells were then resuspended in 60 mL growth media and allowed to recover for 1 to 32 h. At various time points (Figures 7 and 8), a 0.5-μL aliquot of cells was frozen in liquid nitrogen for RNA analysis (see above) and a separate 2.5-mL aliquot was incubated for 1 h with 5 μL BrdU labeling reagent (GE Healthcare). Following BrdU labeling, cells were washed twice in PBS, fixed in 4% (w/v) formaldehyde for 1 h, and labeled (see above) with anti-BrdU antibodies (GE Healthcare), anticatalase antibodies, and DAPI (Sigma-Aldrich). For determination of the S-phase labeling index, the number of BrdU-positive nuclei was divided by the total number of DAPI-labeled nuclei. For determination of the M-phase mitotic index, the number of cells with obvious metaphase or anaphase mitotic profiles was divided by the total number of nuclei.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: PEX11a (At1g47750), PEX11b (At3g47430), PEX11c (At1g01820), PEX11d (At2g45740), PEX11e (At3g61070), ACT2 (At3g18780), FIS1a (At3g57090), FIS1b (At5g12390), DRP3A (At4g33650), and HISTONE H4 (At2g28740).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Colocalization of myc-DRP3A with MITO-GFP in Cotransformed Arabidopsis Suspension Cells.
Supplemental Figure 2. Sequence Alignment of Arabidopsis PEX11a, PEX11b, PEX11c, PEX11d, and PEX11e Coding Sequences and 3′ Untranslated Regions.
Supplemental Table 1. Names of Transient RNAi Cells, dsRNA Constructs, and Plasmid DNA Used for Protoplast Transformations.
Supplemental Table 2. P Values (Two-Tail) for Samples in Figure 4A from Two-Sample t Tests.
Supplemental Table 3. P Values (Two-Tail) for Samples in Figure 5A from Two-Sample t Tests.
Supplemental Table 4. P Values (Two-Tail) for Samples in Figure 5B from Two-Sample t Tests.
Supplemental Table 5. P Values (Two-Tail) for Samples in Figure 6A from Two-Sample t Tests.
Supplemental Table 6. Synthetic Oligonucleotide Primers Used in This Study.
Supplemental Data Set 1. Text File of Alignment Corresponding to Supplemental Figure 2.
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation (Grant MCB-0091826) (R.N.T.), the Natural Sciences and Engineering Research Council (Grant 217291) (R.T.M.), and the Ontario Research and Development Challenge Fund (Ontario Centre for Agriculture Genomics; Grant 046061) (S.J.R.). We thank David Rhoads for the PEX11e cDNA, Stanton Gelvin for BiFC plasmids, Uwe Schumann for constructing GFP-PEROX and pRTL2/RFP-MFP, Rosa Di Leo for constructing MITO-GFP, Wayne Snedden for the Arabidopsis NADK cDNA, Robert Roberson for expert guidance on image presentation and providing FB28, and Bonnie Bartel for donating RT-PCR supplies. We also thank Erin Chase Lingard for direction with the statistical analyses, Graham Smith and Kimberly Gibson for cell culture maintenance, Heather Tardif for cell culture maintenance and assistance with protoplasting experiments, and Bonnie Bartel, Andrew Woodward, Sarah Ratzel, and Elizabeth Poggio for comments on the manuscript. PEX11b, FIS1a, FIS1b, and DRP3A cDNAs were provided by the ABRC.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Richard N. Trelease (trelease.dick@asu.edu).
Online version contains Web-only data.
