Abstract
Brassica napus cultivar Westar is non-embryogenic under all standard protocols for induction of microspore embryogenesis; however, the rare embryos produced in Westar microspore cultures, induced with added brassinosteroids, were found to develop into heritably stable embryogenic lines after chromosome doubling. One of the Westar-derived doubled haploid (DH) lines, DH-2, produced up to 30% the number of embryos as the highly embryogenic B. napus line, Topas DH4079. Expression analysis of marker genes for embryogenesis in Westar and the derived DH-2 line, using real-time reverse transcription-PCR, revealed that the timely expression of embryogenesis-related genes such as LEAFY COTYLEDON1 (LEC1), LEC2, ABSCISIC ACID INSENSITIVE3, and BABY BOOM1, and an accompanying down-regulation of pollen-related transcripts, were associated with commitment to embryo development in Brassica microspores. Microarray comparisons of 7 d cultures of Westar and Westar DH-2, using a B. napus seed-focused cDNA array (10 642 unigenes), identified highly expressed genes related to protein synthesis, translation, and response to stimulus (Gene Ontology) in the embryogenic DH-2 microspore-derived cell cultures. In contrast, transcripts for pollen-expressed genes were predominant in the recalcitrant Westar microspores. Besides being embryogenic, DH-2 plants showed alterations in morphology and architecture as compared with Westar, for example epinastic leaves, non-abscised petals, pale flower colour, and longer lateral branches. Auxin, cytokinin, and abscisic acid (ABA) profiles in young leaves, mature leaves, and inflorescences of Westar and DH-2 revealed no significant differences that could account for the alterations in embryogenic potential or phenotype. Various mechanisms accounting for the increased capacity for embryogenesis in Westar-derived DH lines are considered.
Keywords: Brassica napus, embryogenesis, microarray, microspore, transcript profiling
Introduction
Plant breeding is facilitated by the rapid development of doubled haploid (DH) plant lines, homozygous at all loci. This is most readily accomplished through microspore embryogenesis, where freshly isolated uninucleate or binucleate microspores (male gametophytes) are induced in culture to shift from a gametophytic or pollen pathway to embryo development. After chromosome doubling, the resultant plant lines are maintained as breeding stocks. Successful and efficient microspore-derived embryo production is genotype dependent, and much labour is consumed in developing and optimizing tissue culture conditions. Homozygous DH lines have been integrated into breeding programmes for superior varieties of canola (Brassica spp.), barley (Hordeum vulgare L.), maize (Zea mays), wheat (Triticum aestivum L.), rice (Oryza sativa L.), pepper (Capsicum annum), asparagus (Asparagus officinalis), and several grasses such as Lolium and Festuca spp. (Thomas et al., 2003; Kopecky et al., 2005; Forster et al., 2007). Some selected DH lines also have been used to produce commercial hybrids, or directly as commercial lines (Forster et al., 2007). In addition, DH lines have become an extremely important resource for chromosome mapping studies and also facilitate selection of recessive or polygenic traits (Forster and Thomas, 2005).
Brassica napus is a model system for studies of microspore embryogenesis; however, not all genotypes respond equally well to inducing culture conditions. Brassica napus cv. Topas embryogenic line DH4079 is one of the most responsive genotypes, and >10% of cultured microspores form embryos (Ferrie, 2003). Some other genotypes of B. napus, for example cv. Allons, Garrison, and Westar, are poorly embryogenic, and <0.5% of cultured microspores form embryos using the standard B. napus protocol for microspore embryogenesis (Ferrie et al., 2005). A poor embryogenic response limits the utility of desirable cultivars in breeding programs.
Isolated microspores of suitable cultivars of B. napus can be induced to form embryos in vitro with appropriate culture media and stress treatments, for example heat (32 °C) or osmotic stress (polyethylene glycol) (Ferrie, 2003; Ferrie and Keller, 2007). Several factors are known to be critical for the optimum response of cultured microspores, including donor plant growth conditions, culture conditions, media, genotype, and age of the donor plants. Quantitative trait locus (QTL) analysis in Brassica cultivars for regeneration potential from protoplast cultures has revealed a low number of genes involved in this process (Holme et al., 2004). Zhang and Takahata (2001) have reported that microspore embryogenic ability is controlled by two multiple gene loci in B. napus. Also, random amplified polymorphic DNA (RAPD) markers with additive effects have been linked to microspore embryogenic ability in Chinese cabbage and oilseed rape (Zhang et al., 2003).
Improvements in microspore embryogenesis for poorly embryogenic cultivars of B. napus (i.e. cv. Westar) have been reported following additions of 24-epibrassinolide (EBR) or brassinolide to the culture medium (Ferrie et al., 2005). In addition, there are numerous reports in the literature of positive changes in embryogenic response in plant species and cultivars due to alterations in either donor plant conditions, media composition, or culture conditions (Li and Devaux, 2001; Croser et al., 2006; Kim and Moon, 2007); however, there have been no reports, or molecular studies, of cases of induced, stable, and heritable improvements in embryogenic potential in DH plant lines resulting from such manipulation of previously non-embryogenic, parental material. In the case described here, the rare embryos produced in the Westar microspore cultures following a 3 d exposure to brassinosteroid-supplemented media developed into heritably stable embryogenic lines after chromosome doubling (DH-1, DH-2, DH-3 and DH-4). Embryo development, transformation efficiency, gene expression profiles, hormone concentrations, and phenotypic differences have been compared in the non-embryogenic B. napus cv. Westar parental line and a Westar-derived embryogenic line, DH-2. The molecular characterization utilized a set of well-established marker genes for embryogenesis (Malik et al., 2007) and a newly developed Brassica seed-focused cDNA array (10 642 unigenes; Xiang et al., 2008).
Materials and methods
Plant material
Plants of B. napus cv. Westar were grown in 15 cm pots in a growth cabinet with a 16 h/8 h day/night photoperiod, light intensity of 400 μmol m−2 s−1, and day/night temperatures of 20 °C/15 °C. Following flower bud formation, and in preparation for microspore culture, the day/night temperatures were lowered to 10 °C/5 °C. Microspore collections and cultures were initiated as described by Ferrie and Keller (1995). For comparison purposes, it should be noted that microspores collected from 100 buds were sufficient for ∼20 culture plates. Embryogenesis was induced in microspores isolated at the late-uninucleate to early-binucleate stage (Ferrie and Keller, 1995) using a defined medium containing 13% sucrose and heat stress at 32 °C for 3 d. EBR (OlChemIm) at 10−6 M was added to the medium to improve the response in the poorly embryogenic cultivar Westar (Ferrie et al., 2005), and several of the resulting microspore-derived embryos from those cultures were regenerated into plants. The plantlets were treated with 0.34% colchicine solution for chromosome doubling in order to ensure recovery of DH plants. The colchicine-treated plantlets were transferred to soil, grown under the conditions described above, and used for subsequent microspore cultures. To assess embryogenic potential, microspores isolated from the Westar-derived DH lines (DH-1, DH-2, DH-3 and DH-4) and Westar were cultured on standard NLN-13% sucrose medium without the addition of brassinosteroids. For in-depth studies, microspore cultures of the DH-2 line and Westar were observed at 1, 3, 5, 7, 14, and 21 d for embryo development. The 5 d and 7 d microspore cultures from both lines were collected by centrifugation and stained with 1% acetocarmine to observe divisions during early embryogenesis. Light microscopy images were captured on a Leica DMR microscope.
Transformation
Agrobacterium tumefaciens strain GV3101:pMP90, carrying binary vector pHS723 which includes a β-glucuronidase–neomycin phosphotransferase (GUS–NPT) translational fusion driven by an enhanced 35S promoter with a nopaline synthase (NOS) terminator (Datla et al., 1991; Nair et al., 2000), was used for transformation. Hypocotyl explants were prepared from 5-d-old seedlings of Westar and the Westar-derived DH lines (DH-1, DH-2, DH-3 and DH-4), pre-cultured, and inoculated with Agrobacterium according to DeBlock et al. (1989), with modifications by Zou et al. (1997). Selection was carried out on shoot induction medium with 20 mg l−1 kanamycin. Green regenerants were tested for GUS activity by incubating leaf pieces in X-Gluc substrate (Jefferson et al., 1986).
RNA isolation
Total RNA from 0 h, 1, 3, 5, and 7 d cultured microspores was isolated using the RNeasy Midi kit (Qiagen), including on-column DNase digestion. For semi-quantitative RT-PCR, 3 μg of total RNA was used for first-strand cDNA synthesis with oligo(dT)16 primers (DNA Sequencing Lab, NRC-Plant Biotechnology Institute) and SUPERSCRIPT II reverse transcriptase (Invitrogen) according to the manufacturer's protocol. Gene-specific primer pairs were designed using Primer 3 software (http://gene.pbi.nrc.ca/cgi-bin/primer/primer3_www.cgi) to obtain PCR products that were 350–550 bp in length. PCRs were one cycle at 95 °C for 5 min and 30 cycles at 95 °C for 30 s, 55 °C for 30 s, 72 °C for 45 s using 0.6 μl of template cDNA from the first-strand cDNA synthesis reaction.
