Abstract
The insulin-like growth factor-1 receptor (IGF1R) is a receptor tyrosine kinase (RTK) that has a critical role in mitogenic signalling during embryogenesis and an antiapoptotic role in the survival and progression of many human tumours. Here, we present the crystal structure of the tyrosine kinase domain of IGF1R (IGF1RK), in its unphosphorylated state, in complex with a novel compound, cis-3-[3-(4-methyl-piperazin-l-yl)-cyclobutyl]-1-(2-phenyl-quinolin-7-yl)-imidazo[1,5-a]pyrazin-8-ylamine (PQIP), which we show is a potent inhibitor of both the unphosphorylated (basal) and phosphorylated (activated) states of the kinase. PQIP interacts with residues in the ATP-binding pocket and in the activation loop, which confers specificity for IGF1RK and the highly related insulin receptor (IR) kinase. In this crystal structure, the IGF1RK active site is occupied by Tyr1135 from the activation loop of an symmetry (two-fold)-related molecule. This dimeric arrangement affords, for the first time, a visualization of the initial trans-phosphorylation event in the activation loop of an RTK, and provides a molecular rationale for a naturally occurring mutation in the activation loop of the IR that causes type II diabetes mellitus.
Keywords: autophosphorylation, IGF1 receptor, insulin receptor, small-molecule inhibitor, tyrosine kinase
Introduction
The insulin-like growth factor-1 receptor (IGF1R) is a transmembrane receptor tyrosine kinase (RTK) that transduces IGF1 and IGF2 signals to regulate growth, differentiation and survival of cells (Baserga et al, 1997; Butler et al, 1998; Adams et al, 2000). IGF1R has a particularly important function in prenatal development; IGF1R-deficient mice die at birth, exhibiting severe growth deficiencies (Liu et al, 1993). Like the closely related insulin receptor (IR), IGF1R consists of two extracellular α-subunits disulphide-bonded to two transmembrane-spanning β-subunits containing cytoplasmic tyrosine kinase activity. Binding of IGF1 or IGF2 to the ectodomain of IGF1R triggers a structural rearrangement that results in phosphorylation of specific tyrosine residues in the cytoplasmic domains, stimulating catalytic (tyrosine kinase) activity and generating recruitment sites for IR substrate (IRS) proteins and Shc, among other signalling proteins. Phosphorylation of these substrates by IGF1R leads to activation of mitogen-activated protein kinase and phosphoinositide 3′-kinase signalling cascades (Baserga et al, 1997; Butler et al, 1998; Adams et al, 2000).
The tyrosine kinase activity of IGF1R is tightly controlled. The mechanism for enzyme regulation is similar to that in IR (Hubbard et al, 1994; Hubbard, 1997). In the basal state, the activation loop—a flexible segment within the C lobe of the kinase domain—adopts an autoinhibitory conformation with Tyr1135 bound in the active site (Munshi et al, 2002, 2003). Ligand binding to IGF1R promotes autophosphorylation in trans (one kinase domain phosphorylating the other) of Tyr1135 and two other activation-loop tyrosines, Tyr1131 and Tyr1136. Tyrosines in the juxtamembrane region (between the transmembrane helix and the kinase domain) and the region C-terminal to the kinase domain are also autophosphorylated, with pTyr950 in the juxtamembrane region serving as a recruitment site for the phosphotyrosine-binding domains of IRS proteins and Shc (Kato et al, 1994; LeRoith et al, 1995).
The phosphorylation events in the activation loop destabilize the autoinhibitory conformation of the loop and stabilize the active conformation. Two-dimensional phosphopeptide-mapping studies (Favelyukis et al, 2001) indicate that the first residue to be phosphorylated is predominantly Tyr1135, followed by Tyr1131 and then Tyr1136. The molecular determinants that govern the order of autophosphorylation in the activation loop are not well understood. Steady-state kinetic analyses of the isolated phosphorylated forms of IGF1RK demonstrated that each phosphorylation event increases enzyme turnover number and decreases Km for ATP and peptide substrate (Favelyukis et al, 2001). The crystal structure of IGF1RK-3P showed that the tris-phosphorylated activation loop is well ordered and anchored to the kinase C lobe in a conformation that facilitates substrate binding and catalysis (Favelyukis et al, 2001).
IGF1R is overexpressed in many forms of human cancer, including lung cancer, colon carcinoma, tumours of the central nervous system, cervical cancer and Wilms tumour (Baserga, 1999; Valentinis and Baserga, 2001; LeRoith and Roberts, 2003). The survival of these tumour cells is partially dependent on the antiapoptotic effects of IGF1R. Although no IGF1R mutations have been identified in cancers, loss of IGF2 imprinting is an epigenetic alteration found in colorectal and other tumours (Sakatani et al, 2005). Studies in cell culture have shown that overexpression of IGF1R can lead to morphological transformation, whereas interference with IGF1R expression reverses the transformed phenotype (Baserga et al, 1997). Inhibition of IGF1R can induce apoptosis in tumour cells by repressing tumorigenesis and metastases (Baserga et al, 1997). In breast cancer models in which HER2/ErbB2/Neu is overexpressed, increased levels of IGF1R interfere with the action of the HER2 inhibitor Herceptin (Lu et al, 2001). For these reasons, IGF1R has emerged as a promising target for anticancer therapy.
Currently, several avenues are being pursued for targeting IGF1R, including small-molecule kinase inhibitors, IGF1R ectodomain antibodies, antisense oligonucleotides and RNA interference (Riedemann and Macaulay, 2006). A challenge in the development of IGF1R inhibitors is the potential for off-target inhibition of IR, due to the high similarity between these two receptors; the sequence identity is ∼60% overall and ∼80% in the kinase domains. Several groups have described kinase inhibitors that are effective against IGF1R and show some selectivity for IGF1R over IR in cells. Two IGF1R inhibitors of the pyrrolo[2,3-d]pyrimidine class have been reported. One compound inhibited IGF1R in tissue xenografts (Garcia-Echeverria et al, 2004), and the other compound showed significant antitumour activity in a mouse model of multiple myeloma (Mitsiades et al, 2004). These two compounds show comparable activity against IGF1R and IR in vitro, but show selectivity for IGF1R over IR in cells (27- and 16-fold). A benzimidazole compound is also equipotent against IGF1R and IR in vitro, but shows selectivity in cells (Wittman et al, 2005). The mechanism underlying the in-cell selectivity of these ATP-competitive inhibitors for IGF1R over IR is not currently understood.
The imidazopyrazine cis-3-[3-(4-methyl-piperazin-l-yl)-cyclobutyl]-1-(2-phenyl-quinolin-7-yl)-imidazo[1,5-a]pyrazin-8-ylamine (PQIP) was recently described as a potent ATP-competitive inhibitor of IGF1R, with a cellular IC50 of 19 nM for inhibition of IGF1R autophosphorylation (Ji et al, 2007). PQIP inhibits both IGF1R and IR in vitro, but displays a 14-fold selectivity for IGF1R in intact cells. PQIP has antitumour activity in xenograft models at concentrations that do not cause significant hyperglycaemia, confirming the possibility of selective IGF1R inhibition by this compound. A close derivative of PQIP, OSI-906, is currently in phase I clinical trials.