References
- Abe, I., Okumoto, K., Tamura, S., and Fujiki, Y. (1998). Clofibrate-inducible, 28-kDa peroxisomal integral membrane protein is encoded by PEX11. FEBS Lett. 431 468–472. [DOI] [PubMed] [Google Scholar]
- Afitlhile, M.M., Fukaushige, H., Nishimura, M., and Hildebrand, D.F. (2005). A defect in glyoxysomal fatty acid b-oxidation reduces jasmonic acid accumulation in Arabidopsis. Plant Physiol. Biochem. 43 603–609. [DOI] [PubMed] [Google Scholar]
- Arimura, S., and Tsutsumi, N. (2002). A dynamin-like protein (ADL2b), rather than FtsZ, is involved in Arabidopsis mitochondrial division. Proc. Natl. Acad. Sci. USA 99 5727–5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett, P., Tabak, H.F., and Hettema, E.H. (2000). Nuclear receptors arose from pre-existing protein modules during evolution. Trends Biochem. Sci. 25 227–228. [DOI] [PubMed] [Google Scholar]
- Bhat, R.A., Lahaye, T., and Panstruga, R. (2006). The visible touch: In planta visualization of protein-protein interactions by fluorophore-based methods. Plant Methods 2: 12. [DOI] [PMC free article] [PubMed]
- Borgese, N., Brambillasca, S., and Colombo, S. (2007). How tails guide tail-anchored proteins to their destinations. Curr. Opin. Cell Biol. 19 368–375. [DOI] [PubMed] [Google Scholar]
- Bracha-Drori, K., Shichrur, K., Katz, A., Oliva, M., Angelovici, R., Yalovsky, S., and Ohad, N. (2004). Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J. 40 419–427. [DOI] [PubMed] [Google Scholar]
- Chaubet, N., Flenet, M., Clement, B., Brignon, P., and Gigot, C. (1996). Identification of cis-elements regulating the expression of an Arabidopsis histone H4 gene. Plant J. 10 425–435. [DOI] [PubMed] [Google Scholar]
- Chaumont, F., Silva Filho Mde, C., Thomas, D., Leterme, S., and Boutry, M. (1994). Truncated presequences of mitochondrial F1-ATPase beta subunit from Nicotiana plumbaginifolia transport CAT and GUS proteins into mitochondria of transgenic tobacco. Plant Mol. Biol. 24 631–641. [DOI] [PubMed] [Google Scholar]
- Chuong, S.D., Park, N.I., Freeman, M.C., Mullen, R.T., and Muench, D.G. (2005). The peroxisomal multifunctional protein interacts with cortical microtubules in plant cells. BMC Cell Biol. 6 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong, S.D.X., Mullen, R.T., and Muench, D.G. (2002). Identification of a rice RNA- and microtubule-binding protein as the multifunctional protein, a peroxisomal enzyme involved in the beta-oxidation of fatty acids. J. Biol. Chem. 277 2419–2429. [DOI] [PubMed] [Google Scholar]
- Citovsky, V., Lee, L.Y., Vyas, S., Glick, E., Chen, M.H., Vainstein, A., Gafni, Y., Gelvin, S.B., and Tzfira, T. (2006). Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J. Mol. Biol. 362 1120–1131. [DOI] [PubMed] [Google Scholar]
- Desai, M., and Hu, J. (2008). Light induces peroxisome proliferation in Arabidopsis seedlings through the photoreceptor phytochrome A, the transcription factor HY5 HOMOLOG, and the peroxisomal protein PEROXIN11b. Plant Physiol. 146 1117–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling, J.H., and Pikaard, C.S. (1993). Transient expression in Arabidopsis thaliana protoplasts derived from rapidly established cell-suspension cultures. Plant Cell Rep. 12 241–244. [DOI] [PubMed] [Google Scholar]
- Draviam, V.M., Orrechia, S., Lowe, M., Pardi, R., and Pines, J. (2001). The localization of human cyclins B1 and B2 determines CDK1 substrate specificity and neither enzyme requires MEK to disassemble the Golgi apparatus. J. Cell Biol. 152 945–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagarasanu, A., Fagarasanu, M., and Rachubinski, R.A. (2007). Maintaining peroxisome populations: A story of division and inheritance. Annu. Rev. Cell Dev. Biol. 23 321–344. [DOI] [PubMed] [Google Scholar]
