Abstract
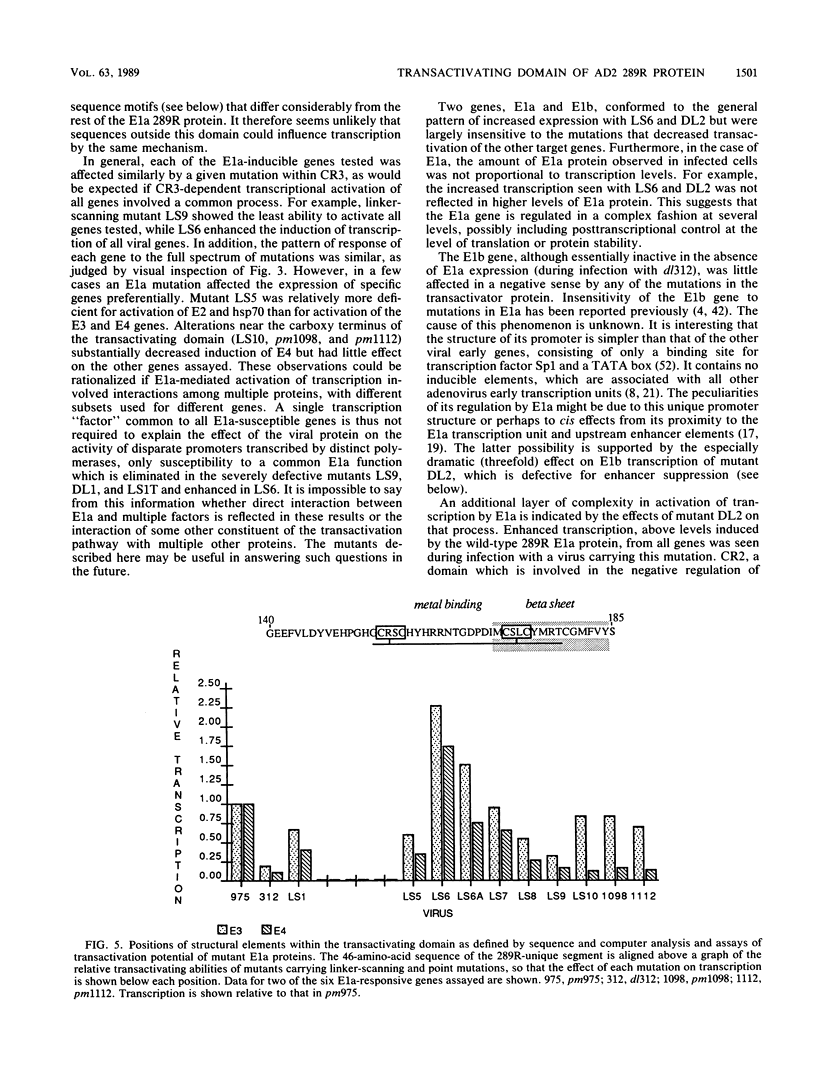
A series of linker-scanning, deletion, and frameshift mutations were made in the pm975 variant of the adenovirus type 2 E1a gene, which expresses only the larger of the two major E1a proteins. Most of these were within the 46-amino-acid segment unique to the larger E1a protein product (the 289R protein), which confers on it the ability to activate in trans the expression of other genes. The mutations were recombined into virus and assayed by in vitro transcription in nuclei isolated from infected cells for their ability to activate the transcription of other viral early genes and of the endogenous hsp70 gene. Mutant E1a proteins from which the 289R-unique segment was removed by deletion or truncation did not completely lose the ability to transactivate by comparison with a virus which makes no E1a at all, indicating that sequences outside this domain are active in the positive regulation of transcription. The E1a mutations tested fell into several classes: those that increased transactivation of virtually all genes, those that severely depressed transactivation of all genes, and those that depressed transactivation only moderately. Each mutation had similar effects on the expression of all transcription units tested, indicating a common process in their transactivation. However, some mutants in the third category decreased transactivation of some induced genes more severely than of others. Such gene-specific defects suggest the existence of subclasses of E1a-responsive transcription units, consistent with the involvement of diverse proteins in the transactivation of different genes. Two specific structural components of the transactivating domain, a putative metal-binding element and a region with high potential for beta-sheet formation at its carboxy-terminus, appear to be important to the transactivation function.