Expression analyses by real-time RT-PCR
Total RNA (150 ng) from each tissue, developmental stage, or cultivar was used for one-step real-time reverse transcription-PCR (RT-PCR) analyses using the QuantiTect SYBR Green RT-PCR Kit (Qiagen Inc.) and gene-specific primers. Primer pairs were designed using the Primer Quest software (Integrated DNA Technologies) to give PCR products from 100 to 400 bp. Real-time RT-PCR was performed on an Mx3000P™ Real-time PCR system (Stratagene, La Jolla, CA, USA). Relative expression was calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001) with 18S rRNA as the internal control gene. The cycling parameters were one cycle at 50 °C for 60 min (reverse transcription reaction), one cycle at 95 °C for 15 min, then 35 cycles of 95 °C for 30 s, 58 °C for 1 min, 72 °C for 45 s. Primer sequences were described in a previous publication (Malik et al., 2007).
Microarray analysis using a B. napus seed-focused cDNA array (Bn10K)
For microarray analyses, the 7 d microspore cultures of Westar and the Westar-derived DH-2 line were size-selected on a mesh screen (Sefar; pore size 35 μm) in order to collect the microspores and/or cell clusters that had enlarged, and to discard the smaller physiologically unresponsive microspores. A 10 μg aliquot of total RNA from each sample was labelled with Cy3 or Cy5 (Amersham) and hybridized to the Brassica seed-focused cDNA microarray (Bn10K; http://brassicagenomics.ca) (Xiang et al., 2008) using the Pronto!™ Plus indirect system according to the manufacturer's instructions (Corning). The slides were scanned using a ScanArray 4000 laser scanner at a resolution of 10 μm. The image analysis and signal quantification were done with QUANTARRAY (GSI Lumonics, Watertown, MA, USA). Data storage and preliminary data processing, including normalization, were done on BioArray Software Environment (BASE 1.0; Saal et al., 2002). Background-subtracted signals were used to identify differentially expressed genes with Significance Analysis of Microarrays (SAM 2.0; Tusher et al., 2001).
Hormone profiling
One gram fresh weight tissue samples from young unexpanded leaves, mature leaves, and inflorescences bearing buds, with 3–4 open flowers, of matched samples of Westar and DH-2 were collected, frozen in liquid nitrogen, and freeze-dried. Three biological replicates (50 mg dry weight) were used for hormone profiling of abscisic acid (ABA) and related metabolites, auxins, and cytokinins using the protocol described in Chiwocha et al. (2005).
Results
Embryogenic potential of the DH lines
The DH lines (DH-1, DH-2, DH-3 and DH-4) were developed from four embryos randomly selected from Westar microspore cultures that had been previously treated with EBR to improve embryogenesis (Ferrie et al., 2005). In the absence of an EBR treatment, embryo development from Westar-derived microspores under standard inducing conditions was extremely rare (Table 1). The selected embryos were regenerated to plantlets, grown to flowering, and microspore embryogenesis was re-initiated in the DH lines. Surprisingly, all four of the lines had greatly improved rates of microspore embryogenesis as compared with the non-embryogenic parental line Westar (Table 1). The embryogenic response was variable among the lines, but always was consistently better than the Westar parent. The most embryogenic line, DH-2, produced thousands of embryos under standard culture conditions where Westar might only produce one embryo, and in this respect it was 30% as productive as a previously characterized highly embryogenic line, B. napus DH4079 (Ferrie et al., 2005; Malik et al., 2007). The DH lines were selfed (to maintain the original homozygosity) and F2 seed was collected separately for each line. Plants (from F2 seed) of the most embryogenic Westar-derived line (DH-2; Table 1) were used as donor plants for further studies of microspore embryogenesis (F3 material). The improved embryogenic potential of the DH-2 line remained stable through at least these two subsequent generations (F2 and F3). Molecular and phenotypic comparisons were made between the non-embryogenic parental line, Westar, and the embryogenic Westar-derived DH-2 line.
Table 1.
Embryogenic response and transformation potential in four independently selected Westar-derived lines
| B. napus, CV and line | Embryogenic responsea | Transformation response | |||
| No. of explants | Green shoots | No. of GUS positives | Transformation efficiency (%) | ||
| Westar | 1±0 | 510 | 72 | 39 | 7.6 |
| Westar DH-1 | 40±11.9 | 484 | 49 | 24 | 3.9 |
| Westar DH-2 | 2235.3±1366.5 | 850 | 8 | 3 | 0.4 |
| Westar DH-3 | 131.0±99.5 | 510 | 33 | 17 | 3.3 |
| Westar DH-4 | 685.3±657.4 | 96 | 26 | 1 | 1.0 |
Embryogenesis was assessed as the average frequency of embryos per plate from three replicate experiments from microspore cultures of Brassica napus cv. Westar and Westar-derived DH (doubled haploid) lines. On average there are ∼1 000 000 microspores transferred to each plate. Transformation success was assessed by determining the number of independent GUS-positive shoots relative to the number of original Agrobacterium-treated explants.
Mean ±SE.
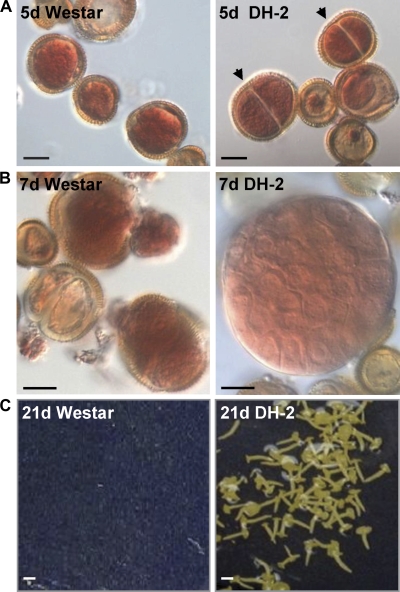
At the time of isolation (0 h), and the beginning of culture, the microspores of both lines had a prominent nucleus and the cytoplasm was restricted to the periphery. In a previous study it was noted that enlargement of the microspore was a prominent feature of 3 d heat-stress-induced embryogenic microspores of B. napus Topas DH4079, and that cell and nuclear divisions were predominant in the 5 d and 7 d induced microspores, with only marginal increases in size occurring during this latter period (Malik et al., 2007). No differences were noted between the microspores of the Westar parental line and the Westar-derived DH-2 line during the first 3 d of culture (data not shown). Following a heat stress treatment for embryo induction (3 d at 32 °C), responding Westar and DH-2 microspores enlarged similarly, to more than double their original size, and the numbers of enlarged microspores in the two lines were equivalent (∼30% of cultured microspores); however, by 5 d the induced microspores of the DH-2 line had undergone a first symmetric mitotic division (Fig. 1). In contrast, the enlarged microspores of the Westar line did not divide, although they were swollen and stained bright red with 1% acetocarmine (Fig. 1). Thereafter, the induced microspores of the DH-2 line continued to undergo random cell divisions, and by 7 d the embryogenic microspores of the DH-2 line appeared as cell clusters (globular and pre-globular embryos) with remnants of the ruptured exine still remaining on the developing embryo (Fig. 1). There were very few dividing structures in 7 d Westar cultures, and some of those dividing structures already had started to degenerate (Fig. 1).
Fig. 1.
Microspore-derived embryo development in Brassica napus cv. Westar and the Westar-derived DH-2 line. (A) Acetocarmine-stained 5 d enlarged microspores. Arrowheads indicate divisions in the microspores of the DH-2 line. (B) Acetocarmine-stained 7 d enlarged microspores in Westar and a dividing pre-globular embryo in the DH-2 line. (C) Twenty-one day mid-maturation stage embryos in the DH-2 line; no embryos developed in this microspore culture plate of Westar. Black bars=10 μm, white bars=35 μm.
Although B. napus is a model system for microspore embryogenesis, there are very few lines that are both embryogenic and transformable, for example B. napus DH12075 (Li et al., 2003) and cv. Lisandra (Fukuoka et al., 1998; Zhang et al., 2003), and thus appropriate for both genetic and reverse genetics applications. Westar is easily transformable (Cardoza and Stewart, 2004) but not highly embryogenic (Table 1; Ferrie et al., 2005), while B. napus Topas DH4079 is highly embryogenic (Malik et al., 2007), but not transformable using standard techniques (J Hammerlindl, unpublished data). Therefore, it was of interest to examine transformability of the Westar-derived DH lines (Table 1). Some of the lines were easily transformed with Agrobacterium (e.g. DH-1 and DH-3); however, the most embryogenic line (DH-2) was the least transformable of the lines developed (Table 1). This line also showed restricted organogenesis (shoot regeneration) and, therefore, perhaps transformability was limited by and could be improved with some focus on regeneration and shoot production.