Here, we describe studies aimed at understanding the mechanism of IGF1R inhibition by PQIP. We report that PQIP displays potent activity against both the unphosphorylated (basal) form of IGF1RK (IGF1RK-0P) and the tris-phosphorylated (activated) form (IGF1RK-3P). The crystal structure of PQIP bound to IGF1RK-0P shows that the compound binds deep in the ATP-binding pocket, with the activation loop adopting an extended conformation that is neither autoinhibitory nor active. The inhibitor-induced expulsion of the activation loop from the active site facilitated formation of a symmetric IGF1RK dimer in the crystal, in which Tyr1135 of the activation loop is bound in the active site of the other IGF1RK molecule, positioned for phosphoryl transfer. This structure affords a visualization of how the first trans-phosphorylation event in the IGF1RK activation loop ensues, and provides a molecular rationale for a mutation in the IR activation loop found in a subset of patients with type II diabetes mellitus (Formisano et al, 1993).
Results
In vitro inhibition of IGF1RK by PQIP
To determine the specificity of PQIP (Figure 1A) towards the activation state of IGF1RK, we tested the compound against purified IGF1RK-0P and -3P, the basal and fully activated forms of the enzyme. We used a continuous spectrophotometric kinase assay (Favelyukis et al, 2001) to measure IGF1RK autophosphorylation at an ATP concentration of 1 mM in the presence of various concentrations of PQIP (Figure 1B). As we have described previously (Favelyukis et al, 2001), the autophosphorylation progress curves are biphasic, due to the activation of IGF1RK by trans-phosphorylation. Analysis of these data gave an IC50 value of 118 nM for PQIP inhibition of IGF1RK-0P. To confirm the effectiveness of PQIP against IGF1RK-0P, we analysed autophosphorylation reactions by native PAGE (Figure 1C). In the absence of PQIP, IGF1RK-0P progressed to the 1P, 2P and 3P forms of the enzyme. In contrast, addition of PQIP to IGF1RK-0P prevented the transition from 0P to 1P. To test PQIP inhibition of the fully activated enzyme, we measured phosphorylation of a synthetic peptide substrate by IGF1RK-3P at 1 mM ATP. From the initial rates of peptide phosphorylation, we obtained an IC50 value of 145 nM (Figure 1D).
Figure 1.
Inhibition of IGF1RK by PQIP. (A) Chemical structure of PQIP: cis-3-[3-(4-methyl-piperazin-l-yl)-cyclobutyl]-1-(2-phenyl-quinolin-7-yl)-imidazo[1,5-a]pyrazin-8-ylamine. (B) Enzyme progress curves from the spectrophotometric assay for autophosphorylation of IGF1RK (1 mM ATP) in the presence of various concentrations of PQIP as indicated on the right-hand side. In the inset, the fitting of the inhibition data gave an IC50 value of 118 nM. (C) Native PAGE analysis of IGF1RK autophosphorylation (5 mM ATP) in the absence and presence (50 μM) of PQIP. The gel was stained with Coomassie blue. The different phosphorylation states of IGF1RK are marked on the left-hand side. (D) Enzyme progress curves from the spectrophotometric assay for peptide substrate phosphorylation by IGF1RK-3P (1 mM ATP) in the presence of various concentrations of PQIP as indicated on the right-hand side. In the inset, the fitting of the inhibition data gave an IC50 value of 145 nM.
To characterize further the inhibitory characteristics of PQIP, we performed in vitro kinase reactions (using a peptide substrate) at various ATP concentrations to determine the type of inhibition and inhibitory constant (Ki) for PQIP against IGF1RK-0P and -3P. The data are consistent with ATP-competitive inhibition of PQIP for both IGF1RK-0P and -3P, with Ki values of 157 and 140 nM, respectively (Supplementary Figure S1). Thus, PQIP is a potent inhibitor of both the basal and activated forms of IGF1RK.
Crystal structure of PQIP bound to IGF1RK-0P
We obtained crystals of PQIP bound to IGF1RK-0P with one PQIP–IGF1RK complex in the asymmetric unit, and determined the structure by molecular replacement, using the structure of IGF1RK-0P (PDB code 1P4O) (Munshi et al, 2003) as the search model. Data collection and refinement statistics at 2.3-Å resolution are given in Table I.
Table 1.
X-ray data collection and refinement statistics
| Data collection | |
| Resolution (Å) | 50–2.3 |
| Observations | 73 243 |
| Unique reflections | 18 251 |
| Redundancy | 4.0 |
| Completenessa (%) | 97.5 (99.2) |
| Rsyma,b (%) | 7.8 (39.4) |
| <I/σI>a | 12.3 (2.1) |
| Refinementc | |
| Resolution (Å) | 50–2.3 |
| Reflections | 17 319 |
| Rcrystd/Rfree (%) | 19.7/23.5 |
| r.m.s.d. bond lengths (Å) | 0.010 |
| r.m.s.d. bond angles (deg) | 1.29 |
| Average B-factors (Å2) | |
| All atoms | 34.2 |
| IGF1RK | 34.3 |
| PQIP | 21.6 |
| Solvent | 35.1 |
| aValue in parentheses is for the highest resolution shell: 2.38–2.30 Å. | |
| bRsym=100 × ∑∣I–〈I〉∣/∑I. | |
| cAtomic model includes 2324 protein atoms, 37 PQIP atoms, 1 Ca2+ and 165 water molecules. | |
| dRcryst=100 × ∑∣∣Fo∣–∣Fc∣∣/∑∣Fo∣, where Fo and Fc are the observed and calculated structure factors, respectively (Fo>0σ). Rfree was determined from 5% of the data. | |
PQIP binds in the ATP-binding pocket of the kinase domain (Figure 2A) and makes extensive interactions (Figure 2B and C), burying a total (inhibitor+kinase) surface area of 1053 Å2. The imidazopyrazinyl core of the compound binds in the position of the adenine base of ATP (Supplementary Figure S2A) and makes hydrogen bonds to the same two backbone atoms of the kinase hinge region (connecting the N and C lobes) as the adenine base (Figure 2B and C). The quinolinyl substituent is sandwiched between activation-loop residues Gly1122 and Asp1123 (of the kinase-conserved DFG motif) in the C lobe and Met1049 (‘gatekeeper' residue), Met1024 and Val1033 in the N lobe (Figure 2B and C). Protein kinase-conserved Lys1003 (β-strand 3) is hydrogen-bonded to the quinolinyl nitrogen atom, to Ser979 in the nucleotide-binding loop and to a water molecule that bridges to the imidazopyrazinyl substituent. The phenyl substituent on the quinoline makes van der Waals contacts with Phe980 (nucleotide-binding loop), Phe1124 and Gly1125 (DFG motif, activation loop), and Phe1017 and Ala1021 (α-helix C).