- Flynn, C.R., Heinze, M., Schumann, U., Gietl, C., and Trelease, R.N. (2005). Compartmentalization of the plant peroxin, AtPex10p, within subdomain(s) of ER. Plant Sci. 168 635–652. [Google Scholar]
- Fritze, C.E., and Anderson, T.R. (2000). Epitope tagging: General method for tracking recombinant proteins. Methods Enzymol. 327 3–16. [DOI] [PubMed] [Google Scholar]
- Haan, G.-J., Baerends, R.J.S., Krikken, A.M., Otzen, M., Veenhuis, M., and Klei, I.J. (2006). Reassembly of peroxisomes in Hansenula polymorpha pex3 cells on reintroduction of Pex3p involves the nuclear envelope. FEMS Yeast Res. 6 186–194. [DOI] [PubMed] [Google Scholar]
- Hoepfner, D., Schidknegt, D., Braakman, I., Philippsen, P., and Tabak, H.F. (2005). Contribution of the endoplasmic reticulum to peroxisome formation. Cell 122 85–95. [DOI] [PubMed] [Google Scholar]
- Hoepfner, D., van den Berg, M., Philippsen, P., Tabak, H.F., and Hettema, E.H. (2001). A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae. J. Cell Biol. 155 979–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, P.K., Mullen, R.T., Schumann, U., and Lippincott-Schwartz, J. (2006). The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J. Cell Biol. 173 521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, S., Tanaka, A., and Fujiki, Y. (2007). Fis1, DLP1, and Pex11p coordinately regulate peroxisome morphogenesis. Exp. Cell Res. 313 1675–1686. [DOI] [PubMed] [Google Scholar]
- Koch, A., Schneider, G., Luers, G.H., and Schrader, M. (2004). Peroxisome elongation and constriction but not fission can occur independently of dynamin-like protein 1. J. Cell Sci. 117 3995–4006. [DOI] [PubMed] [Google Scholar]
- Koch, A., Thiemann, M., Grabenbauer, M., Yoon, Y., McNiven, M.A., and Schrader, M. (2003). Dynamin-like protein 1 is involved in peroxisomal fission. J. Biol. Chem. 278 8597–8605. [DOI] [PubMed] [Google Scholar]
- Koch, A., Yoon, Y., Bonekamp, N.A., McNiven, M.A., and Schrader, M. (2005). A role for Fis1 in both mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell 16 5077–5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragt, A., Voorn-Brouwer, T., van den Berg, M., and Distel, B. (2005). Endoplasmic reticulum-directed Pex3p routes to peroxisomes and restores peroxisome formation in a Saccharomyces cerevisiae pex3Δ strain. J. Biol. Chem. 280 34350–34357. [DOI] [PubMed] [Google Scholar]
- Kunce, C.M., Trelease, R.N., and Turley, R.B. (1988). Purification and biosynthesis of cottonseed (Gossypium hirsutum L) catalase. Biochem. J. 251 147–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuravi, K., Nagotu, S., Krikken, A.M., Sjollema, K., Deckers, M., Erdmann, R., Veenhuis, M., and van der Klei, I.J. (2006). Dynamin-related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisiae. J. Cell Sci. 119 3994–4001. [DOI] [PubMed] [Google Scholar]
- Leber, B., Lin, J., and Andrews, D.W. (2007). Embedded together: the life and death consequences of interaction of the Bcl-2 family with membranes. Apoptosis 12 897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M.S., Mullen, R.T., and Trelease, R.N. (1997). Oilseed isocitrate lyases lacking their essential type 1 peroxisomal targeting signal are piggybacked to glyoxysomes. Plant Cell 9 185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Baumgart, E., Dong, G.X., Morrell, J.C., Jimenez-Sanchez, G., Valle, D., Smith, K.D., and Gould, S.J. (2002. b). PEX11α is required for peroxisome proliferation in response to 4-phenylbutyrate but is dispensable for peroxisome