Expression of embryogenesis-related genes
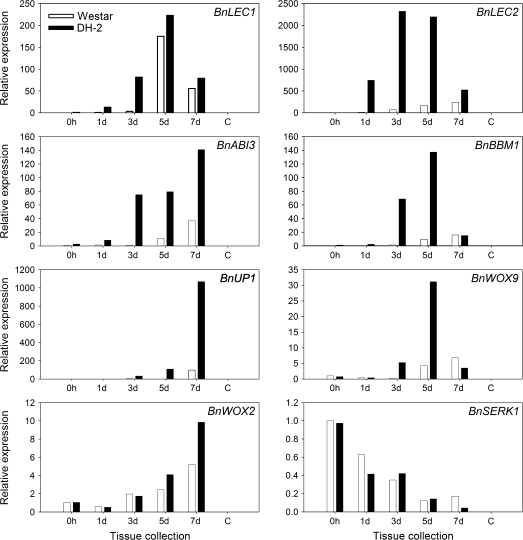
Previously, several clusters of differentially expressed genes marking the developmental transitions from freshly isolated microspores (0 h) to committed 7 d embryogenic cell clusters (globular and pre-globular embryos), as well as a set of 16 unambiguous marker genes for the induction of microspore embryogenesis, were identified (Malik et al., 2007). These molecular marker genes were not expressed in microspores at the time of culture (0 h) in the highly embryogenic B. napus line Topas DH4079, nor in microspores cultured under non-inductive conditions (18 °C), and thus can be used both quantitatively and qualitatively to measure a cultivar's responsiveness to embryogenesis-inducing conditions (Malik et al., 2007). Based on real-time RT-PCR, the transcript abundance for the marker gene BnLEC1 was not significantly different between the two lines at 5 d and 7 d (Fig. 2), confirming the acquisition of a certain level of embryogenic competence in cv. Westar (Fig. 1). However, BnLEC1 expression in Westar was markedly delayed at 1 d and 3 d as compared with the DH-2 cultures (Fig. 2). Expression of the other marker genes, namely BnLEC2, BnABI3, BnBBM1, BnUP1, and BnWOX9, was uniformly much reduced at all time points in Westar cultures as compared with DH-2 cultures (Fig. 2), reflecting the differences in embryogenic potential between the two lines. There were no significant differences in the level of expression of BnSERK1 between Westar and DH-2 during the early stages of microspore culture (Fig. 2), despite the marked differences in embryogenic potential.
Fig. 2.
Real time RT-PCR analyses of embryo-specific marker genes (BnLEC1, BnLEC2, BnABI3, BnBBM1, BnUP1, BnWOX9, and BnWOX2) and BnSERK1 in microspore cultures of non-embryogenic B. napus cv. Westar and the embryogenic Westar-derived DH-2 line. Stages of microspore-derived embryo (MDE) development (0 h, 1, 3, 5, and 7 d) are indicated for each of the lines (Westar, DH-2). Expression was calculated according to the 2-ΔΔCT method (Livak and Schmittgen, 2001). Relative expression was based on comparisons with transcript levels in 0 h microspores of cv. Westar with 18S rRNA as the internal control for normalization.
Expression of pollen-related genes
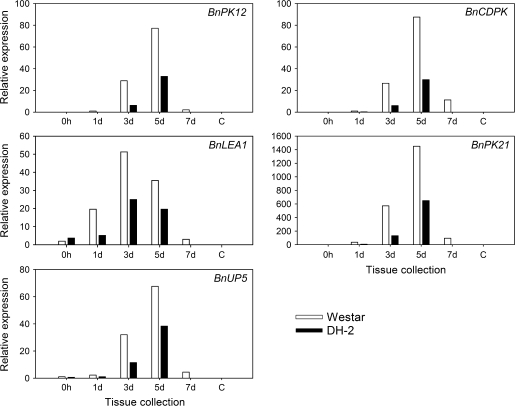
Putative pollen-expressed genes have been identified from cDNA libraries made from 3 d and 5 d embryogenic cultures and cDNA libraries representing in vitro pollen (Joosen et al., 2007; Malik et al., 2007; M R Malik et al., unpublished data). Subsequent RT-PCR analyses have confirmed a number of predominantly pollen-expressed genes, for example BnPK12, BnCDPK, BnLEA1, BnPK21, and BnUP5 (see Supplementary Table S1 available at JXB online). Expression analyses by real-time RT-PCR showed at least a 2-fold greater expression of these pollen-related genes in the recalcitrant Westar cultivar, particularly at the 3 d and 5 d stages of microspore culture, as compared with the embryogenic Westar-derived DH-2 line (Fig. 3). Although the expression of these pollen-related genes diminished in cultures of both lines by 7 d, expression was still higher in the Westar microspores (Fig. 3).
Fig. 3.
Real time RT-PCR analyses of pollen-specific genes BnPK12 (At3g18810), BnCDPK (At2g31500), BnLEA1 (At4g13230), BnPK21 (At2g24370), and BnUP5 in microspore cultures of non-embryogenic B. napus cv. Westar and the embryogenic Westar-derived DH-2 line (the closest Arabidopsis match for each B. napus pollen-specific gene is given in parentheses). Stages of microspore-derived embryo (MDE) development (0 h, 1, 3, 5, and 7 d) are indicated for each of the lines (Westar, DH-2). Expression was calculated according to the 2−ΔΔCT method (Livak and Schmittgen, 2001). Relative expression was based on comparisons with transcript levels in 0 h microspores of cv. Westar with 18S rRNA as the internal control for normalization.
Transcript profiling using the Bn10K seed cDNA microarray
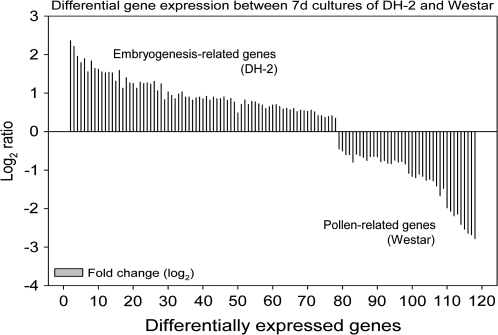
Microarray analyses of gene expression using the B. napus 10K seed cDNA array (Xiang et al., 2008) identified significant expression of 637 genes in 7 d enlarged microspores of Westar and 456 genes in 7 d enlarged and dividing microspores of the DH-2 line (both samples >35 μm) and, of these, 314 genes that were expressed in both 7 d Westar and DH-2 (signal intensity >500 in two or more replicates). Using the software program SAM (Tusher et al., 2001), 117 differentially expressed genes were identified that mark the developmental differences between Westar and Westar-derived DH-2 (77 genes up-regulated in 7 d enlarged and dividing DH-2 microspores, 40 genes up-regulated in 7 d enlarged Westar microspores) (Fig. 4). Many of the genes up-regulated in the 7 d enlarged and dividing DH-2 microspores were related to either protein biosynthesis, response to stimulus, or cellular transport (Table 2). Notable amongst these are genes encoding 40S (RPS2C, RPS15A, RPS9B, RPSaA, RPS9B, RPS17A) and 60S (RPL10aB, RPL8A, RPL21E, RPP1B, RPL23C, RPL28C, RPL30C, RPL36aA, RPL3A) ribosomal proteins, elongation factor 2, heat shock cognate 70 kDa protein1 (HSC70-1), lipid transfer proteins, and seed storage proteins (Table 2). No differences were detected on the microarray in the levels of expression of the transcription factor genes LEC1 and LEC2 between the two lines, although these differences were demonstrated by real-time RT-PCR (Fig. 2), perhaps because microarray expression levels per se were very low for these genes (data not shown). Nonetheless, some known targets of the LEC2 transcription factor were up-regulated and differentially expressed in the 7 d enlarged and dividing microspores of the DH-2 line, for example 2S seed storage protein 1 (At4g27140), oleosin (At4g25140), and cysteine proteinase (At3g54940) (Table 2; Braybrook et al., 2006).
Fig. 4.
Microarray analysis of differentially expressed genes between 7 d enlarged (dividing) embryogenic microspores of DH-2 and 7 d induced (non-dividing) microspores of the parental line, Westar. Labelled total RNA (10 μg) was used for hybridization to the Bn10K seed cDNA array. Signal intensities were normalized and gene lists extracted using SAM (minimum 1.5-fold change in expression). A total of 117 differentially expressed genes were identified: 77 genes up-regulated in DH-2 and 40 genes up-regulated in Westar (negative log2 values). Gene identifications are listed in Tables 2 and 3.
Table 2.