Full text
PDF


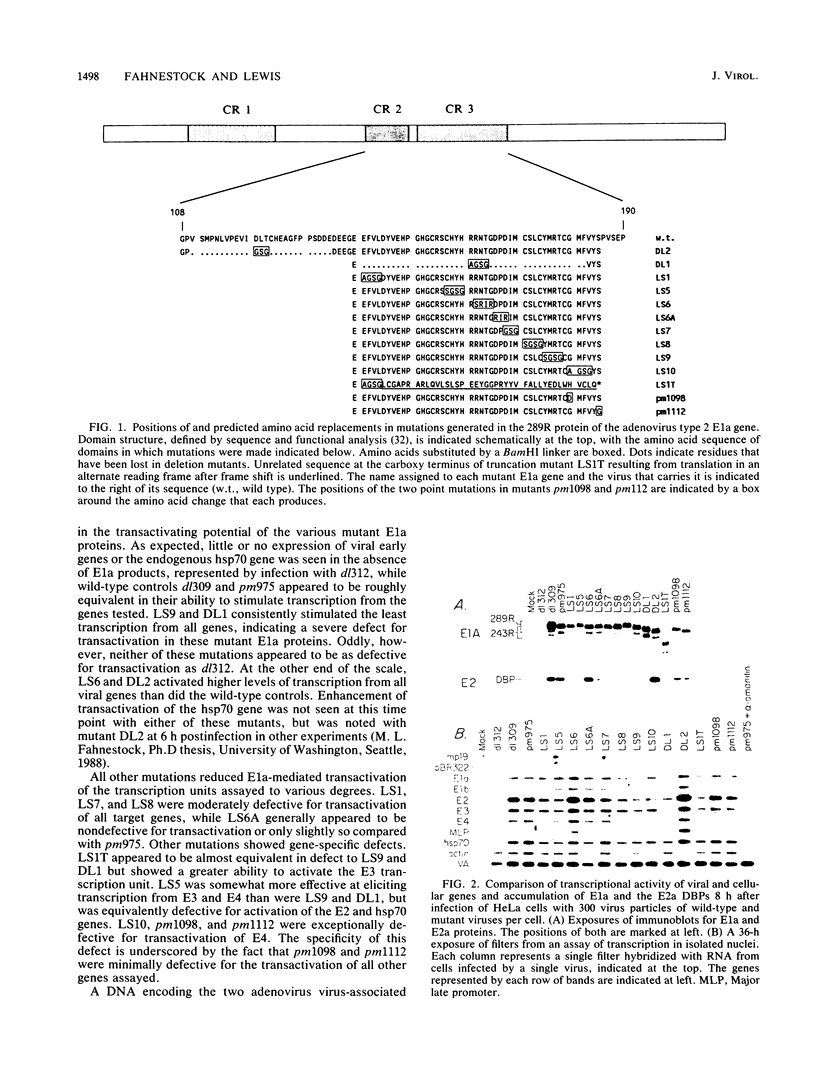
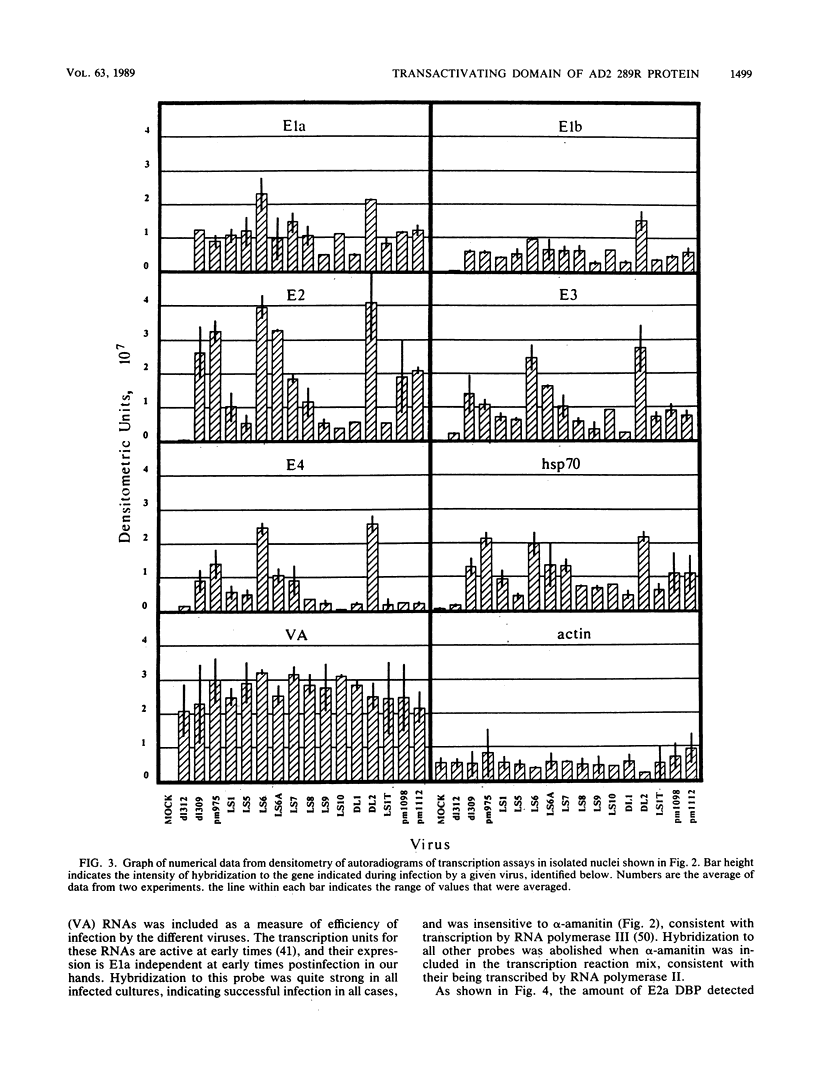
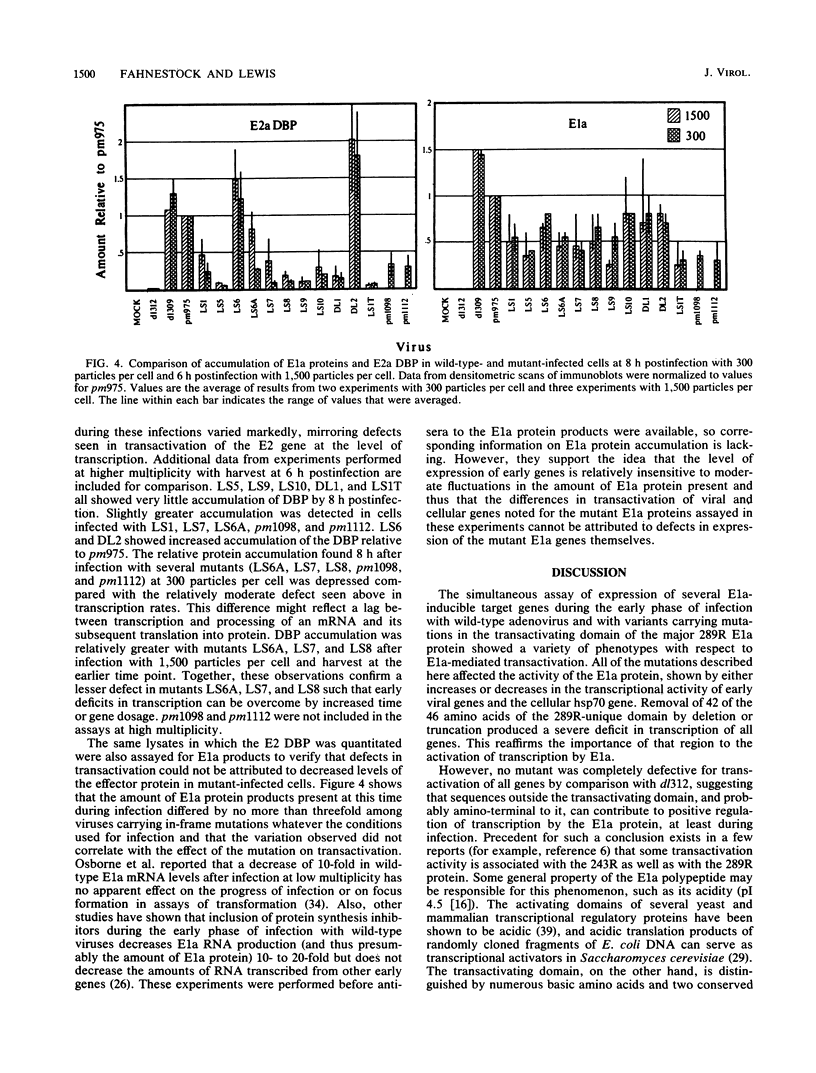



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berg J. M. Potential metal-binding domains in nucleic acid binding proteins. Science. 1986 Apr 25;232(4749):485–487. doi: 10.1126/science.2421409. [DOI] [PubMed] [Google Scholar]
- Berk A. J. Adenovirus promoters and E1A transactivation. Annu Rev Genet. 1986;20:45–79. doi: 10.1146/annurev.ge.20.120186.000401. [DOI] [PubMed] [Google Scholar]
- Berk A. J. Functions of adenovirus E1A. Cancer Surv. 1986;5(2):367–387. [PubMed] [Google Scholar]
- Carlock L. R., Jones N. C. Transformation-defective mutant of adenovirus type 5 containing a single altered E1a mRNA species. J Virol. 1981 Dec;40(3):657–664. doi: 10.1128/jvi.40.3.657-664.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- Ferguson B., Krippl B., Andrisani O., Jones N., Westphal H., Rosenberg M. E1A 13S and 12S mRNA products made in Escherichia coli both function as nucleus-localized transcription activators but do not directly bind DNA. Mol Cell Biol. 1985 Oct;5(10):2653–2661. doi: 10.1128/mcb.5.10.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowlkes D. M., Shenk T. Transcriptional control regions of the adenovirus VAI RNA gene. Cell. 1980 Nov;22(2 Pt 2):405–413. doi: 10.1016/0092-8674(80)90351-7. [DOI] [PubMed] [Google Scholar]
- Garcia J., Wu F., Gaynor R. Upstream regulatory regions required to stabilize binding to the TATA sequence in an adenovirus early promoter. Nucleic Acids Res. 1987 Oct 26;15(20):8367–8385. doi: 10.1093/nar/15.20.8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W. Genes-in-pieces revisited. Science. 1985 May 17;228(4701):823–824. doi: 10.1126/science.4001923. [DOI] [PubMed] [Google Scholar]
- Gingeras T. R., Sciaky D., Gelinas R. E., Bing-Dong J., Yen C. E., Kelly M. M., Bullock P. A., Parsons B. L., O'Neill K. E., Roberts R. J. Nucleotide sequences from the adenovirus-2 genome. J Biol Chem. 1982 Nov 25;257(22):13475–13491. [PubMed] [Google Scholar]
- Glenn G. M., Ricciardi R. P. Adenovirus 5 early region 1A host range mutants hr3, hr4, and hr5 contain point mutations which generate single amino acid substitutions. J Virol. 1985 Oct;56(1):66–74. doi: 10.1128/jvi.56.1.66-74.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., Smiley J., Russell W. C., Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977 Jul;36(1):59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- Green M., Loewenstein P. M., Pusztai R., Symington J. S. An adenovirus E1A protein domain activates transcription in vivo and in vitro in the absence of protein synthesis. Cell. 1988 Jun 17;53(6):921–926. doi: 10.1016/s0092-8674(88)90429-1. [DOI] [PubMed] [Google Scholar]