proliferator-activated receptor alpha-mediated peroxisome proliferation. Mol. Cell. Biol. 22 8226–8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Baumgart, E., Morrell, J.C., Jimenez-Sanchez, G., Valle, D., and Gould, S.J. (2002. a). PEX11β deficiency is lethal and impairs neuronal migration but does not abrogate peroxisome function. Mol. Cell. Biol. 22 4358–4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., and Gould, S.J. (2002). PEX11 promotes peroxisome division independently of peroxisome metabolism. J. Cell Biol. 156 643–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., and Gould, S.J. (2003). The dynamin-like GTPase DLP1 is essential for peroxisome division and is recruited to peroxisomes in part by PEX11. J. Biol. Chem. 278 17012–17020. [DOI] [PubMed] [Google Scholar]
- Lingard, M.J., and Trelease, R.N. (2006). Five Arabidopsis peroxin 11 homologs individually promote peroxisome elongation, duplication or aggregation. J. Cell Sci. 119 1961–1972. [DOI] [PubMed] [Google Scholar]
- Lisenbee, C.S., Heinze, M., and Trelease, R.N. (2003). Peroxisomal ascorbate peroxidase resides within a subdomain of rough endoplasmic reticulum in wild-type Arabidopsis cells. Plant Physiol. 132 870–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisenbee, C.S., Lingard, M.J., and Trelease, R.N. (2005). Arabidopsis peroxisomes possess functionally redundant membrane and matrix isoforms of monodehydroascorbate reductase. Plant J. 43 900–914. [DOI] [PubMed] [Google Scholar]
- Lorenz, P., Maier, A.G., Baumgart, E., Erdmann, R., and Clayton, C. (1998). Elongation and clustering of glycosomes in Trypanosoma brucei overexpressing the glycosomal Pex11p. EMBO J. 17 3542–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe, M., Gonatas, N.K., and Warren, G. (2000). The mitotic phosphorylation cycle of the cis-Golgi matrix protein GM130. J. Cell Biol. 149 341–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe, M., Rabouille, C., Nakamura, N., Watson, R., Jackman, M., Jamsa, E., Rahman, D., Pappin, D.J., and Warren, G. (1998). Cdc2 kinase directly phosphorylates the cis-Golgi matrix protein GM130 and is required for Golgi fragmentation in mitosis. Cell 94 783–793. [DOI] [PubMed] [Google Scholar]
- Mano, S., Nakamori, C., Kondo, M., Hayashi, M., and Nishimura, M. (2004). An Arabidopsis dynamin-related protein, DRP3A, controls both peroxisomal and mitochondrial division. Plant J. 38 487–498. [DOI] [PubMed] [Google Scholar]
- Mano, S., and Nishimura, M. (2005). Plant peroxisomes. Vitam. Horm. 72 111–154. [DOI] [PubMed] [Google Scholar]
- Marshall, P.A., Dyer, J.M., Quick, M.E., and Goodman, J.M. (1996). Redox-sensitive homodimerization of Pex11p: A proposed mechanism to regulate peroxisomal division. J. Cell Biol. 135 123–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, P.A., Krimkevich, Y.I., Lark, R.H., Dyer, J.M., Veenhuis, M., and Goodman, J.M. (1995). Pmp27 promotes peroxisomal proliferation. J. Cell Biol. 129 345–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur, J., Koncz, C., and Szabados, L. (1995). A simple method for isolation, liquid culture, transformation and regeneration of Arabidopsis thaliana protoplasts. Plant Cell Rep. 14 221–226. [DOI] [PubMed] [Google Scholar]
- McCartney, A.W., Greenwood, J.S., Fabian, M.R., White, K.A., and Mullen, R.T. (2005). Localization of the tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway. Plant Cell 17 3513–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges, M., and Murray, J.A.H. (2002). Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J. 30 203–212. [DOI] [PubMed] [Google Scholar]