Genes up-regulated in 7 d microspore cultures of embryogenic Westar-derived DH-2 line
| Accession no. | Genes up-regulated in Westar DH-2 | Best match to Arabidopsis | E-value | Biological process | Broad functional category |
| EE541057 | 60S ribosomal protein L32 (RPL32A) | AT4G18100.1 | 4e-71 | Ribosome biogenesis and assembly | Cell organization and biogenesis |
| CN735630 | Histone H2B, putative | AT2G37470.1 | 1e-41 | Chromosome organization and biogenesis | Cell organization and biogenesis |
| DY010080 | 60S ribosomal protein L5 (RPL5B) | AT5G39740.1 | e-123 | Ribosome biogenesis and assembly | Cell organization and biogenesis |
| EE542594 | 60S ribosomal protein L37a (RPL37aC) | AT3G60245.1 | 3e-49 | Ribosome biogenesis and assembly | Cell organization and biogenesis |
| EE541600 | MEE26 (maternal effect embryo arrest 26) | AT2G34870.1 | 4e-9 | Embryonic development ending in seed dormancy | Developmental processes |
| DY009433 | EMBRYO DEFECTIVE 2386; identical to 60S ribosomal protein L19-1 (RPL19A) | AT1G02780.1 | 1e-93 | Embryonic development ending in seed dormancy; translation | Developmental processes; protein metabolism |
| CN734060 | EMBRYO DEFECTIVE 2171; 60S ribosomal protein L23 (RPL23A) | AT3G04400.1 | 2e-76 | Embryonic development ending in seed dormancy; translation | Developmental processes; protein metabolism |
| CN727564 | EMBRYO DEFECTIVE 2171; 60S ribosomal protein L23 (RPL23A) | AT3G04400.1 | 4e-77 | Embryonic development ending in seed dormancy | Developmental processes |
| EE542973 | Histone H2A, putative | AT5G59870.1 | 8e-52 | Nucleosome assembly | DNA or RNA metabolism |
| EE550299 | NAD2B; encodes subunit of mitochondrial NAD(P)H dehydrogenase | ATMG01320.1 | 3e-23 | Electron transport | Electron transport or energy pathways |
| EE550879 | COB; mitochondrial apocytochrome b | ATMG00220.1 | e-114 | Aerobic respiration | Electron transport or energy pathways |
| DY007371 | Mitochondrial NADH dehydrogenase 5 | ATMG00665.1 | 7e-41 | Electron transport | Electron transport or energy pathways |
| CN732202 | LIPID TRANSFER PROTEIN 3 | AT5G59320.1 | 1e-52 | Response to abscisic acid stimulus | Other biological processes |
| EE548239 | Fructose-bisphosphate aldolase, putative | AT3G52930.1 | e-117 | Pentose-phosphate shunt | Other cellular processes |
| CN737059 | GLUTATHIONE S-TRANSFERASE 29, ATGSTU18 | AT1G10360.1 | 4e-72 | Toxin catabolic process | Other cellular processes |
| EE541185 | MATK; encodes a maturase located in the trnK intron in the chloroplast genome | ATCG00040.1 | 8e-70 | RNA splicing | Other cellular processes |
| CN727662 | GAPC-2; glyceraldehyde-3-phosphate dehydrogenase | AT1G13440.1 | 4e-72 | Gluconeogenesis | Other cellular processes |
| CN733306 | THI1 (THIAZOLE REQUIRING) | AT5G54770.1 | 7e-96 | Thiamine biosynthetic process | Other cellular processes |
| EE462458 | ATP synthase beta chain 2 | AT5G08690.1 | 1e-52 | ATP biosynthetic process | Other cellular processes |
| CN726197 | OLEO1 (OLEOSIN1) | AT4G25140.1 | 6e-62 | Sequestering of lipid | Other metabolic processes |
| EE548291 | TPI; ATCTIMC (CYTOSOLIC TRIOSEPHOSPHATE ISOMERASE | AT3G55440.1 | 1e-97 | Metabolic process | Other metabolic processes |
| DY010101 | Calmodulin binding/elongation factor 1-alpha/EF-1-alpha | AT5G60390.2 | e-138 | Translational elongation | Protein metabolism |
| No Acc. No. | Elongation factor 1-alpha/EF-1-alpha | AT1G07920 | Translational elongation | Protein metabolism | |
| EE551198 | RPS3; ribosomal protein S3 | ATMG00090.1 | e-127 | Translation | Protein metabolism |
| EE461044 | RPS3; encodes a chloroplast ribosomal protein S3 | ATCG00800.1 | 4e-68 | Translation | Protein metabolism |
| EE439665 | Elongation factor 1-alpha/EF-1-alpha | AT5G60390.1 | 7e-94 | Translational elongation | Protein metabolism |
| EE542027 | Elongation factor 1-alpha/EF-1-alpha | AT5G60390.1 | e-133 | Translational elongation | Protein metabolism |
| EE550930 | CLPP1; encodes the only ClpP (caseinolytic protease) encoded within the plastid genome | ATCG00670.1 | 2e-38 | Proteolysis | Protein metabolism |
| CN725880 | Cysteine proteinase, putative | AT3G54940.3 | 3e-85 | Proteolysis | Protein metabolism |
| CN730121 | 40S ribosomal protein S4 (RPS4B) | AT5G07090.1 | e-114 | Translation | Protein metabolism |
| EE543924 | RPS7, RPS7.1; encodes a chloroplast ribosomal protein S7 | ATCG00900.1 | 1e-82 | Translation | Protein metabolism |
| EE542577 | 60S ribosomal protein L36 (RPL36B) | AT3G53740.4 | 7e-50 | Translation | Protein metabolism |
| EE550742 | RPS6 (RIBOSOMAL PROTEIN S6) | AT4G31700.1 | 4e-95 | Translation | Protein metabolism |
| CN737473 | RPS15 (RIBOSOMAL PROTEIN S15) | AT1G04270.1 | 3e-82 | Translation | Protein metabolism |
| CX270671 | RPL2.2; encodes a chloroplast ribosomal protein L2 | ATCG01310.1 | 1e-67 | Translation | Protein metabolism |
| CN726155 | 60S ribosomal protein L10A (RPL10aB) | AT2G27530.2 | e-102 | Translation | Protein metabolism |
| EE542556 | 60S ribosomal protein L4/L1 (RPL4A) | AT3G09630.1 | 5e-89 | Translation | Protein metabolism |
| CN737233 | 60S ribosomal protein L29 (RPL29B) | AT3G06680.1 | 7e-31 | Translation | Protein metabolism |
| EE551248 | RPL14; encodes a chloroplast ribosomal protein L14 | ATCG00780.1 | 4e-43 | Translation | Protein metabolism |
| EE543194 | CCB452; cytochrome c biogenesis orf452 | ATMG00180.1 | 5e-61 | Translation | Protein metabolism |
| DY009980 | RPL2.2; encodes a chloroplast ribosomal protein L2 | ATCG01310.1 | e-102 | Translation | Protein metabolism |
| CN731939 | 60S acidic ribosomal protein P1 | AT4G00810.2 | 2e-29 | Translational elongation | Protein metabolism |
| EE541984 | RPS11-BETA (putative ribosomal protein S11-beta) | AT5G23740.1 | 6e-75 | Translation | Protein metabolism |
| EE541830 | 60S ribosomal protein L10 (RPL10C) | AT1G66580.1 | e-125 | Translation | Protein metabolism |
| CN735269 | Eukaryotic translation initiation factor 5A, putative/eIF-5A, putative | AT1G26630.1 | 3e-70 | Translational initiation | Protein metabolism |
| EE569706 | ATP1; ATPase subunit 1 | ATMG01190.1 | e-142 | Response to oxidative stress | Response to stress |
| EE462567 | CRT1 (CALRETICULIN 1); calcium ion binding | AT1G56340.2 | e-132 | Response to oxidative stress | Response to stress |
| DY013565 | HSP70-1, HEAT SHOCK COGNATE 70 KDA PROTEIN 1 | AT5G02500.1 | e-116 | Response to cold | Response to stress |
| CN730192 | AT1G56075.1, LOS1 (low expression of osmotically responsive genes 1) | AT1G56070.1 | 7e-52 | Response to cold | Response to stress |
| EE439511 | AT1G56075.1, LOS1 (low expression of osmotically responsive genes 1) | AT1G56070.1 | e-144 | Response to cold | Response to stress |
| CN737456 | 60S ribosomal protein L26 (RPL26A) | AT3G49910.1 | 4e-58 | Response to cold | Response to stress |
| EE541625 | ATCYP1, ROC5 (ROTAMASE CYP 5) | AT4G34870.1 | 5e-68 | Aignal transduction | Signal transduction |
| EE541128 | Protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | AT5G38195.1 | 2e-32 | Lipid transport | Transport |
| EE541584 | LTP12 (LIPID TRANSFER PROTEIN 12) | AT3G51590.1 | 3e-51 | Lipid transport | Transport |
| DY003849 | 2S seed storage protein 1 | AT4G27140.1 | 1e-46 | Lipid transport | Transport |
| DY013108 | LTP2 (LIPID TRANSFER PROTEIN 2). | AT2G38530.1 | 2e-45 | Phospholipid transfer to membrane | Transport |
| CN735143 | LTP5 (LIPID TRANSFER PROTEIN 5) | AT3G51600.1 | 2e-52 | Lipid transport | Transport |
| EE543261 | Protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | AT1G55260.1 | 2e-73 | Lipid transport | Transport |
| CN728941 | AAC2 (ADP/ATP CARRIER 2) | AT5G13490.2 | 1e-67 | Transport | Transport |
| EE550070 | Unknown protein | AT1G49310.1 | 3e-23 | Biological process unknown | Unknown biological processes |
| EE541862 | Unknown protein | AT1G49310.1 | 9e-7 | Biological process unknown | Unknown biological processes |
| EE543031 | BURP domain-containing protein | AT1G49320.1 | e-101 | Biological process unknown | Unknown biological processes |
| EE548292 | Glycine-rich protein | AT3G24250.1 | 9e-8 | Biological process unknown | Unknown biological processes |
| CX271266 | BURP domain-containing protein | AT1G49320.1 | 5e-21 | Biological process unknown | Unknown biological processes |
| ES265407 | Unknown protein | AT1G75870.1 | 4e-29 | Biological process unknown | Unknown biological processes |
| EE549026 | Glycine-rich protein | AT2G30560.1 | 2e-10 | Biological process unknown | Unknown biological processes |
| EE542023 | UInknown protein | AT3G06090.1 | 2e-7 | Biological process unknown | Unknown biological processes |
| EE550161 | Similar to unknown protein (Arabidopsis thaliana) | AT1G49290.1 | 3e-9 | Biological process unknown | Unknown biological processes |
| EE548723 | No hits found | ||||
| EE569070 | No hits found | ||||
| EE569674 | No hits found | ||||
| CN735696 | No hits found | ||||
| CN730585 | No hits found | ||||
| EE544345 | No hits found | ||||
| CN726406 | No hits found | ||||
| DY011515 | No hits found | ||||
| EE569173 | No hits found |
Locus identifiers for the best gene match in Arabidopsis are based on BlastX against the TAIR7_pep database. Biological process is taken from the Gene Ontology (GO) annotation on TAIR for each locus identifier (http://www.arabidopsis.org/tools/bulk/go/index.jsp). GenBank accession numbers identify the longest EST sequence for each gene from the collection of ESTs (∼67 000 ESTs) examined to construct this cDNA array (see Xiang et al., 2008), and these are included in the GAL file as descriptors for each reporter (Brassica gene) on the microarray (http://www.brassicagenomics.ca/cdnaarray.html).