Figure 2.
Crystal structure of PQIP–IGF1RK. (A) Ribbon diagram of the PQIP–IGF1RK crystal structure. PQIP is shown in stick representation with carbon atoms coloured yellow and nitrogen atoms coloured blue. An Fo–Fc omit map (PQIP omitted, followed by positional refinement) in the vicinity of PQIP is shown in purple mesh, calculated to 2.3 Å and contoured at 4σ. The N lobe of IGF1RK is coloured dark grey and the C lobe is coloured light grey. Within the N lobe, α-helix C (residues 1011–1026) is coloured pink and the nucleotide-binding loop (residues 978–981) is coloured blue. Within the C lobe, the activation loop (residues 1123–1144) is coloured green and the catalytic loop (residues 1103–1110) is coloured orange. The N and C termini of IGF1RK are indicated. (B) Stereo view of the interactions between PQIP and IGF1RK. IGF1RK residues that interact with PQIP are shown explicitly on the backdrop of a semitransparent ribbon diagram. Carbon atoms in IGF1RK are coloured according to the colouring scheme in (A), oxygen atoms are coloured red, nitrogen atoms are coloured blue and sulphur atoms are coloured green. An ordered water molecule is shown by a red sphere, and select hydrogen bonds are shown by dashed lines. (C) Schematic representation of the interactions of PQIP with IGF1RK residues. Residues (side chains, except for Gly1122 and Gly1125) making van der Waals (⩽3.9 Å) contacts with PQIP (coloured blue) are coloured according to the scheme in (A, B). Hydrogen-bonding interactions are represented by dashed lines. The red circle represents a water molecule. Figures 2, 3 and 4 were rendered using PyMOL (http://pymol.sourceforge.net).
The activation loop (residues 1122–1144) in this IGF1RK-0P structure adopts an extended rather than an autoinhibitory conformation (Figure 3A). Because of the phenylquinolinyl group, binding of PQIP is not compatible with the autoinhibitory conformation of the activation loop, distinguishing this binding mode from that of the benzimidazole compounds (see below). The activation loop in the PQIP–IGF1RK structure is also not in an active conformation—nor could it adopt it—owing to a steric clash between the phenyl substituent of the compound and Phe1124 of the DFG sequence (Figure 3B). Further C-terminal in the activation loop, Arg1128 makes a hydrogen bond with a backbone carbonyl group, which may help to stabilize this activation-loop conformation (Figure 2B).
Figure 3.
Comparison of IGF1RK activation-loop conformations. (A) The activation loop (light green), catalytic loop (orange) and PQIP (yellow carbon atoms, semitransparent molecular surface) from the PQIP–IGF1RK structure are shown. Superimposed (by their catalytic loops) is the unphosphorylated activation loop (dark green) from the autoinhibited IGF1RK structure (PDB code 1P4O) (Munshi et al, 2003). The N and C termini (Gly1122 and Pro1145) of the activation loops are indicated by arrows. (B) Same as (A), except that superimposed (by their catalytic loops) is the phosphorylated activation loop (olive) from the IGF1RK-3P structure (Favelyukis et al, 2001).
A crystal structure of IGF1RK-0P in complex with a small-molecule inhibitor of the benzimidazole class has been reported (Velaparthi et al, 2007). The total buried surface area upon binding of this inhibitor to IGF1RK is 786 Å2 (versus 1053 Å2 for PQIP–IGF1RK). In this crystal structure, the inhibitor binds in the ATP-binding pocket of the kinase domain, but does not engage residues in α-helix C, and the activation loop adopts the autoinhibitory conformation with Tyr1135 bound in the active site (Supplementary Figure S2B). Although this binding mode is sterically compatible with binding to IGF1RK-3P, the loss of interactions with the re-positioned, phosphorylated activation loop presumably makes this inhibitor much less effective against the active state of IGF1RK.
The Abl kinase inhibitor imatinib (STI-571/Gleevec) also binds deep in the ATP-binding pocket (Schindler et al, 2000; Nagar et al, 2002), engaging residues in α-helix C, with a total buried surface of 1174 Å2 (Supplementary Figure S2D). Imatinib binds exclusively to the unphosphorylated (autoinihibited) form of Abl, due to steric clashes between the compound and the activation loop in its phosphorylated, active conformation (Schindler et al, 2000; Nagar et al, 2002). In contrast, the Src and Abl inhibitor dasatinib (BMS-3354825) will bind to both the autoinhibited and activated forms of Abl (Supplementary Figure S2C) (Tokarski et al, 2006). Imatinib and dasatinib are ineffective against IGF1RK, in large part because the ‘gatekeeper' residue in IGF1RK (and IRK) is a bulkier methionine (Met1049) rather than threonine (Thr315 in Abl).
Trans-phosphorylation of Tyr1135
In the PQIP–IGF1RK structure, a crystallographic two-fold axis relates two interacting IGF1RK molecules. Strikingly, in this symmetric dimer, Tyr1135, the first tyrosine in the activation loop to be phosphorylated in trans (Favelyukis et al, 2001), is bound in the active site of the other kinase domain, and vice versa (Figure 4A). The total surface area buried in the dimer interface is 2072 Å2, and the residues involved in dimerization are limited to the activation loop of one molecule and the active site region of the other molecule.
Figure 4.
Trans-phosphorylation in the IGF1RK dimer. (A) The two IGF1RK-0P molecules related by a crystallographic two-fold axis (perpendicular to the page) are shown as a ribbon diagram. The colouring scheme is the same as in Figure 2, with the activation loop coloured light or dark green. The activation-loop tyrosine in the active site (Y1135) and the catalytic aspartic acid (D1105) are shown in ball-and-stick representation. An apostrophe denotes residues in the symmetry-related molecule. The N and C termini of the two IGF1RK molecules are labelled. (B) View in the IGF1RK active site. The activation loop (residues 1128–1142 only) acting as substrate is coloured dark green, with residues labelled with an apostrophe. Select hydrogen bonds are shown by dashed lines, and select water molecules are shown as red spheres. (C) View in the active site of autoinhibited IGF1RK-0P (Munshi et al, 2003), with the same orientation as in (B) (catalytic loops aligned). Here, Tyr1135 in the activation loop binds in the active site in cis. (D) View in the active site of IGF1RK-3P with bound peptide substrate and ATP analogue (Favelyukis et al, 2001), the carbon atoms of which are coloured yellow. Semitransparent surfaces are shown for the side chains of V(P+1), I(P+3) and F(P+5) of the peptide substrate. Here, the peptide substrate contains an isoleucine at P+3 rather than the canonical methionine (YΦXM).
