Abstract
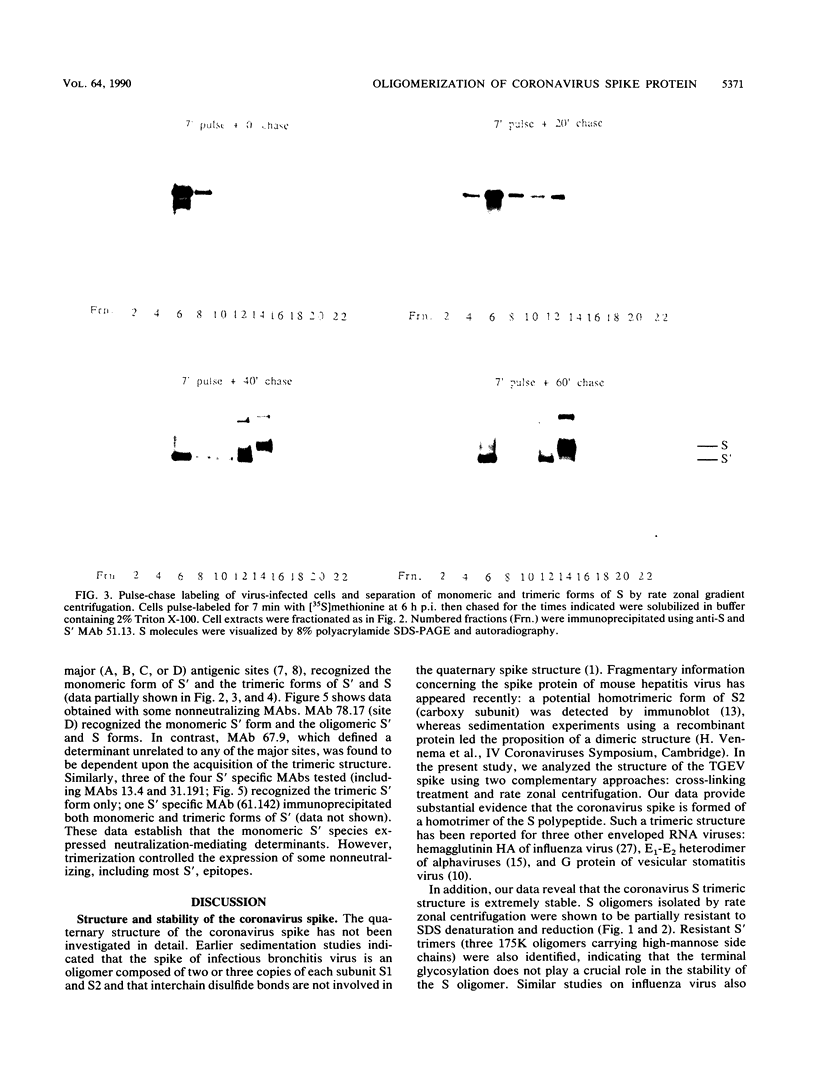
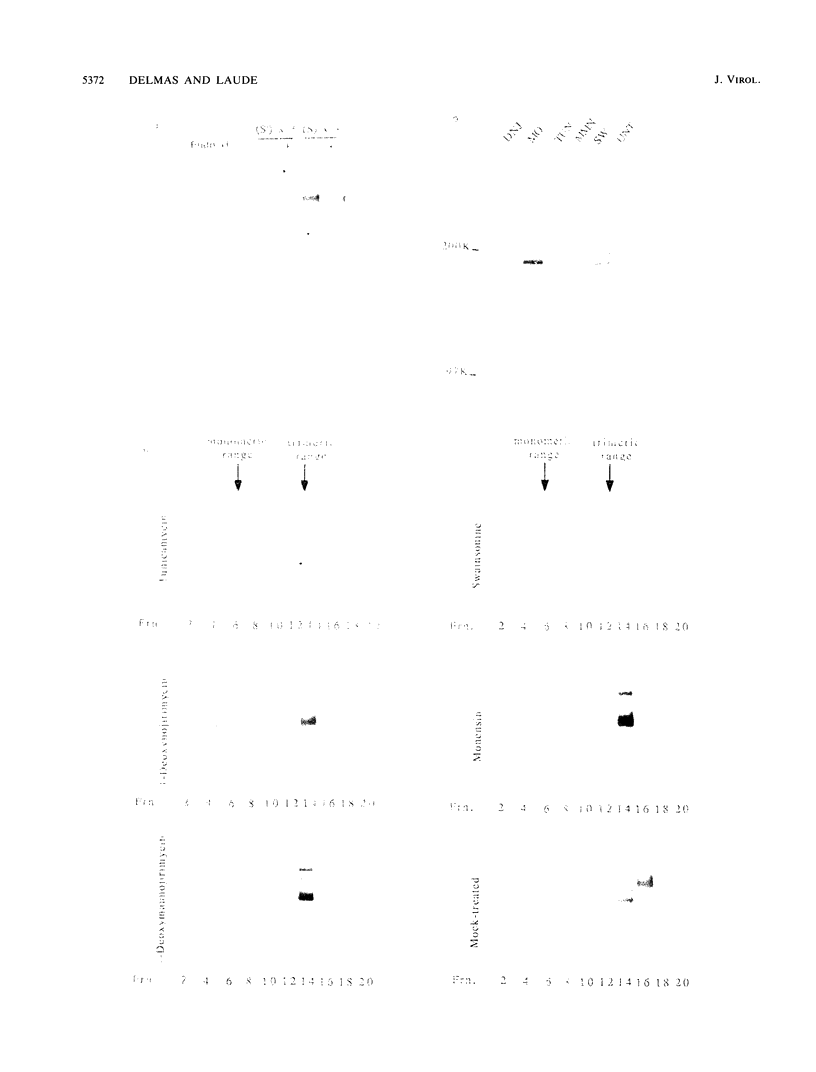
The folding and oligomerization of coronavirus spike protein were explored using a panel of monoclonal antibodies. Chemical cross-linking and sedimentation experiments showed that the spike of transmissible gastroenteritis virus is a homotrimer of the S membrane glycoprotein. The spike protein was synthesized as a 175,000-apparent-molecular-weight (175K) monomer subunit that is sensitive to endo-beta-N-acetylglucosaminidase H. Assembly of monomers into a trimeric structure was found to occur on a partially trimmed polypeptide and to be a rate-limiting step, since large amounts of monomers failed to trimerize 1 h after completion of synthesis. Terminal glycosylation of newly assembled trimers, resulting in the biosynthesis of three 220K oligomers, occurred with a half time of approximately 20 min. Monomeric (230K to 240K) processed forms were also observed in cells and in virions. The 175K monomeric form expressed four major antigenic sites previously localized within the amino-terminal half of the S polypeptide chain; however, two classes of trimer-restricted epitopes (borne by three 220K and/or three 175K oligomers) were identified. The S glycoprotein of coronavirus might be a valuable model system for discovering new aspects of the maturation of membrane glycoproteins.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cavanagh D. Coronavirus IBV: structural characterization of the spike protein. J Gen Virol. 1983 Dec;64(Pt 12):2577–2583. doi: 10.1099/0022-1317-64-12-2577. [DOI] [PubMed] [Google Scholar]
- Clegg J. C. Sequential translation of capsid and membrane protein genes of alphaviruses. Nature. 1975 Apr 3;254(5499):454–455. doi: 10.1038/254454a0. [DOI] [PubMed] [Google Scholar]
- Copeland C. S., Doms R. W., Bolzau E. M., Webster R. G., Helenius A. Assembly of influenza hemagglutinin trimers and its role in intracellular transport. J Cell Biol. 1986 Oct;103(4):1179–1191. doi: 10.1083/jcb.103.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland C. S., Zimmer K. P., Wagner K. R., Healey G. A., Mellman I., Helenius A. Folding, trimerization, and transport are sequential events in the biogenesis of influenza virus hemagglutinin. Cell. 1988 Apr 22;53(2):197–209. doi: 10.1016/0092-8674(88)90381-9. [DOI] [PubMed] [Google Scholar]
- Davies G. E., Stark G. R. Use of dimethyl suberimidate, a cross-linking reagent, in studying the subunit structure of oligomeric proteins. Proc Natl Acad Sci U S A. 1970 Jul;66(3):651–656. doi: 10.1073/pnas.66.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas B., Gelfi J., Laude H. Antigenic structure of transmissible gastroenteritis virus. II. Domains in the peplomer glycoprotein. J Gen Virol. 1986 Jul;67(Pt 7):1405–1418. doi: 10.1099/0022-1317-67-7-1405. [DOI] [PubMed] [Google Scholar]