Genes listed in bold were identified previously, based on EST abundance, as up-regulated during early stages of microspore embryogenesis in B. napus Topas DH4079 (Malik et al., 2007).
Organellar-derived genes are underlined.
The genes that were up-regulated in the 7 d enlarged Westar microspores (down-regulated during embryogenesis in the DH-2 line) were mostly related to pollen function, for example late embryogenesis abundant domain-containing protein/LEA (At4g13560.1), pectinesterase family protein (At3g05610), polygalacturonase (At3g07840.1), and SNF7 family protein (At5g63880.1) (Table 3). The developmental expression profile of the closest match in Arabidopsis for each of the 40 up-regulated genes in Westar was determined using the electronic Fluorescent Protein (eFP) Browser (http://bbc.botany.utoronto.ca/) and Genevestigator Gene Atlas (Zimmermann et al., 2004) on the The Arabidopsis Information Resource database (http://www.arabidopsis.org). The results of this search showed that at least 19 of these genes are highly expressed in stamen and/or pollen (Table 3).
Table 3.
Genes up-regulated in 7 d microspore cultures of poorly embryogenic Westar
| Accession no. | Genes up-regulated in Westar (non-embryogenic) | Best match to Arabidopsis | E-value | Biological process | Broad functional category |
| ES264806 | Pectinesterase family protein | AT3G05610.1* | 2e-58 | Cell wall modification | Cell organization and biogenesis |
| ES264447 | UNE15 (unfertilized embryo sac 15) | AT4G13560.1 | 7e-29 | Double fertilization forming a zygote and endosperm | Developmental processes |
| ES264881 | Similar to BCP1 (Brassica campestris pollen protein 1) | AT3G26110.1* | 1e-15 | Pollen tube growth | Developmental processes |
| CN728581 | RIC5 (ROP-INTERACTIVE CRIB MOTIF-CONTAINING PROTEIN 5) | AT3G23380.1 | 4e-23 | Pollen tube growth | Developmental processes |
| ES265290 | UNE15 (unfertilized embryo sac 15) | AT4G13560.1* | 5e-43 | Double fertilization forming a zygote and endosperm | Other biological processes |
| CN728588 | NHL repeat-containing protein | AT5G14890.1* | 2e-84 | Double fertilization forming a zygote and endosperm | Other biological processes |
| ES264208 | Similar to unknown protein (Arabidopsis thaliana) | AT5G39870.1* | 2e-31 | Double fertilization forming a zygote and endosperm | Other biological processes |
| CN727705 | Family II extracellular lipase | AT1G20130.1* | 4e-75 | Lipid metabolic process | Other metabolic processes |
| CN728416 | ATBETAFRUCT4, VAC-INV | AT1G12240.1 | e-107 | Carbohydrate metabolic process | Other metabolic processes |
| ES264826 | GDSL-motif lipase | AT5G42160.1 | 2e-27 | Lipid metabolic process | Other metabolic processes |
| ES265298 | Exopolygalacturonase | AT3G14040.1* | 3e-43 | Carbohydrate metabolic process | Other metabolic processes |
| ES264781 | Exopolygalacturonase | AT3G14040.1* | e-101 | Carbohydrate metabolic process | Other metabolic processes |
| ES264609 | LCR1 (low-molecular-weight cysteine-rich 1) | AT5G48543.1 | 4e-9 | Carbohydrate metabolic process | Other metabolic processes |
| CN728494 | Polygalacturonase | AT3G07840.1* | e-110 | Carbohydrate metabolic process | Other metabolic processes |
| CN727751 | Polygalacturonase, putative | AT5G48140.1* | 1e-76 | Carbohydrate metabolic process | Other metabolic processes |
| CN727871 | Protein kinase family protein | AT3G01085.1 | 8e-76 | Protein amino acid phosphorylation | Protein metabolism |
| CN728491 | DNAJ heat shock N-terminal domain-containing protein | AT3G04980.1 | 2e-53 | Protein folding | Protein metabolism |
| CN728503 | Protein kinase family protein | AT3G01085.1 | 2e-86 | Protein amino acid phosphorylation | Protein metabolism |
| CN728527 | BTB/POZ domain-containing protein | AT4G08455.1 | 3e-78 | Transport | Transport |
| CN727745 | LCR11 (low-molecular-weight cysteine-rich 11) | AT4G11485.1 | 4e-7 | Transport | Transport |
| CN728424 | GAMMA-TIP3/TIP1;3 | AT4G01470.1* | e-117 | Transport | Transport |
| CN727761 | Amino acid permease | AT1G71680.1* | 9e-87 | Amino acid transport | Transport |
| CN728482 | Encodes a maternally expressed gene (MEG) family protein | AT2G16535.1 | 4e-17 | Amino acid transport | Transport |
| CN728496 | SNF7 family protein | AT5G63880.1* | e-101 | Protein transport | Transport |
| CN728126 | Encodes a defensin-like (DEFL) family protein | AT4G10603.1 | 6e-8 | Transport | Transport |
| CN728594 | Phytochrome kinase substrate-related | AT1G18810.1 | 6e-12 | Biological process unknown | Unknown biological processes |
| ES264229 | Invertase/pectin methylesterase inhibitor | AT3G17220.1* | 1e-63 | Biological process unknown | Unknown biological processes |
| CN728235 | Unknown protein | AT1G15415.1 | 7e-27 | Biological process unknown | Inknown biological processes |
| CN728176 | Similar to unknown protein (Arabidopsis thaliana) | AT3G28840.1* | 4e-34 | Biological process unknown | Inknown biological processes |
| CN728423 | Similar to unknown protein (Arabidopsis thaliana) | AT3G28790.1* | 2e-31 | Biological process unknown | Inknown biological processes |
| CN728566 | Similar to unknown protein (Arabidopsis thaliana) | AT3G28780.1* | 2e-63 | Biological process unknown | Unknown biological processes |
| CN728544 | Similar to unknown protein (Arabidopsis thaliana) | AT3G28790.1* | 1e-16 | Biological process unknown | Unknown biological processes |
| CN727786 | Similar to unknown protein (Arabidopsis thaliana) | AT3G28790.1* | 2e-41 | Biological process unknown | Unknown biological processes |
| ES264118 | No hits found | ||||
| ES265176 | No hits found | ||||
| CN728230 | No hits found | ||||
| ES264291 | No hits found | ||||
| CN728497 | No hits found | ||||
| ES264343 | No hits found | ||||
| CN728523 | No hits found |
Locus identifiers for the best gene match in Arabidopsis are based on BlastX against the TAIR7_pep database. Biological process is taken from the Gene Ontology (GO) annotation on TAIR for each locus identifier (http://www.arabidopsis.org/tools/bulk/go/index.jsp). GenBank accession numbers identify the longest EST sequence for each gene from the collection of ESTs (∼67 000 ESTs) examined to construct this cDNA array (see Xiang et al., 2008), and these are included in the GAL file as descriptors for each reporter (Brassica gene) on the microarray (http://www.brassicagenomics.ca/cdnaarray.html).