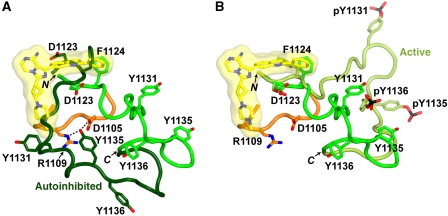
Tyr1135' (an apostrophe denotes residues in the symmetry-related molecule) is hydrogen-bonded to catalytic residues Asp1105 and Arg1109 (Figure 4B). Proximal to Tyr1135', Asp1134' makes water-mediated hydrogen bonds with Arg1109 and Gln1181, and Tyr1136' is hydrogen-bonded to Glu1189. Tyr1135' and Tyr1136' are hydrogen-bonded (backbone–backbone) to Pro1144 and Leu1143 at the end of the activation loop. Although the B-factors for Arg1137' are higher than for other activation-loop residues, indicating some local disorder (see Supplementary Figure S3 for 2Fo–Fc and B-factor maps), the side chain of Arg1137' is in position to salt bridge with Glu1132' across the loop (Figure 4B and Supplementary Figure S3).
The interactions mediated by Arg1137 are of special interest because a naturally occurring mutation in the corresponding residue in IR, R1164Q (R1152Q, exon 11- numbering), has been identified in a small number of patients with type II diabetes (Cocozza et al, 1992). Biochemical studies of R1164Q in cultured fibroblasts demonstrated that this mutated receptor was unable to undergo insulin-stimulated autophosphorylation, although phosphorylation of other substrates was not impaired (Formisano et al, 1993; Miele et al, 1999). Because the activation-loop sequences in IGF1R and IR are identical, the crystal structure of PQIP–IGF1RK provides a structural rationale for the autophosphorylation deficiency of R1164Q: Arg1164 (Arg1137 in IGF1RK) stabilizes a conformation of the activation loop that is favourable for presentation of Tyr1162 (Tyr1135 in IGF1RK) into the active site of the other kinase domain for trans-phosphorylation.
The binding mode of Tyr1135 in trans in the present structure is similar to that of Tyr1135 in cis in the IGF1RK-0P autoinhibited structure (Munshi et al, 2003) (Figure 4C). The interactions involving Tyr1135 and Tyr1136 are essentially identical in trans and in cis. In the autoinhibited structure, Asp1134 is hydrogen-bonded directly to residues of the C lobe, whereas in trans, Asp1134 makes only water-mediated hydrogen bonds. The difference in the positioning of Asp1134 is probably due to the divergent paths of the activation loop in trans versus cis distal to Tyr1135.
Significant differences exist between the binding mode of Tyr1135 in trans as a substrate and that of a typical tyrosine substrate of IGF1RK (e.g. as found in IRS1) (Figure 4D), which is contained in a YΦXM motif (Φ, hydrophobic; X, any residue). As shown in the ternary IGF1RK-3P structure (Favelyukis et al, 2001), binding of a YΦXM substrate involves packing of hydrophobic residues at the +1, +3 and (in some cases) +5 positions (relative to the substrate tyrosine) into the peptide binding groove that forms beneath the phosphorylated activation loop (Figure 4D). Furthermore, more extensive backbone hydrogen bonding occurs between the YΦXM substrate and the end of the activation loop than for Tyr1135 in trans. Thus, Tyr1135 in the activation loop is a suboptimal substrate for IGF1RK and thus may require the stabilizing Arg1137–Glu1132 interaction for efficient presentation of Tyr1135 into the IGF1RK active site.
Autophosphorylation of IGF1R mutants in cells
To test whether Arg1137 in IGF1R serves a similar function as Arg1164 in IR in the autophosphorylation process, we generated two mutations in full-length IGF1R at this position: R1137Q and R1137A. In addition, we mutated Glu1132, an interacting partner of Arg1137 in the crystal structure, to alanine. Experiments were conducted in murine fibroblasts derived from IGF1R-deficient mice. Cells were transfected with wild-type or mutant forms of IGF1R and treated with IGF1. Receptor autophosphorylation was assayed by Western blotting of IGF1R immunoprecipitates with an anti-pTyr1135/1136 site-specific antibody. Wild-type IGF1R was robustly phosphorylated on the activation loop upon IGF1 stimulation, but R1137Q and R1137A were poorly phosphorylated (Figure 5A), consistent with the results for R1164Q in IR (Formisano et al, 1993). To rule out the possibility that mutation of Arg1137 adversely affected binding of the anti-pTyr1135/1136 antibody, we also performed Western blotting with a pan-phosphotyrosine antibody (4G10), which yielded very similar results (data not shown). E1132A was also impaired in its ability to undergo activation-loop autophosphorylation (Figure 5A).
Figure 5.
Analysis of IGF1R activation-loop mutants. (A) Western blotting of anti-IGF1R immunoprecipitates from IGF1R-deficient fibroblasts transfected with wild-type or mutant IGF1R and stimulated (or not) with IGF1. (B) Enzyme progress curves from the spectrophotometric assay for autophosphorylation of wild-type and mutant IGF1RK. Analysis of the initial rates of autophosphorylation yields a normalized (with respect to wild-type, 100%) rate of 25% for R1137Q and 16% for E1132A.
Autophosphorylation of IGF1RK mutants in vitro
To examine in more detail the properties of R1137Q and E1132A, we expressed the mutant kinase domains in baculovirus-infected insect cells and purified them to near homogeneity. Native PAGE analysis confirmed that these proteins were in their unphosphorylated (0P) form after purification from insect cells (data not shown). We compared autophosphorylation of wild-type and mutant forms of IGF1RK using the continuous spectrophotometric assay. The mutant kinases R1137Q and E1132A were clearly impaired in their autophosphorylation activity as deduced from the decreased slopes and longer lag times (Figure 5B), with rates of autophosphorylation calculated to be 25 and 16%, respectively, of wild-type IGF1RK. These in-cell and in vitro data support a role for Arg1137 and Glu1132 in stabilizing an activation-loop conformation conducive to trans-phosphorylation of Tyr1135.
Discussion
Mechanism and specificity of PQIP inhibition of IGF1RK
As shown previously (Ji et al, 2007), PQIP is highly selective for IGF1R and IR versus other tyrosine and serine/threonine kinases. The crystal structure indicates that binding specificity derives in large part from the interaction of the phenylquinolinyl moiety of PQIP with IGF1RK residues in α-helix C, deep in the ATP-binding pocket (Figure 2B and C). The ability of the IGF1RK activation loop to adopt the conformation observed in the crystal structure is also likely to be an important specificity determinant. All of the IGF1RK residues contacted by PQIP are conserved in IRK, which is consistent with the comparable IC50 values for PQIP against the soluble kinases in vitro (Ji et al, 2007 and data not shown). The basis for the in-cell selectivity of PQIP (and other small-molecule inhibitors) for IGF1R over IR is unknown. It is conceivable that the conformational dynamics of the two kinase domains within the holoreceptor, which could affect inhibitor binding, might be somewhat different in IGF1R versus IR.
