Abstract
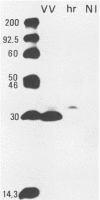
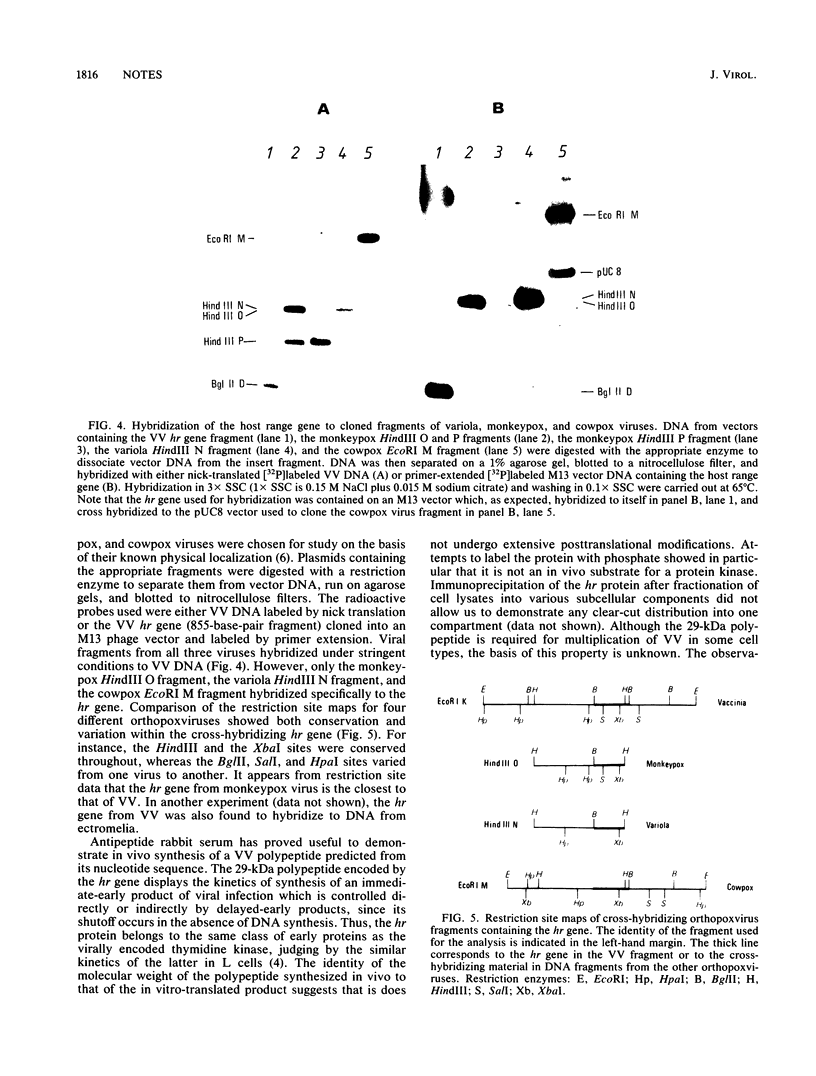
A vaccinia virus gene required for multiplication in some cell lines but not in others has been previously isolated and sequenced. A synthetic peptide predicted from the nucleotide sequence and corresponding to the carboxy-terminal 18 amino acids was used to raise antibodies in rabbits. The immune serum enabled detection of a 29-kilodalton (kDa) polypeptide by either immunoprecipitation or Western immunoblot assays. Synthesis of the 29-kDa polypeptide occurred immediately after infection and lasted for about 3 h. Shutoff of its synthesis was concomitant with the appearance of a delayed early polypeptide that may be antigenically related to the 29-kDa polypeptide. Analysis of cloned segments of the genomes of other orthopoxviruses by hybridization with the vaccinia virus host range gene demonstrates that it is well conserved within this genus.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Drillien R., Koehren F., Kirn A. Host range deletion mutant of vaccinia virus defective in human cells. Virology. 1981 Jun;111(2):488–499. doi: 10.1016/0042-6822(81)90351-2. [DOI] [PubMed] [Google Scholar]
- Gillard S., Spehner D., Drillien R., Kirn A. Localization and sequence of a vaccinia virus gene required for multiplication in human cells. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5573–5577. doi: 10.1073/pnas.83.15.5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillard S., Spehner D., Drillien R. Mapping of a vaccinia host range sequence by insertion into the viral thymidine kinase gene. J Virol. 1985 Jan;53(1):316–318. doi: 10.1128/jvi.53.1.316-318.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruby D. E., Ball L. A. Control of expression of the vaccinia virus thymidine kinase gene. J Virol. 1981 Nov;40(2):456–464. doi: 10.1128/jvi.40.2.456-464.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake J. R., Cooper P. D. Deletions of the terminal sequences in the genomes of the white pock (u) and host-restricted (p) mutants of rabbitpox virus. J Gen Virol. 1980 May;48(1):135–147. doi: 10.1099/0022-1317-48-1-135. [DOI] [PubMed] [Google Scholar]
- Mackett M., Archard L. C. Conservation and variation in Orthopoxvirus genome structure. J Gen Virol. 1979 Dec;45(3):683–701. doi: 10.1099/0022-1317-45-3-683. [DOI] [PubMed] [Google Scholar]
- Moss B., Winters E., Cooper J. A. Deletion of a 9,000-base-pair segment of the vaccinia virus genome that encodes nonessential polypeptides. J Virol. 1981 Nov;40(2):387–395. doi: 10.1128/jvi.40.2.387-395.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer R. W., Rothe C. T. The white pock mutants of rabbit poxvirus. I. Spontaneous host range mutants contain deletions. Virology. 1980 Apr 15;102(1):119–132. doi: 10.1016/0042-6822(80)90075-6. [DOI] [PubMed] [Google Scholar]
- Paez E., Dallo S., Esteban M. Generation of a dominant 8-MDa deletion at the left terminus of vaccinia virus DNA. Proc Natl Acad Sci U S A. 1985 May;82(10):3365–3369. doi: 10.1073/pnas.82.10.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panicali D., Davis S. W., Mercer S. R., Paoletti E. Two major DNA variants present in serially propagated stocks of the WR strain of vaccinia virus. J Virol. 1981 Mar;37(3):1000–1010. doi: 10.1128/jvi.37.3.1000-1010.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkus M. E., Panicali D., Mercer S., Paoletti E. Insertion and deletion mutants of vaccinia virus. Virology. 1986 Jul 30;152(2):285–297. doi: 10.1016/0042-6822(86)90132-7. [DOI] [PubMed] [Google Scholar]