These genes are highly expressed in the pollen and/or stamen of Arabidopsis (electronic Fluorescent Protein Browser; http://bbc.botany.utoronto.ca/, Winter et al., 2007).
The data indicate that induced microspores of DH-2 show up-regulation of many genes related to protein biosynthesis, while the heat-treated non-dividing microspores of Westar instead express genes related to carbohydrate and cell wall metabolism, indicative of a continued pollen developmental pathway. Additionally, some of the differentially expressed genes in induced DH-2 microspores are organellar transcripts suggestive of increased growth and multiplication of the mitochondria and chloroplasts, while no organellar transcripts were included in the list of differentially expressed genes representing Westar (pollen-like) microspores (Tables 2 and 3).
Phenotypic differences between Westar and Westar-derived DH-2 plants
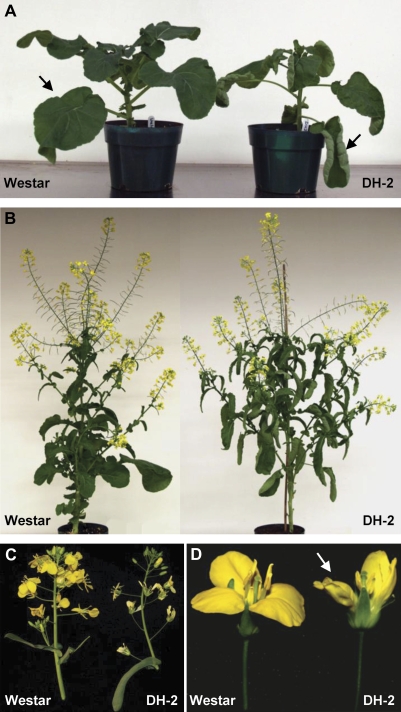
In addition to the differences in embryogenic potential between Westar and DH-2, there are some striking differences in general plant morphology between these two lines (Fig. 5). The leaves of the DH-2 line are epinastic compared with leaves of Westar (Fig. 5A), and lateral branches in DH-2 are longer and more advanced than those of Westar plants of the same age, giving DH-2 plants a much bushier appearance (Fig. 5B). The petals of DH-2 plants are a paler yellow than petals of Westar flowers, and two petals of the DH-2 flowers are curled, thus imparting an asymmetry to the flower (Fig. 5C, D). The petals of DH-2 do not abscise normally, and a majority of the mature and dried siliques present retained petals, now white in colour, at the time of harvesting (Fig 5B). In addition, the uppermost portions of the inflorescences of DH-2 are looser and more elongated than the normally compact inflorescences of Westar (Fig. 5C). These features impart a different architecture to the plants and inflorescences of Westar and DH-2 lines; however, there are no differences in seed set and pollen fertility between the two lines.
Fig. 5.
Differences in morphology and architecture between plants of B. napus cv. Westar and the Westar-derived DH-2 line. (A) Young plants. The arrows indicate differences in leaf expansion between the two lines. (B) Plants during flowering and silique development. (C) Inflorescences. (D) Flowers on the day of anthesis. The arrow indicates wrinkled petals of the DH-2 line.
Hormone profiles
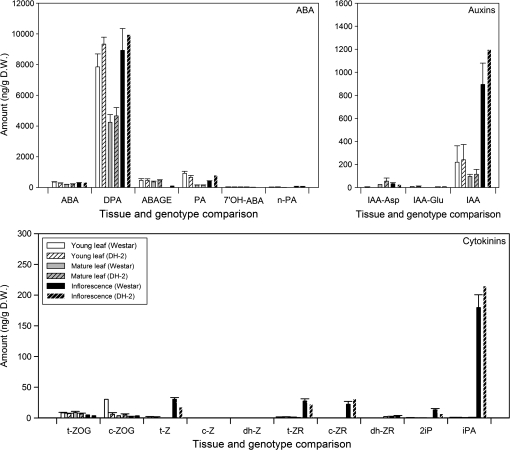
It was considered that the morphological differences between the Westar and Westar-derived DH-2 plants may be related to hormone levels and, therefore, ABA, auxins, and cytokinins were profiled in young and mature leaves and inflorescences of the two lines. Determination of ABA and its metabolites showed that the highest concentrations were of dihydrophaseic acid, an oxidation product of ABA, in all three tissues examined, but especially in young leaves and inflorescences of both lines (Fig. 6). There were no notable differences in the amounts of ABA or any of its intermediates between Westar and DH-2 in the tissues examined (Fig. 6). Isopentenyladenosine (iPA) was the most abundant cytokinin detected in any of the tissues, and showed the greatest accumulation in inflorescences of Westar and DH-2 (Fig. 6). Trans-zeatin (t-Z), trans-zeatin riboside (t-ZR), cis-zeatin riboside (c-ZR), and isopentenyladenine (2iP) were also present in higher amounts in the inflorescences as compared with the leaves, but at much lower concentrations than iPA (Fig. 6). IAA (indole-3-acetic acid) was present in high amounts in the inflorescences, at much lower levels in young leaves, and the least amount was found in mature leaves (Fig. 6). The auxin peptide conjugates, IAA-Asp (indole-3-aspartate) and IAA-Glu (indole-3-glutamate), were detected at low levels in the leaves and the inflorescences (Fig. 6). In summary, no notable differences in concentrations of any of these phytohormones were found in Westar and DH-2 tissues sampled that would account for the phenotypic differences and altered embryogenic response (Fig. 6). Methods are being developed that require less tissue in order to permit routine hormone profiling of microspores, developing embryos, and microspore-derived embryos.
Fig. 6.
Hormone profiling of metabolites and related compounds of ABA, auxin, and cytokinin in young and mature leaves and inflorescences (buds with 2–3 flowers) of B. napus cv. Westar and Westar-derived DH-2. Histograms indicate mean values (±SE) for each of the measured compounds, in three replicate tissue samples, each harvested from a different set of plants.
Discussion
The rescue of stable DH embryogenic lines from the rare embryos obtained from brassinosteroid-induced microspore cultures of the recalcitrant B. napus cultivar, Westar, has been described. The cultured haploid microspores bear the effects of the previous meiotic recombination, and thus collectively represent a population of all possible combinations of shuffled gene alleles prescribed within each parental genotype. Four of the rare embryos that developed from brassinosteroid-induced microspore cultures of cv. Westar were selected randomly to grow into F1 plants, and all of these showed greatly increased rates of microspore embryogenesis in culture (Table 1) and in the later generations, as compared with the originating Westar parent. Thus, there was a direct relationship between microspores that formed embryos in the initial EBR-induced cultures, and the ability to express this characteristic heritably in succeeding generations.
The Westar-derived DH lines might have resulted from related genetic (or perhaps non-genetic) heritable changes, such as common re-arrangements or phenotypic exposure of recessive gene alleles due to fixed homozygosity at all loci. Alternatively, gene mutations should be considered. Uppermost estimates of the spontaneous mutation rate in gene-coding non-neutral DNA in plants are at 0.1–0.9 mutations per haploid genome per generation (Johnston and Schoen, 1995; Drake et al., 1998; Schultz et al., 1999). Taking the average (0.5), one would expect that out of 1 000 000 microspores per plate, there could conceivably be a single nucleotide mutation in one gene (out of >50 000 genes) in up to half of the cultured microspores in each plate. The level could even be higher if the heat exposure induced mutations. Regardless of the mechanism, it seems reasonable to speculate that there might be a limited number of ways to overcome the block in embryogenesis genetically in cv. Westar. In addition, it seems likely that the newly acquired heritable embryogenic potential of the Westar-derived DH lines was fixed early during the first microspore culture of cv. Westar, and that it is this same capability for embryogenesis that facilitated the first rare embryogenic events in culture, and later underlies the heritable improvements in embryo production for each of the Westar-derived DH lines.
The mechanism(s) by which the applications of brassinosteroids increased rates of embryogenesis in the original microspore cultures are still unknown (Ferrie et al., 2005). Brassinosteroids have been shown to have several effects on tissue culture material, including stimulating cell elongation, cell division, ethylene production, adventitious tissue formation, and increased resistance to abiotic stress (Miyazawa et al., 2003; Mussig and Altmann, 2003; Hardtke et al., 2007; Kagale et al., 2007). Brassinosteroids have also been used to improve somatic embryogenesis in conifers (Pullman et al., 2003). Brassinosteroid additions were most effective in improving microspore embryogenesis in various Brassica species and cultivars when included in the initial media during the heat stress treatment, and were relatively ineffective when included in media after heat stress induction (A M R Ferrie et al., unpublished data). A lingering question is whether the brassinosteroid additions in some way caused, or affected, the heritable increase in embryogenic potential, or whether these compounds merely permitted the expression, or rescue, of those alterations.