Our in vitro biochemical results (Figure 1B and D; Supplementary Figure S1) demonstrate that PQIP is a potent inhibitor of IGF1RK in both its basal (0P) and activated (3P) states. Although PQIP inhibits both states, the binding mode of the compound is not compatible with either the ‘native' autoinhibitory (0P) or active (3P) conformation of the activation loop (Figure 3). For this reason, it is very probable that PQIP will inhibit the intermediate phosphorylation states (1P and 2P) as well. That PQIP is likely to be effective against all phosphorylated forms of IGF1R, yet retains specificity for IGF1R (and IR), is highly advantageous as a potential drug, and contrasts with many protein kinase inhibitors that target a restricted number of kinase sub-states (e.g. imatinib). The ability to inhibit multiple activation-loop conformations is also true for dasatinib, and this property is believed to underlie its higher potency versus imatinib (Tokarski et al, 2006).
Mechanism and specificity of activation-loop trans-phosphorylation
Autophosphorylation of the activation loops in IGF1R and IR (identical in sequence) is a critical and early event in the receptor activation process, preceding autophosphorylation of tyrosines in the juxtamembrane and C-terminal regions. For both receptors, the second tyrosine in the activation loop—Tyr1135/1162 (IGF1R/IR)—is predominantly the first to be phosphorylated in trans (Wei et al, 1995; Favelyukis et al, 2001). This same tyrosine is bound in the active site (in cis) in the autoinhibited forms of IGF1RK (Munshi et al, 2003) and IRK (Hubbard et al, 1994). Thus, autophosphorylation of this tyrosine is essential for relief of autoinhibition and kinase activation.
In the PQIP–IGF1RK crystal, a crystallographic two-fold axis relates two interacting IGF1RK-0P molecules, in which Tyr1135 in the activation loop of one kinase is inserted into the active site of the other kinase, and vice versa (Figure 4A). Formation of this dimer is dependent upon binding of PQIP, which prevents the unphosphorylated activation loop from adopting its normal autoinhibitory conformation. Binding of ATP to IGF1RK-0P presumably elicits a similar, but likely more transient (because of lower affinity) expulsion of the activation loop from the active site. We do not see evidence of a PQIP-mediated IGF1RK dimer in solution (data not shown). This is not troubling, though, because the two kinase domains exist in vivo in the context of a dimeric transmembrane receptor, and IGF1 binding to the ectodomain juxtaposes them for autophosphorylation in trans (Favelyukis et al, 2001).
Although the dimer in the crystal structure is exactly two-fold symmetric (imposed crystallographic symmetry), trans-phosphorylation of Tyr1135 in the holoreceptor presumably does not require both kinase active sites to be occupied simultaneously by Tyr1135. Yet because the activation loop is of finite length (limiting the ensemble of sterically feasible arrangements), the head-to-tail configuration of the two kinase domains as viewed in the crystal structure is probably a reasonable approximation to their spatial arrangement during actual activation-loop trans-phosphorylation.
Previous structural studies on checkpoint kinase-2 (Chk2) (Oliver et al, 2006) and several related protein serine/threonine kinases (Pike et al, 2008) revealed kinase dimers formed through activation-loop exchange, in which the activation loop of one kinase interacts extensively with the C lobe of the other kinase, and vice versa. This dimeric configuration is thought to facilitate trans-phosphorylation of the activation loop by stabilizing the active state of the unphosphorylated (on the activation loop) kinases. However, in this case, the activation loop must undergo a rearrangement to position properly the threonine (Thr383 in Chk2) in the active site for trans-phosphorylation (see Supplementary Figure S4), whereas in the IGF1RK structure, Tyr1135 is exactly poised for phosphoryl transfer, hydrogen-bonded to Asp1105 and Arg1109 in the catalytic loop. To the best of our knowledge, our dimeric IGF1RK structure is the first visualization of a protein tyrosine kinase ‘in the act' of activation-loop trans-phosphorylation.
What are the determinants that select for Tyr1135 to be the first tyrosine phosphorylated in the activation loop? For steric reasons, the position of the tyrosine within the loop is likely to be important. The ‘tether points' of the loop are approximately Gly1122, just before the kinase-conserved DFG sequence, and Pro1145, with the C-terminal end more surface-exposed. These attributes will favour phosphorylation of Tyr1135 and Tyr1136 over Tyr1131. The sequence proximal to the tyrosine is also likely to be a determinant (intrinsic sequence specificity). As noted above, the kinase domains of IGF1R and IR prefer tyrosine substrates with a YΦXM motif preceded by acidic residues (Shoelson et al, 1992; Zhou et al, 1995) (Figure 4D). Although none of the activation-loop tyrosines is present in such a motif, an acidic residue (Asp1134) precedes Tyr1135 and Tyr1136 is observed to bind in the P+1 pocket in cis (Munshi et al, 2003).
It should be noted that residues in the activation loops of IGF1R and IR (and in other RTKs) serve multiple functions, including autoinhibition (cis) and substrate recognition (trans) prior to activation-loop phosphorylation, and activation-loop stabilization (cis) post-phosphorylation. For example, Asp1161 in IR (Asp1134 in IGF1R) is important for autoinhibition (Ablooglu et al, 2001; Till et al, 2001) (Figure 4C), substrate recognition of Tyr1162 in trans (Figure 4B) and recruitment of the adaptor protein APS upon activation (Hu et al, 2003). Thus, it is not surprising that activation-loop residues proximal to tyrosine autophosphorylation sites are not fully optimized for substrate presentation.
Because the intrinsic sequence specificity only weakly favours phosphorylation of Tyr1135 over the other two sites, additional determinants may exist. In the crystal structure of PQIP–IGF1RK, Arg1137 in the activation loop (two residues C-terminal to Tyr1135) is positioned for salt-bridge formation with Glu1132 across the loop (Figure 4B). By stabilizing an activation-loop conformation that facilitates insertion of Tyr1135 into the active site of the adjacent kinase domain, the Arg1137–Glu1132 interaction may constitute a third molecular determinant (in addition to accessibility and intrinsic sequence) for the initial trans-phosphorylation of Tyr1135.
After phosphorylation of Tyr1135, Tyr1131 and Tyr1136 are subsequently trans-phosphorylated. It is not clear why Tyr1131 is predominantly the second site to be phosphorylated (Favelyukis et al, 2001), but phosphorylation of Tyr1136 is likely ‘primed' by prior phosphorylation of Tyr1135, as pTyr1135 can serve as a P-1 acidic residue for Tyr1136. The Arg1137–Glu1132 interaction is probably applicable only to phosphorylation of Tyr1135. Indeed, in the fully (tris-phosphorylated) activated form of IGF1RK, Arg1137 is salt-bridged to pTy1135 (Favelyukis et al, 2001).