- Motley, A.M., and Hettema, E.H. (2007). Yeast peroxisomes multiply by growth and division. J. Cell Biol. 178 399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen, R.T., Lisenbee, C.S., Flynn, C.R., and Trelease, R.N. (2001). Stable and transient expression of chimeric peroxisomal membrane proteins induces an independent “zippering” of peroxisomes and an endoplasmic reticulum subdomain. Planta 213 849–863. [DOI] [PubMed] [Google Scholar]
- Mullen, R.T., and Trelease, R.N. (2006). The ER-peroxisome connection in plants: Development of the “ER semi-autonomous peroxisome maturation and replication” model for plant peroxisome biogenesis. Biochim. Biophys. Acta 1763 1655–1668. [DOI] [PubMed] [Google Scholar]
- Murphy, M.A., Phillipson, B.A., Baker, A., and Mullen, R.T. (2003). Characterization of the targeting signal of the Arabidopsis 22-kD integral peroxisomal membrane protein. Plant Physiol. 133 813–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenführ, A., Gallagher, L.A., Dunahay, T.G., Frohlick, J.A., Mazurkiewicz, A.M., Meehl, J.B., and Staehelin, L.A. (1999). Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiol. 121 1127–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nito, K., Kamigaki, A., Kondo, M., Hayashi, M., and Nishimura, M. (2007). Functional classification of Arabidopsis peroxisome biogenesis factors proposed from analyses of knockdown mutants. Plant Cell Physiol. 48 763–774. [DOI] [PubMed] [Google Scholar]
- Nyathi, Y., and Baker, A. (2006). Plant peroxisomes as a source of signalling molecules. Biochim. Biophys. Acta 1763 1478–1495. [DOI] [PubMed] [Google Scholar]
- Orth, T., Reumann, S., Zhang, X., Fan, J., Wenzel, D., Quan, S., and Hu, J. (2007). The PEROXIN11 protein family controls peroxisome proliferation in Arabidopsis. Plant Cell 19 333–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud, C., Perennes, C., Reuzeau, C., Catrice, O., Brown, S., and Bergounioux, C. (2005). Cell and plastid division are coordinated through the prereplication factor AtCDT1. Proc. Natl. Acad. Sci. USA 102 8216–8221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann, S., and Weber, A.P.M. (2006). Plant peroxisomes respire in the light: Some gaps of the photorespiratory C-2 cycle have become filled - Others remain. Biochim. Biophys. Acta 1763 1496–1510. [DOI] [PubMed] [Google Scholar]
- Rottensteiner, H., Stein, K., Sonnenhol, E., and Erdmann, R. (2003). Conserved function of Pex11p and the novel Pex25p and Pex27p in peroxisome biogenesis. Mol. Biol. Cell 14 4316–4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai, Y., Marshall, P.A., Saiganji, A., Takabe, K., Saiki, H., Kato, N., and Goodman, J.M. (1995). The Candida boidinii peroxisomal membrane protein Pmp30 has a role in peroxisomal proliferation and is functionally homologous to Pmp27 from Saccharomyces cerevisiae. J. Bacteriol. 177 6773–6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader, M., and Fahimi, H.D. (2006). Growth and division of peroxisomes. Int. Rev. Cytol. 255 237–289. [DOI] [PubMed] [Google Scholar]
- Schrader, M., Krieglstein, K., and Fahimi, H.D. (1998. a). Tubular peroxisomes in HepG2 cells: Selective induction by growth factors and arachidonic acid. Eur. J. Cell Biol. 75 87–96. [DOI] [PubMed] [Google Scholar]
- Schrader, M., Reuber, B.E., Morrell, J.C., Jimenez-Sanchez, G., Obie, C., Stroh, T.A., Valle, D., Schroer, T.A., and Gould, S.J. (1998. b). Expression of PEX11 beta mediates peroxisome proliferation in the absence of extracellular stimuli. J. Biol. Chem. 273 29607–29614. [DOI] [PubMed] [Google Scholar]