Iterative modifications to tissue culture protocols are frequently used to improve embryogenic responses for microspores from poorly embryogenic or recalcitrant varieties, cultivars, or species; however, there have been relatively few investigations as to whether the resulting microspore-derived embryos (and perhaps newly represented genotypes) maintain the same capacity to form embryos in the next generation. In cases where microspore-derived embryos result from the acquisition of such a potential, this may be a significant way to obtain stable embryogenic lines from recalcitrant varieties. Previous reports on Medicago and interspecific crosses of Helianthus have shown that the regenerated explants from tissue culture acquired and retained the ability to respond to in vitro conditions and form somatic embryos in multiple subcultures (Nolan et al., 1989; Fambrini et al., 1997). In the case of Westar and the Westar-derived DH lines, 100% of the DH embryos tested (four of four) showed increased embryogenesis in the next generation as compared with Westar (Table 1).
Westar DH-2 also showed several striking differences in morphology and architecture as compared with the Westar parental cultivar. These included leaf form, axillary branching patterns, petal colour, flower symmetry, and petal abscision (Fig. 5). These abnormal phenotypes were not associated with altered concentrations of ABA, auxins, or cytokinins, or their related metabolites between Westar and DH-2 (Fig. 6). Another notable difference between the two lines was a persistent difficulty in the quantitative isolation of RNA from both microspores and early microspore-derived embryos of the DH-2 line as compared with cv. Westar and all other Brassica species and cultivars previously investigated (Malik et al., 2007). The RNA yields were up to five times less from microspores and microspore cultures of DH-2, especially from the 5 d and 7 d stages, as compared with the parent line (data not shown). There were no differences in the extractability of RNA from any other tissues of DH-2, and DNA isolation was normal in all tissues examined (data not shown).
Future studies will be necessary to determine whether any of these phenotypic characteristics between DH-2 and Westar reflect pleiotropic effects linked directly to the embryogenic potential, or instead result from additional linked genes and/or somaclonal variations induced by tissue culture. Earlier reports in B. napus have identified two multiple gene loci with direct effects on microspore embryogenic ability (Zhang and Takahata, 2001; Zhang et al., 2003). Loci with additive gene effects on in vitro regeneration systems have been reported for other species, Brassica oleracea (Holme et al., 2004), Arabidopsis thaliana (Schianteralli et al., 2001), Oryza sativa (Taguchi-Shiobara et al., 1997), Solanum lycopersicum (Koorneef et al., 1993), Zea mays (Armstrong et al., 1992), and Hordeum vulgare (Komatsuda et al., 1995). Genes with additive functions might account for changes in embryogenic potential as well as the accompanying phenotypic variability due to dosage effects. Similarly, chromosomal rearrangements occurring during meiosis, including translocation and homologous and homeologous recombination events, might alter gene and allelic complements in a qualitative or dosage-dependent manner, thus affecting downstream morphological and physiological outcomes. Chromosomal rearrangements during meiosis have been well documented in B. napus cultivars and lines, including cv. Westar (Osborn et al., 2003; Udall et al., 2005). Additionally, there are reports of other genetic- or epigenetically regulated variations caused by tissue culture (Kaeppler et al., 2000; Guo et al., 2007). Tissue culture-induced variations in barley, studied with the aid of amplified fragment length polymorphism (AFLP)-based approaches, for example, have been linked to the genotype of the donor plants, medium composition, and the length of time in culture (Bednarek et al., 2007, and references within). These phenotypic variations may be detrimental in micropropagation experiments, but can be exploited to good effect in other instances where they give rise to useful traits.
Microscopic observations have shown that microspores of Westar become enlarged and swollen during the first 3 d of heat stress treatment; however, only a few of the induced microspores subsequently undergo cell divisions, thus accounting for the poor embryogenic response in cv. Westar (Fig. 1). The observations suggest a block to further embryogenic development in enlarged microspores of Westar by the 5 d stage, because similar-appearing enlarged microspores of the DH-2 line show cell divisions at 5 d and continue on through embryo development (Fig. 1). Quantitative real-time RT-PCR data revealed expression of LEC1, LEC2, ABI3, BBM1, UP1, and WOX9 in both Westar and DH-2, but notably that there was restricted transcription of all of these genes in the Westar cultivar, with the exception of LEC1 (Fig. 2). Moreover, transcription of LEC1 in 1 d and 3 d Westar cultures was delayed/inhibited, as compared with DH-2 (Fig. 2). These results provide molecular confirmation that microspores of Westar have attained some level of embryogenic competence under inductive conditions; however, this was not sufficient to drive the Westar microspores through to commitment to embryogenesis. Additionally, in conjunction with the timely and optimal expression of embryogenesis-related genes in competent microspores, pollen-expressed genes are down-regulated in Westar-derived DH-2, but not in cv. Westar, during the progression through to embryogenesis (Fig. 3).
So far these experiments have not revealed the nature of the molecular impediments that prevent the progression to embryogenesis in the original Westar material. To et al. (2006) have shown that LEC1, LEC2, ABI3, and FUS3 are major interacting and redundant regulators of embryogenesis and seed development, and each of these regulators is involved in embryo-specific transcriptional cascades affecting embryo identity and storage product accumulation (Wang et al., 2007). Ectopic overexpression of LEC1, LEC2, or BBM1 is sufficient to induce embryogenesis in somatic tissues (Lotan et al., 1998; Stone et al., 2001; Boutilier et al., 2002), and recent studies have further demonstrated that LEC1, LEC2, and FUS3 are required for zygotic and somatic embryogenesis in Arabidopsis (Gaj et al., 2005). LEC1 and LEC2 up-regulate the expression of ABI3 and FUS3 (Kagaya et al., 2005; To et al., 2006; Wang et al., 2007), while the regulon for LEC2 includes storage protein genes (Braybrook et al., 2006). The factors initially responsible for the transcriptional activation of LEC1, LEC2, and ABI3 during heat stress-induced microspore embryogenesis in B. napus are not known, although there are some data to implicate alkalinization, calcium signalling, and GTPase regulation (Pauls et al., 2006; Chan and Pauls, 2007). More recently, a downstream involvement of auxin pools and carbohydrate metabolism in the embryogenic response, correlated with the expression of some of these transcription factors, has emerged (Casson and Lindsey, 2006). Nonetheless, there is still little information on the role of LEC1 and LEC2 (and BBM1) in potentiating an environment conducive to the induction of an embryogenic programme, or the upstream events triggering the initial transcription of these embryogenesis-related genes.
Transcript profiling using expressed sequence tag (EST) frequencies from cDNA libraries and microarray analyses have provided considerable gene expression data for various developmental stages of microspore embryogenesis in B. napus (Joosen et al., 2007; Malik et al., 2007; Tsuwamoto et al., 2007; Xiang et al., 2008). In the present case, microarray comparisons of Westar and Westar-derived DH-2 have provided an opportunity to examine contrasting embryogenic responses of genetically related material cultured under identical conditions. The lists of differentially expressed genes compiled for 7 d DH-2 and Westar material clearly indicate there has been a major shift in metabolism in the DH-2 tissues during commitment to embryogenesis, involving increased translation and protein biosynthesis (Tables 2 and 3). The genes identified through the microarray comparison of Westar and DH-2 include some of the genes previously identified by EST profiling (Malik et al., 2007) and/or microarray analyses of highly embryogenic B. napus Topas DH4079 (Xiang et al., 2008), as well as many genes previously not listed or annotated (see Tables 2 and 3). These latter include mitochondrial- and chloroplast-derived genes, annotated embryo-defective genes, and several genes implicated in calcium responses.
In contrast, genes up-regulated in Westar include many pollen-related genes, thus underscoring the poorly embryogenic characteristics of this material (Table 3). Transcriptomic and proteomic profiling data for developing and mature pollen of Arabidopsis have shown a functional skew towards cell wall metabolism, carbohydrate metabolism, and cell structure (see Grennan, 2007, and references within). Surprisingly, in those same studies, genes related to protein biosynthesis were not detected in the pollen transcriptome, although their products were found in the proteome (Honys and Twell, 2004; Holmes-Davis et al., 2005), thereby indicating that some pollen proteins were formed early during pollen development, possibly in the uninucleate microspores which are enriched in genes related to protein biosynthesis (Malik et al., 2007).