The Arg1137–Glu1132 interaction illuminates previous studies of a heterozygous mutation in the IR activation loop, R1164Q. IR autophosphorylation (but not kinase activity) was markedly decreased in this mutant (Formisano et al, 1993). Our biochemical experiments on the corresponding R1137Q mutation in IGF1R, both in the context of the full-length receptor in cells (Figure 5A) and in vitro with the purified kinase domain (Figure 5B), showed that this mutation also impairs IGF1R autophosphorylation. The structural and biochemical data on Arg1137 in IGF1R support the hypothesis that Arg1164 in IR has an analogous function in trans-phosphorylation of Tyr1162 in the activation loop, and that this role is compromised in type II diabetic patients who harbour the R1164Q mutation.
The activation loops of RTKs contain between one and three tyrosine autophosphorylation sites. In addition to IGF1R and IR, the vascular endothelial growth factor (VEGF) receptors, fibroblast growth factor receptors-1 and -3 (FGFR3), TrkA-C, Axl/Mer/Tyro3, Ror1, Ret and MuSK all contain an arginine or lysine two residues C-terminal to an activation-loop tyrosine, and either an acidic residue or another hydrogen-bonding acceptor at the Glu1132-equivalent position. Mutation of the corresponding lysine in FGFR3 (K649E) also results in impaired receptor autophosphorylation (Webster et al, 1996). Thus, it may be more generally true in RTK activation that a local conformational constraint in the activation loop is critical for the efficiency of the initial trans-phosphorylation event.
Materials and methods
Protein expression and purification
We expressed and purified IGF1RK in its unphosphorylated (0P) and tris-phosphorylated (3P) forms essentially as described (Favelyukis et al, 2001). Spodoptera frugiperda (Sf9) cells were infected with a recombinant baculovirus encoding the kinase domain of human IGF1R (residues 956–1256). IGF1RK mutants were produced with the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocols. The mutants were made in a pFastBac-1 baculovirus transfer vector (Invitrogen) encoding IGF1RK. Cells were harvested 72 h post-infection and lysed in 20 mM Tris–HCl, pH 7.5, 5 mM EDTA, 2 mM dithiothreitol, 0.2% (v/v) Triton X-100, 5 mg/ml leupeptin, 5 mg/ml aprotinin and 2 mM phenylmethylsulphonyl fluoride. IGF1RK-0P was purified by three FPLC chromatographic steps: Source-Q, Superdex-75 and Mono-Q (GE Healthcare). IGF1RK-3P was generated from IGF1RK-0P as described (Favelyukis et al, 2001).
In vitro kinase assays
IGF1R kinase activity was measured using a continuous spectrophotometric assay (Barker et al, 1995). The reactions (in 50 μl) were carried out at 30°C in 100 mM Tris–HCl, pH 7.5, 10 mM MgCl2, 1 mM phosphoenolpyruvate, 0.28 mM NADH, 89 U/ml pyruvate kinase, 124 U/ml lactate dehydrogenase and 2% DMSO. Reactions were initiated by the addition of ATP to mixtures containing enzyme and various concentrations of PQIP. Assays of IGF1RK-0P autophosphorylation were carried out at 6 μM enzyme concentration and 1 mM ATP. The IGF1RK-3P peptide phosphorylation assays were carried out with 150 nM enzyme, 1 mM ATP and 500 μM peptide substrate (KKEEEEYMMMM). Data were recorded every 6 s on a VersaMax microplate reader (Molecular Devices). The IC50 values were determined by fitting to the Michaelis–Menten equation. For Ki measurements, initial velocities of peptide phosphorylation were determined at a series of PQIP concentrations and at ATP concentrations of 100, 500 and 1000 μM. Ki values were determined by a graphical method (Dixon, 1953).
For native PAGE analysis, IGF1RK (20 μM) was autophosphorylated at room temperature by incubation with 5 mM ATP in 20 mM Tris pH 7.5, 30 mM MgCl2 and 6% DMSO with or without 50 μM PQIP. Reactions were terminated at various time points by addition of 100 mM EDTA and analysed by native PAGE using 10% Tris–HCl gels. The phosphorylated forms of IGF1RK were visualized by staining with Coomassie blue.
Crystallization, data collection and structure determination
To facilitate crystallization, we used an IGF1RK variant that contained two point mutations in the kinase-insert region (E1067A/E1069A) (Munshi et al, 2003). These mutations do not alter the catalytic properties of IGF1RK (data not shown). IGF1RK was concentrated to 10 mg/ml in a buffer containing 20 mM Tris–HCl, pH 7.5 and 100 mM NaCl. PQIP was solubilized in DMSO and mixed with IGF1RK in a 4:1 molar ratio. Crystals of PQIP–IGF1RK were obtained at 20°C by vapour diffusion in hanging drops containing 1 μl of protein–compound mixture and 1 μl of reservoir solution (12% PEG8000, 0.1 M imidazole, pH 7.5 and 0.2 M calcium acetate). The crystals belong to space group R32 with one complex per asymmetric unit (60% solvent content) and with unit cell dimensions of a=b=208.2 Å, c=50.85 Å. Crystals were soaked in a cryo-solvent (12% PEG8000, 0.1 M imidazole, pH 7.5, 0.2 M calcium acetate and 20% ethylene glycol) and flash-frozen in liquid nitrogen.
Data were collected at beamline X4A at the National Synchrotron Light Source (Brookhaven National Laboratory) and were processed using HKL2000 (Otwinowski and Minor, 1997). The structure of PQIP–IGF1RK was solved by molecular replacement using MOLREP (Vagin and Teplyakov, 1997) with the structure of IGF1RK-0P (PDB code 1P4O) (Munshi et al, 2003) as the search model. The compound and the reorganized activation loop of IGF1RK were built into 2Fo–Fc and Fo–Fc electron density maps using COOT (Emsley and Cowtan, 2004). Included in the atomic model are residues 956–1255, except for residues 1066–1076 in the kinase-insert region (between α-helices D and E), which are disordered. The stereochemical library description for PQIP was created using Sketcher in the CCP4 suite (CCP4 (Collaborative Computational Project, 1994). The atomic model was refined using REFMAC (Murshudov et al, 1997). Data statistics are listed in Table I.