- Gunning P., Ponte P., Okayama H., Engel J., Blau H., Kedes L. Isolation and characterization of full-length cDNA clones for human alpha-, beta-, and gamma-actin mRNAs: skeletal but not cytoplasmic actins have an amino-terminal cysteine that is subsequently removed. Mol Cell Biol. 1983 May;3(5):787–795. doi: 10.1128/mcb.3.5.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison T., Graham F., Williams J. Host-range mutants of adenovirus type 5 defective for growth in HeLa cells. Virology. 1977 Mar;77(1):319–329. doi: 10.1016/0042-6822(77)90428-7. [DOI] [PubMed] [Google Scholar]
- Harter M. L., Lewis J. B. Adenovirus type 2 early proteins synthesized in vitro and in vivo: identification in infected cells of the 38,000- to 50,000- molecular-weight protein encoded by the left end of the adenovirus type 2 genome. J Virol. 1978 Jun;26(3):736–749. doi: 10.1128/jvi.26.3.736-749.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearing P., Shenk T. The adenovirus type 5 E1A enhancer contains two functionally distinct domains: one is specific for E1A and the other modulates all early units in cis. Cell. 1986 Apr 25;45(2):229–236. doi: 10.1016/0092-8674(86)90387-9. [DOI] [PubMed] [Google Scholar]
- Hearing P., Shenk T. The adenovirus type 5 E1A transcriptional control region contains a duplicated enhancer element. Cell. 1983 Jul;33(3):695–703. doi: 10.1016/0092-8674(83)90012-0. [DOI] [PubMed] [Google Scholar]
- Hen R., Borrelli E., Sassone-Corsi P., Chambon P. An enhancer element is located 340 base pairs upstream from the adenovirus-2 E1A capsite. Nucleic Acids Res. 1983 Dec 20;11(24):8747–8760. doi: 10.1093/nar/11.24.8747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffler W. K., Roeder R. G. Enhancement of RNA polymerase III transcription by the E1A gene product of adenovirus. Cell. 1985 Jul;41(3):955–963. doi: 10.1016/s0092-8674(85)80076-3. [DOI] [PubMed] [Google Scholar]
- Jones N. C., Rigby P. W., Ziff E. B. Trans-acting protein factors and the regulation of eukaryotic transcription: lessons from studies on DNA tumor viruses. Genes Dev. 1988 Mar;2(3):267–281. doi: 10.1101/gad.2.3.267. [DOI] [PubMed] [Google Scholar]