- Schrader, M., Wodopia, R., and Fahimi, H.D. (1999). Induction of tubular peroxisomes by UV irradiation and reactive oxygen species in HepG2 cells. J. Histochem. Cytochem. 47 1141–1148. [DOI] [PubMed] [Google Scholar]
- Schrader, M., and Yoon, Y. (2007). Mitochondria and peroxisomes: Are the ‘big brother’ and the ‘little sister’ closer than assumed? Bioessays 29 1105–1114. [DOI] [PubMed] [Google Scholar]
- Scott, I., Tobin, A.K., and Logan, D.C. (2006). BIGYIN, an orthologue of human and yeast FIS1 genes functions in the control of mitochondrial size and number in Arabidopsis thaliana. J. Exp. Bot. 57 1275–1280. [DOI] [PubMed] [Google Scholar]
- Shockey, J.M., Gidda, S.K., Chapital, D.C., Kuan, J.C., Dhanoa, P.K., Bland, J.M., Rothstein, S.J., Mullen, R.T., and Dyer, J.M. (2006). Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 18 2294–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J.J., Marelli, M., Christmas, R.H., Vizeacoumar, F.J., Dilworth, D.J., Ideker, T., Galitski, T., Dimitrov, K., Rachubinski, R.A., and Aitchison, J.D. (2002). Transcriptome profiling to identify genes involved in peroxisome assembly and function. J. Cell Biol. 158 259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterlin, C., Hsu, P., Mallabiabarrena, A., and Malhotra, V. (2002). Fragmentation and dispersal of the pericentriolar Golgi complex is required for entry into mitosis in mammalian cells. Cell 109 359–369. [DOI] [PubMed] [Google Scholar]
- Tabak, H.F., Murk, J.L., Braakman, I., and Geuze, H.J. (2003). Peroxisomes start their life in the endoplasmic reticulum. Traffic 4 512–518. [DOI] [PubMed] [Google Scholar]
- Tam, Y.Y.C., Torres-Guzman, J.C., Vizeacoumar, F.J., Smith, J.J., Marelli, M., Aitchison, J.D., and Rachubinski, R.A. (2003). Pex11-related proteins in peroxisome dynamics: a role for novel peroxin Pex27p in controlling peroxisome size and number in Saccharomyces cerevisiae. Mol. Biol. Cell 14 4089–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoulou, F.L., Holdsworth, M., and Baker, A. (2006). Peroxisomal ABC transporters. FEBS Lett. 580 1139–1155. [DOI] [PubMed] [Google Scholar]
- Thoms, S., and Erdmann, R. (2005). Dynamin-related proteins and Pex11 proteins in peroxisome division and proliferation. FEBS J. 272 5169–5181. [DOI] [PubMed] [Google Scholar]
- Titorenko, V.I., and Mullen, R.T. (2006). Peroxisome biogenesis: The peroxisomal endomembrane system and the role of the ER. J. Cell Biol. 174 11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trelease, R.N., and Lingard, M.J. (2006). Particpation of the plant ER in peroxisomal biogenesis. In The Plant Endoplasmic Reticulum, D.G. Robinson, ed (Heidelberg, Germany: Springer-Verlag), pp. 205–232.
- Walter, M., Chaban, C., Schutze, K., Batistic, O., Weckermann, K., Nake, C., Blazevic, D., Grefen, C., Schumacher, K., Oecking, C., Harter, K., and Kudla, J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40 428–438. [DOI] [PubMed] [Google Scholar]
- Yan, M., Rayapuram, N., and Subramani, S. (2005). The control of peroxisome number and size during division and proliferation. Curr. Opin. Cell Biol. 17 1–8. [DOI] [PubMed] [Google Scholar]
- Yoon, Y., Krueger, E.W., Oswald, B.J., and McNiven, M.A. (2003). The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol. Cell. Biol. 23 5409–5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, T., Fox, R.J., Burwell, L.S., and Yoon, Y. (2005). Regulation of mitochondrial fission and apoptosis by the mitochondrial outer membrane protein hFis1. J. Cell Sci. 118 4141–4151. [DOI] [PubMed] [Google Scholar]
- Zolman, B.K., Yoder, A., and Bartel, B. (2000). Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156 1323–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.