- Delmas B., Rasschaert D., Godet M., Gelfi J., Laude H. Four major antigenic sites of the coronavirus transmissible gastroenteritis virus are located on the amino-terminal half of spike glycoprotein S. J Gen Virol. 1990 Jun;71(Pt 6):1313–1323. doi: 10.1099/0022-1317-71-6-1313. [DOI] [PubMed] [Google Scholar]
- Doms R. W., Helenius A. Quaternary structure of influenza virus hemagglutinin after acid treatment. J Virol. 1986 Dec;60(3):833–839. doi: 10.1128/jvi.60.3.833-839.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doms R. W., Keller D. S., Helenius A., Balch W. E. Role for adenosine triphosphate in regulating the assembly and transport of vesicular stomatitis virus G protein trimers. J Cell Biol. 1987 Nov;105(5):1957–1969. doi: 10.1083/jcb.105.5.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doms R. W., Ruusala A., Machamer C., Helenius J., Helenius A., Rose J. K. Differential effects of mutations in three domains on folding, quaternary structure, and intracellular transport of vesicular stomatitis virus G protein. J Cell Biol. 1988 Jul;107(1):89–99. doi: 10.1083/jcb.107.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbein A. D. Inhibitors of the biosynthesis and processing of N-linked oligosaccharide chains. Annu Rev Biochem. 1987;56:497–534. doi: 10.1146/annurev.bi.56.070187.002433. [DOI] [PubMed] [Google Scholar]
- Gallagher T. M., Parker S. E., Buchmeier M. J. Neutralization-resistant variants of a neurotropic coronavirus are generated by deletions within the amino-terminal half of the spike glycoprotein. J Virol. 1990 Feb;64(2):731–741. doi: 10.1128/jvi.64.2.731-741.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gething M. J., McCammon K., Sambrook J. Expression of wild-type and mutant forms of influenza hemagglutinin: the role of folding in intracellular transport. Cell. 1986 Sep 12;46(6):939–950. doi: 10.1016/0092-8674(86)90076-0. [DOI] [PubMed] [Google Scholar]
- Hurtley S. M., Bole D. G., Hoover-Litty H., Helenius A., Copeland C. S. Interactions of misfolded influenza virus hemagglutinin with binding protein (BiP). J Cell Biol. 1989 Jun;108(6):2117–2126. doi: 10.1083/jcb.108.6.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld R., Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- Kreis T. E., Lodish H. F. Oligomerization is essential for transport of vesicular stomatitis viral glycoprotein to the cell surface. Cell. 1986 Sep 12;46(6):929–937. doi: 10.1016/0092-8674(86)90075-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laude H., Chapsal J. M., Gelfi J., Labiau S., Grosclaude J. Antigenic structure of transmissible gastroenteritis virus. I. Properties of monoclonal antibodies directed against virion proteins. J Gen Virol. 1986 Jan;67(Pt 1):119–130. doi: 10.1099/0022-1317-67-1-119. [DOI] [PubMed] [Google Scholar]