- Kao H. T., Nevins J. R. Transcriptional activation and subsequent control of the human heat shock gene during adenovirus infection. Mol Cell Biol. 1983 Nov;3(11):2058–2065. doi: 10.1128/mcb.3.11.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimelman D., Miller J. S., Porter D., Roberts B. E. E1a regions of the human adenoviruses and of the highly oncogenic simian adenovirus 7 are closely related. J Virol. 1985 Feb;53(2):399–409. doi: 10.1128/jvi.53.2.399-409.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J. L., Dalie B. L., Goldman E., Harter M. L. Adenovirus-2 early region IA protein synthesized in Escherichia coli extracts indirectly associates with DNA. EMBO J. 1986 Jul;5(7):1645–1651. doi: 10.1002/j.1460-2075.1986.tb04407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leong K., Berk A. J. Adenovirus early region 1A protein increases the number of template molecules transcribed in cell-free extracts. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5844–5848. doi: 10.1073/pnas.83.16.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. B., Mathews M. B. Control of adenovirus early gene expression: a class of immediate early products. Cell. 1980 Aug;21(1):303–313. doi: 10.1016/0092-8674(80)90138-5. [DOI] [PubMed] [Google Scholar]
- Lillie J. W., Green M., Green M. R. An adenovirus E1a protein region required for transformation and transcriptional repression. Cell. 1986 Sep 26;46(7):1043–1051. doi: 10.1016/0092-8674(86)90704-x. [DOI] [PubMed] [Google Scholar]
- Lillie J. W., Loewenstein P. M., Green M. R., Green M. Functional domains of adenovirus type 5 E1a proteins. Cell. 1987 Sep 25;50(7):1091–1100. doi: 10.1016/0092-8674(87)90175-9. [DOI] [PubMed] [Google Scholar]
- Ma J., Ptashne M. A new class of yeast transcriptional activators. Cell. 1987 Oct 9;51(1):113–119. doi: 10.1016/0092-8674(87)90015-8. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Kingsbury R. Transcriptional control signals of a eukaryotic protein-coding gene. Science. 1982 Jul 23;217(4557):316–324. doi: 10.1126/science.6283634. [DOI] [PubMed] [Google Scholar]
- Montell C., Fisher E. F., Caruthers M. H., Berk A. J. Resolving the functions of overlapping viral genes by site-specific mutagenesis at a mRNA splice site. Nature. 1982 Feb 4;295(5848):380–384. doi: 10.1038/295380a0. [DOI] [PubMed] [Google Scholar]
- Moran E., Grodzicker T., Roberts R. J., Mathews M. B., Zerler B. Lytic and transforming functions of individual products of the adenovirus E1A gene. J Virol. 1986 Mar;57(3):765–775. doi: 10.1128/jvi.57.3.765-775.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran E., Mathews M. B. Multiple functional domains in the adenovirus E1A gene. Cell. 1987 Jan 30;48(2):177–178. doi: 10.1016/0092-8674(87)90418-1. [DOI] [PubMed] [Google Scholar]