IGF1R phosphorylation in cells
Wild-type and mutant forms of IGF1R were expressed in IGF1R-deficient R cells (a gift of Dr R Baserga, Thomas Jefferson University) as described previously (Craddock et al, 2007). Cells were transfected using TransIT polyamine transfection reagent (Mirus) according to the manufacturer's instructions. The cells were starved for 16 h, then stimulated with 40 ng/ml IGF1 (Calbiochem) for 10 min at 37°C. Cells were lysed in a buffer containing 25 mM Tris–HCl, pH 8.0, 2 mM EDTA, 140 mM NaCl, 1% NP40, 1 mg/ml aprotinin, 1 mg/ml leupeptin and 2 mM activated sodium orthovanadate. Immunoprecipitation reactions were carried out by treatment with anti-IGF1R antibody (clone JBW902; Millipore) for 1 h at 4°C, followed by Protein A-conjugated agarose beads (Sigma) at 4°C for 3 h with gentle rocking. After centrifugation, the beads were washed with lysis buffer. Precipitated proteins were eluted by treatment with Laemmli sample buffer, separated by 7.5% SDS–PAGE and transferred to PVDF membranes (Millipore). The blots were probed with anti-IGF1R (pTyr1135/1136) antibody (Biosource International) and visualized with the SuperSignal West Femto Maximum Sensitivity substrate system (Pierce). The blots were then stripped and reprobed with anti-IGF1R antibody.
Accession numbers
Atomic coordinates and structure factors for the PQIP–IGF1RK structure have been deposited in the Protein Data Bank with accession code 3D94.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Acknowledgments
This study was supported by National Institutes of Health grants DK052916 (to SRH) and CA058530 and CA122091 (to WTM) and an Irma T Hirschl-Monique Weill-Caulier Career Scientist Award (to SRH). Beamline X4A at the National Synchrotron Light Source, Brookhaven National Laboratory, a DOE facility, is supported by the New York Structural Biology Consortium.
References
- Ablooglu AJ, Frankel M, Rusinova E, Ross JB, Kohanski RA (2001) Multiple activation loop conformations and their regulatory properties in the insulin receptor's kinase domain. J Biol Chem 276: 46933–46940 [DOI] [PubMed] [Google Scholar]
- Adams TE, Epa VC, Garrett TP, Ward CW (2000) Structure and function of the type 1 insulin-like growth factor receptor. Cell Mol Life Sci 57: 1050–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker SC, Kassel DB, Weigl D, Huang X, Luther MA, Knight WB (1995) Characterization of pp60c-src tyrosine kinase activities using a continuous assay: autoactivation of the enzyme is an intermolecular autophosphorylation process. Biochemistry 34: 14843–14851 [DOI] [PubMed] [Google Scholar]
- Baserga R (1999) The IGF-I receptor in cancer research. Exp Cell Res 253: 1–6 [DOI] [PubMed] [Google Scholar]
- Baserga R, Hongo A, Rubini M, Prisco M, Valentinis B (1997) The IGF-I receptor in cell growth, transformation and apoptosis. Biochim Biophys Acta 1332: F105–F126 [DOI] [PubMed] [Google Scholar]
- Butler AA, Yakar S, Gewolb IH, Karas M, Okubo Y, LeRoith D (1998) Insulin-like growth factor-I receptor signal transduction: at the interface between physiology and cell biology. Comp Biochem Physiol B Biochem Mol Biol 121: 19–26 [DOI] [PubMed] [Google Scholar]
- CCP4 (Collaborative Computational Project, N) (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Cocozza S, Porcellini A, Riccardi G, Monticelli A, Condorelli G, Ferrara A, Pianese L, Miele C, Capaldo B, Beguinot F, Varrones S (1992) NIDDM associated with mutation in tyrosine kinase domain of insulin receptor gene. Diabetes 41: 521–526 [DOI] [PubMed] [Google Scholar]
- Craddock BP, Cotter C, Miller WT (2007) Autoinhibition of the insulin-like growth factor I receptor by the juxtamembrane region. FEBS Lett 581: 3235–3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon M (1953) The determination of enzyme inhibitor constants. Biochem J 55: 170–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Favelyukis S, Till JH, Hubbard SR, Miller WT (2001) Structure and autoregulation of the insulin-like growth factor 1 receptor kinase. Nat Struct Biol 8: 1058–1063 [DOI] [PubMed] [Google Scholar]
- Formisano P, Sohn KJ, Miele C, Di Finizio B, Petruzziello A, Riccardi G, Beguinot L, Beguinot F (1993) Mutation in a conserved motif next to the insulin receptor key autophosphorylation sites de-regulates kinase activity and impairs insulin action. J Biol Chem 268: 5241–5248 [PubMed] [Google Scholar]
- Garcia-Echeverria C, Pearson MA, Marti A, Meyer T, Mestan J, Zimmermann J, Gao J, Brueggen J, Capraro HG, Cozens R, Evans DB, Fabbro D, Furet P, Porta DG, Liebetanz J, Martiny-Baron G, Ruetz S, Hofmann F (2004) In vivo antitumor activity of NVP-AEW541––a novel, potent, and selective inhibitor of the IGF-IR kinase. Cancer Cell 5: 231–239 [DOI] [PubMed] [Google Scholar]
- Hu J, Liu J, Ghirlando R, Saltiel AR, Hubbard SR (2003) Structural basis for recruitment of the adaptor protein APS to the activated insulin receptor. Mol Cell 12: 1379–1389 [DOI] [PubMed] [Google Scholar]
- Hubbard SR (1997) Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. EMBO J 16: 5572–5581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard SR, Wei L, Ellis L, Hendrickson WA (1994) Crystal structure of the tyrosine kinase domain of the human insulin receptor. Nature 372: 746–754 [DOI] [PubMed] [Google Scholar]
- Ji QS, Mulvihill MJ, Rosenfeld-Franklin M, Cooke A, Feng L, Mak G, O'Connor M, Yao Y, Pirritt C, Buck E, Eyzaguirre A, Arnold LD, Gibson NW, Pachter JA (2007) A novel, potent, and selective insulin-like growth factor-I receptor kinase inhibitor blocks insulin-like growth factor-I receptor signaling in vitro and inhibits insulin-like growth factor-I receptor dependent tumor growth in vivo. Mol Cancer Ther 6: 2158–2167 [DOI] [PubMed] [Google Scholar]
- Kato H, Faria TN, Stannard B, Roberts CT Jr, LeRoith D (1994) Essential role of tyrosine residues 1131, 1135, and 1136 of the insulin-like growth factor-I (IGF-I) receptor in IGF-I action. Mol Endocrinol 8: 40–50 [DOI] [PubMed] [Google Scholar]
- LeRoith D, Roberts CT Jr (2003) The insulin-like growth factor system and cancer. Cancer Lett 195: 127–137 [DOI] [PubMed] [Google Scholar]