- Machamer C. E., Rose J. K. Vesicular stomatitis virus G proteins with altered glycosylation sites display temperature-sensitive intracellular transport and are subject to aberrant intermolecular disulfide bonding. J Biol Chem. 1988 Apr 25;263(12):5955–5960. [PubMed] [Google Scholar]
- Niemann H., Boschek B., Evans D., Rosing M., Tamura T., Klenk H. D. Post-translational glycosylation of coronavirus glycoprotein E1: inhibition by monensin. EMBO J. 1982;1(12):1499–1504. doi: 10.1002/j.1460-2075.1982.tb01346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasschaert D., Laude H. The predicted primary structure of the peplomer protein E2 of the porcine coronavirus transmissible gastroenteritis virus. J Gen Virol. 1987 Jul;68(Pt 7):1883–1890. doi: 10.1099/0022-1317-68-7-1883. [DOI] [PubMed] [Google Scholar]
- Rottier P. J., Rose J. K. Coronavirus E1 glycoprotein expressed from cloned cDNA localizes in the Golgi region. J Virol. 1987 Jun;61(6):2042–2045. doi: 10.1128/jvi.61.6.2042-2045.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaan W., Cavanagh D., Horzinek M. C. Coronaviruses: structure and genome expression. J Gen Virol. 1988 Dec;69(Pt 12):2939–2952. doi: 10.1099/0022-1317-69-12-2939. [DOI] [PubMed] [Google Scholar]
- Tooze S. A., Tooze J., Warren G. Site of addition of N-acetyl-galactosamine to the E1 glycoprotein of mouse hepatitis virus-A59. J Cell Biol. 1988 May;106(5):1475–1487. doi: 10.1083/jcb.106.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennema H., Heijnen L., Zijderveld A., Horzinek M. C., Spaan W. J. Intracellular transport of recombinant coronavirus spike proteins: implications for virus assembly. J Virol. 1990 Jan;64(1):339–346. doi: 10.1128/jvi.64.1.339-346.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley D. C., Skehel J. J., Waterfield M. Evidence from studies with a cross-linking reagent that the haemagglutinin of influenza virus is a trimer. Virology. 1977 Jun 15;79(2):446–448. doi: 10.1016/0042-6822(77)90371-3. [DOI] [PubMed] [Google Scholar]
- Wilson I. A., Skehel J. J., Wiley D. C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981 Jan 29;289(5796):366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- Yewdell J. W., Yellen A., Bächi T. Monoclonal antibodies localize events in the folding, assembly, and intracellular transport of the influenza virus hemagglutinin glycoprotein. Cell. 1988 Mar 25;52(6):843–852. doi: 10.1016/0092-8674(88)90426-6. [DOI] [PubMed] [Google Scholar]
- de Groot R. J., Luytjes W., Horzinek M. C., van der Zeijst B. A., Spaan W. J., Lenstra J. A. Evidence for a coiled-coil structure in the spike proteins of coronaviruses. J Mol Biol. 1987 Aug 20;196(4):963–966. doi: 10.1016/0022-2836(87)90422-0. [DOI] [PMC free article] [PubMed] [Google Scholar]