- Osborne T. F., Gaynor R. B., Berk A. J. The TATA homology and the mRNA 5' untranslated sequence are not required for expression of essential adenovirus E1A functions. Cell. 1982 May;29(1):139–148. doi: 10.1016/0092-8674(82)90098-8. [DOI] [PubMed] [Google Scholar]
- Palmer D. K., O'Day K., Wener M. H., Andrews B. S., Margolis R. L. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J Cell Biol. 1987 Apr;104(4):805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perricaudet M., le Moullec J. M., Tiollais P., Pettersson U. Structure of two adenovirus type 12 transforming polypeptides and their evolutionary implications. Nature. 1980 Nov 13;288(5787):174–176. doi: 10.1038/288174a0. [DOI] [PubMed] [Google Scholar]
- Schneider J. F., Fisher F., Goding C. R., Jones N. C. Mutational analysis of the adenovirus E1a gene: the role of transcriptional regulation in transformation. EMBO J. 1987 Jul;6(7):2053–2060. doi: 10.1002/j.1460-2075.1987.tb02470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenk T., Jones N., Colby W., Fowlkes D. Functional analysis of adenovirus-5 host-range deletion mutants defective for transformation of rat embryo cells. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):367–375. doi: 10.1101/sqb.1980.044.01.041. [DOI] [PubMed] [Google Scholar]
- Sigler P. B. Transcriptional activation. Acid blobs and negative noodles. Nature. 1988 May 19;333(6170):210–212. doi: 10.1038/333210a0. [DOI] [PubMed] [Google Scholar]
- Simon M. C., Kitchener K., Kao H. T., Hickey E., Weber L., Voellmy R., Heintz N., Nevins J. R. Selective induction of human heat shock gene transcription by the adenovirus E1A gene products, including the 12S E1A product. Mol Cell Biol. 1987 Aug;7(8):2884–2890. doi: 10.1128/mcb.7.8.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solnick D., Anderson M. A. Transformation-deficient adenovirus mutant defective in expression of region 1A but not region 1B. J Virol. 1982 Apr;42(1):106–113. doi: 10.1128/jvi.42.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangler R., Bruner M., Dalie B., Harter M. L. Activation of adenovirus promoters by the adenovirus E1A protein in cell-free extracts. Science. 1987 Aug 28;237(4818):1044–1046. doi: 10.1126/science.2956686. [DOI] [PubMed] [Google Scholar]