- LeRoith D, Werner H, Beitner-Johnson D, Roberts CT Jr (1995) Molecular and cellular aspects of the insulin-like growth factor I receptor. Endocr Rev 16: 143–163 [DOI] [PubMed] [Google Scholar]
- Liu JP, Baker J, Perkins AS, Robertson EJ, Efstratiadis A (1993) Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r). Cell 75: 59–72 [PubMed] [Google Scholar]
- Lu Y, Zi X, Zhao Y, Mascarenhas D, Pollak M (2001) Insulin-like growth factor-I receptor signaling and resistance to trastuzumab (Herceptin). J Natl Cancer Inst 93: 1852–1857 [DOI] [PubMed] [Google Scholar]
- Miele C, Caruso M, Calleja V, Auricchio R, Oriente F, Formisano P, Condorelli G, Cafieri A, Sawka-Verhelle D, Van Obberghen E, Beguinot F (1999) Differential role of insulin receptor substrate (IRS)-1 and IRS-2 in L6 skeletal muscle cells expressing the Arg1152 → Gln insulin receptor. J Biol Chem 274: 3094–3102 [DOI] [PubMed] [Google Scholar]
- Mitsiades CS, Mitsiades NS, McMullan CJ, Poulaki V, Shringarpure R, Akiyama M, Hideshima T, Chauhan D, Joseph M, Libermann TA, Garcia-Echeverria C, Pearson MA, Hofmann F, Anderson KC, Kung AL (2004) Inhibition of the insulin-like growth factor receptor-1 tyrosine kinase activity as a therapeutic strategy for multiple myeloma, other hematologic malignancies, and solid tumors. Cancer Cell 5: 221–230 [DOI] [PubMed] [Google Scholar]
- Munshi S, Hall DL, Kornienko M, Darke PL, Kuo LC (2003) Structure of apo, unactivated insulin-like growth factor-1 receptor kinase at 1.5 Å resolution. Acta Crystallogr D Biol Crystallogr 59: 1725–1730 [DOI] [PubMed] [Google Scholar]
- Munshi S, Kornienko M, Hall DL, Reid JC, Waxman L, Stirdivant SM, Darke PL, Kuo LC (2002) Crystal structure of the Apo, unactivated insulin-like growth factor-1 receptor kinase. Implication for inhibitor specificity. J Biol Chem 277: 38797–38802 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Nagar B, Bornmann WG, Pellicena P, Schindler T, Veach DR, Miller WT, Clarkson B, Kuriyan J (2002) Crystal structures of the kinase domain of c-Abl in complex with the small molecule inhibitors PD173955 and imatinib (STI-571). Cancer Res 62: 4236–4243 [PubMed] [Google Scholar]
- Oliver AW, Paul A, Boxall KJ, Barrie SE, Aherne GW, Garrett MD, Mittnacht S, Pearl LH (2006) Trans-activation of the DNA-damage signalling protein kinase Chk2 by T-loop exchange. EMBO J 25: 3179–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Pike AC, Rellos P, Niesen FH, Turnbull A, Oliver AW, Parker SA, Turk BE, Pearl LH, Knapp S (2008) Activation segment dimerization: a mechanism for kinase autophosphorylation of non-consensus sites. EMBO J 27: 704–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedemann J, Macaulay VM (2006) IGF1R signalling and its inhibition. Endocr Relat Cancer 13 (Suppl 1): S33–S43 [DOI] [PubMed] [Google Scholar]
- Sakatani T, Kaneda A, Iacobuzio-Donahue CA, Carter MG, de Boom Witzel S, Okano H, Ko MS, Ohlsson R, Longo DL, Feinberg AP (2005) Loss of imprinting of Igf2 alters intestinal maturation and tumorigenesis in mice. Science 307: 1976–1978 [DOI] [PubMed] [Google Scholar]
- Schindler T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J (2000) Structural mechanism for STI-571 inhibition of Abelson tyrosine kinase. Science 289: 1938–1942 [DOI] [PubMed] [Google Scholar]
- Shoelson SE, Chatterjee S, Chaudhuri M, White MF (1992) YMXM motifs of IRS-1 define substrate specificity of the insulin receptor kinase. Proc Natl Acad Sci USA 89: 2027–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till JH, Ablooglu AJ, Frankel M, Bishop SM, Kohanski RA, Hubbard SR (2001) Crystallographic and solution studies of an activation loop mutant of the insulin receptor tyrosine kinase: insights into kinase mechanism. J Biol Chem 276: 10049–10055 [DOI] [PubMed] [Google Scholar]
- Tokarski JS, Newitt JA, Chang CY, Cheng JD, Wittekind M, Kiefer SE, Kish K, Lee FY, Borzillerri R, Lombardo LJ, Xie D, Zhang Y, Klei HE (2006) The structure of dasatinib (BMS-354825) bound to activated ABL kinase domain elucidates its inhibitory activity against imatinib-resistant ABL mutants. Cancer Res 66: 5790–5797 [DOI] [PubMed] [Google Scholar]
- Vagin AA, Teplyakov A (1997) MOLREP: an automated program for molecular replacement. J Appl Cryst 30: 1022–1025 [Google Scholar]
- Valentinis B, Baserga R (2001) IGF-I receptor signalling in transformation and differentiation. Mol Pathol 54: 133–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velaparthi U, Wittman M, Liu P, Stoffan K, Zimmermann K, Sang X, Carboni J, Li A, Attar R, Gottardis M, Greer A, Chang CY, Jacobsen BL, Sack JS, Sun Y, Langley DR, Balasubramanian B, Vyas D (2007) Discovery and initial SAR of 3-(1H-benzo[d]imidazol-2-yl)pyridin-2(1H)-ones as inhibitors of insulin-like growth factor 1-receptor (IGF-1R). Bioorg Med Chem Lett 17: 2317–2321 [DOI] [PubMed] [Google Scholar]
- Webster MK, d'Avis PY, Robertson SC, Donoghue DJ (1996) Profound ligand-independent kinase activation of fibroblast growth factor receptor 3 by activation loop mutation responsible for a lethal skeletal dysplasia, thanatophoric dysplasia type II. Mol Cell Biol 16: 4081–4087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Hubbard SR, Hendrickson WA, Ellis L (1995) Expression, characterization, and crystallization of the catalytic core of the human insulin receptor protein-tyrosine kinase domain. J Biol Chem 270: 8122–8130 [DOI] [PubMed] [Google Scholar]
- Wittman M, Carboni J, Attar R, Balasubramanian B, Balimane P, Brassil P, Beaulieu F, Chang C, Clarke W, Dell J, Eummer J, Frennesson D, Gottardis M, Greer A, Hansel S, Hurlburt W, Jacobson B, Krishnananthan S, Lee FY, Li A (2005) Discovery of a (1H-benzoimidazol-2-yl)-1H-pyridin-2-one (BMS-536924) inhibitor of insulin-like growth factor I receptor kinase with in vivo antitumor activity. J Med Chem 48: 5639–5643 [DOI] [PubMed] [Google Scholar]
- Zhou S, Carraway KL III, Eck MJ, Harrison SC, Feldman RA, Mohammadi M, Schlessinger J, Hubbard SR, Smith DP, Eng C, Lorenzo MJ, Ponder BAJ, Mayer BJ, Cantley LC (1995) Catalytic specificity of protein-tyrosine kinases is critical for selective signalling. Nature 373: 536–539 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4