- Spindler K. R., Rosser D. S., Berk A. J. Analysis of adenovirus transforming proteins from early regions 1A and 1B with antisera to inducible fusion antigens produced in Escherichia coli. J Virol. 1984 Jan;49(1):132–141. doi: 10.1128/jvi.49.1.132-141.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stow N. D. Cloning of a DNA fragment from the left-hand terminus of the adenovirus type 2 genome and its use in site-directed mutagenesis. J Virol. 1981 Jan;37(1):171–180. doi: 10.1128/jvi.37.1.171-180.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson C., Akusjärvi G. Adenovirus 2 early region 1A stimulates expression of both viral and cellular genes. EMBO J. 1984 Apr;3(4):789–794. doi: 10.1002/j.1460-2075.1984.tb01886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderlund H., Pettersson U., Vennström B., Philipson L., Mathews M. B. A new species of virus-coded low molecular weight RNA from cells infected with adenovirus type 2. Cell. 1976 Apr;7(4):585–593. doi: 10.1016/0092-8674(76)90209-9. [DOI] [PubMed] [Google Scholar]
- Tokunaga O., Yaegashi T., Lowe J., Dobbs L., Padmanabhan R. Sequence analysis in the E1 region of adenovirus type 4 DNA. Virology. 1986 Dec;155(2):418–433. doi: 10.1016/0042-6822(86)90204-7. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Weinheimer S. P., McKnight S. L. Transcriptional and post-transcriptional controls establish the cascade of herpes simplex virus protein synthesis. J Mol Biol. 1987 Jun 20;195(4):819–833. doi: 10.1016/0022-2836(87)90487-6. [DOI] [PubMed] [Google Scholar]
- Weinmann R., Brendler T. G., Raskas H. J., Roeder R. G. Low molecular weight viral RNAs transcribed by RNA polymerase III during adenovirus 2 infection. Cell. 1976 Apr;7(4):557–566. doi: 10.1016/0092-8674(76)90206-3. [DOI] [PubMed] [Google Scholar]
- Winberg G., Shenk T. Dissection of overlapping functions within the adenovirus type 5 E1A gene. EMBO J. 1984 Aug;3(8):1907–1912. doi: 10.1002/j.1460-2075.1984.tb02066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Rosser D. S., Schmidt M. C., Berk A. A TATA box implicated in E1A transcriptional activation of a simple adenovirus 2 promoter. Nature. 1987 Apr 2;326(6112):512–515. doi: 10.1038/326512a0. [DOI] [PubMed] [Google Scholar]
- Yoshinaga S., Dean N., Han M., Berk A. J. Adenovirus stimulation of transcription by RNA polymerase III: evidence for an E1A-dependent increase in transcription factor IIIC concentration. EMBO J. 1986 Feb;5(2):343–354. doi: 10.1002/j.1460-2075.1986.tb04218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]