It is well established that plants possess more plasticity in their genomes than most animal cells, thus allowing reprogramming of differentiated cells and expression of totipotency/pluripotency (Gutierrez, 2005; Costa and Shaw, 2007). Little is known about the details of genomic reprogramming in plant cells, but protein complexes including the Polycomb group of proteins (PcG) may be involved in the maintenance of silenced states and cellular memory. It is possible that the ability to aquire embryogenic potential or a totipotent state depends on the capacity of plant cells to modify gene expression in response to some external cues. Kinases, for example SERK (Schmidt et al., 1997), may be involved in the upstream perception of external stimuli, and transcription factors such as LEC1, LEC2, and BBM1 may act downstream to confer embryogenic potential (Lotan et al., 1998; Stone et al., 2001; Boutilier et al., 2002); however, the details of how these genes are de-repressed and the nature of their interactions to permit the expression of embryogenic potential are still not clear. Recent research progress on the stem cell potential of somatic animal tissues has taken a giant leap forward, and surged ahead of some studies in plant systems, with recent discoveries and demonstrations that expression of cassettes of four genes can transform skin cells into embryonic stem cells (Takahashi et al., 2007; Yu et al., 2007). Advances in our understanding of the cellular conditions associated with embryogenic competence and improved protocols for inducing embryogenic potential in a wide selection of plant cells will require further detailed studies of cell biology, proteomics, and metabolomics, in addition to transcript profiling. The development of genetically related lines differing in embryogenic responses, for example Westar and Westar-derived DH-2, provides an ideal opportunity for in-depth molecular studies of embryogenic potential and embryo development in plants.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Accession numbers for embryo- and pollen-specific marker genes.
Supplementary Material
Acknowledgments
This work was supported by the Genome Prairie program ‘Enhancing Canola Through Genomics’ through Genome Canada, a not-for-profit corporation that is leading a national strategy on genomics with $560 million in funding from the Government of Canada. We also acknowledge support from the NRC Genomics and Health Initiative II for FW and an NSERC Strategic Grant (STPGP 258143-02) to JEK. The hormone profiling studies were carried out by the Hormone Profiling Team at NRC-PBI (Sue Abrams, Irina Zaharia, Vera Cekic, Steve Ambrose; http://pbi-ibp.nrc-cnrc.gc.ca/en/research/planthormoneprofiling.htm). The authors are grateful for the critical reviews of this manuscript by Don Palmer (NRC-PBI) and Carrie-Ann Whittle (NRC-PBI). This is a National Research Council of Canada publication (NRCC #50100).
References
- Armstrong CL, Romero-Severson J, Hodges TK. Improved tissue culture response of an elite maize inbred through backcross breeding, and identification of chromosomal regions important for regeneration by RFLP analysis. Theoretical and Applied Genetics. 1992;54:755–762. doi: 10.1007/BF00224181. [DOI] [PubMed] [Google Scholar]
- Bednarek PT, Orlowska R, Koebner R, Zimny J. Quantification of the tissue-culture induced variation in barley (Hordeum vulgare L.) BMC Plant Biology. 2007;7:10. doi: 10.1186/1471-2229-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutilier K, Offringa R, Sharma VK, et al. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. The Plant Cell. 2002;14:1737–1749. doi: 10.1105/tpc.001941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braybrook SA, Stone SL, Park S, Bui AQ, Le BH, Fischer RL, Goldberg RB, Harada JJ. Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proceedings of the National Academy of Sciences, USA. 2006;103:3468–3473. doi: 10.1073/pnas.0511331103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson SA, Lindsey K. The turnip mutant of Arabidopsis reveals that LEAFY COTYLEDON1 expression mediates the effects of auxin and sugars to promote embryonic cell identity. Plant Physiology. 2006;142:526–541. doi: 10.1104/pp.106.080895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoza V, Stewart CN. Brassica biotechnology: progress in cellular and molecular biology. In Vitro and Cellular Developmental Biology–Plant. 2004;40:542–551. [Google Scholar]
- Chan J, Pauls KP. Brassica napus Rop GTPases and their expression in microspore cultures. Planta. 2007;225:469–484. doi: 10.1007/s00425-006-0362-5. [DOI] [PubMed] [Google Scholar]
- Chiwocha SD, Cutler AJ, Abrams SR, Ambrose SJ, Yang J, Ross ARS, Kermode AR. The etr1-2 mutation in Arabidopsis thaliana affects the abscisic acid, auxin, cytokinin and gibberellin metabolic pathways during maintenance of seed dormancy, moist-chilling and germination. The Plant Journal. 2005;42:35–38. doi: 10.1111/j.1365-313X.2005.02359.x. [DOI] [PubMed] [Google Scholar]
- Costa S, Shaw P. ‘Open minded’ cells: how cells can change fate. Trends in Cell Biology. 2007;17:101–106. doi: 10.1016/j.tcb.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Croser JS, Lulsdorf MM, Davies PA, Clarke HJ, Bayliss KL, Mallikarjuna N, Siddique KHM. Toward doubled haploid production in the Fabaceae: progress, constraints, and opportunities. Critical Reviews in Plant Sciences. 2006;25:139–157. [Google Scholar]
- Datla RSS, Hammerlindl JK, Pelcher LE, Crosby WL, Selvaraj G. A bifunctional fusion between beta-glucuronidase and neomycin phosphotransferase: a broad-spectrum marker enzyme for plants. Gene. 1991;101:239–246. doi: 10.1016/0378-1119(91)90417-a. [DOI] [PubMed] [Google Scholar]
- DeBlock M, DeBrouwer D, Tenning P. Transformation of Brassica napus and Brassica oleracea using Agrobacterium tumefaciens and the expression of the bar and neo genes in the transgenic plants. Plant Physiology. 1989;91:694–701. doi: 10.1104/pp.91.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JW, Charlesworth B, Charlesworth D, Crow JE. Rates of spontaneous mutation. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fambrini M, Cionini G, Pugliesi C. Acquisition of high embryogenic potential in regenerated plants of Helianthus annuus×H. tuberosus. Plant Cell, Tissue and Organ Culture. 1997;51:103–110. [Google Scholar]
- Ferrie AMR. Microspore culture of Brassica species. In: Maluszynski M, Kasha KJ, Forster BP, Szarejko I, editors. Doubled haploid production in crop plants. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2003. pp. 195–204. [Google Scholar]
- Ferrie AMR, Dirpaul J, Krishna P, Krochko J, Keller WA. Effects of brassinosteroids on microspore embryogenesis in Brassica species. In Vitro Cell and Developmental Biology–Plant. 2005;41:742–745. [Google Scholar]
- Ferrie AMR, Keller WA. Microspore culture for haploid plant production. In: Gamborg OL, Philips GC, editors. Plant cell, tissue and organ culture: fundamental methods. Berlin: Springer; 1995. pp. 155–164. [Google Scholar]
- Ferrie AMR, Keller WA. Optimization of methods for using polyethylene glycol as a non-permeating osmoticum for the induction of microspore embryogenesis in the Brassicaceae. In Vitro Cellular and Developmental Biology–Plant. 2007;43:348–355. [Google Scholar]
- Forster BP, Heberle-Bors E, Kasha KJ, Touraev A. The resurgence of haploids in higher plants. Trends in Plant Science. 2007;12:368–375. doi: 10.1016/j.tplants.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Forster BP, Thomas WTB. Doubled haploids in genetics and plant breeding. Plant Breeding Reviews. 2005;25:57–88. [Google Scholar]
- Fukuoka H, Ogawa T, Matsuoka M, Ohkawa Y, Yano H. Direct gene delivery into isolated microspores of rapeseed (Brassica napus L.) and the production of fertile transgenic plants. Plant Cell Reports. 1998;17:323–328. doi: 10.1007/s002990050401. [DOI] [PubMed] [Google Scholar]
- Gaj MD, Zhang S, Harada JJ, Lemaux PG. Leafy cotyledon genes are essential for induction of somatic embryogenesis of Arabidopsis. Planta. 2005;222:977–988. doi: 10.1007/s00425-005-0041-y. [DOI] [PubMed] [Google Scholar]
- Grennan AK. An analysis of the Arabidopsis pollen transcriptome. Plant Physiology. 2007;145:3–4. doi: 10.1104/pp.104.900237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo WL, Wu R, Zhang YF, Liu XM, Wang HY, Gong L, Zhang ZH, Liu B. Tissue culture-induced locus-specific alteration in DNA methylation and its correlation with genetic variation in Codonopsis lanceolata Benth. et Hook. F. Plant Cell Reports. 2007;26:1297–1307. doi: 10.1007/s00299-007-0320-0. [DOI] [PubMed] [Google Scholar]
- Gutierrez C. Coupling cell proliferation and development in plants. Nature Cell Biology. 2005;7:535–541. doi: 10.1038/ncb0605-535. [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Dorcey E, Osmont KS, Sibout R. Phytohormone collaboration: zooming in on auxin–brassinosteroid interactions. Trends in Cell Biology. 2007;17:485–492. doi: 10.1016/j.tcb.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Holme IB, Torp AM, Hansen LN, Andersen SB. Quantitative trait loci affecting plant regeneration from protoplasts of Brassica oleracea. Theoretical and Applied Genetics. 2004;108:1513–1520. doi: 10.1007/s00122-003-1570-z. [DOI] [PubMed] [Google Scholar]
- Holmes-Davis R, Tanaka CK, Vensel WH, Hurkman WJ, McCormick S. Proteome mapping of mature pollen of Arabidopsis thaliana. Proteomics. 2005;5:4864–4884. doi: 10.1002/pmic.200402011. [DOI] [PubMed] [Google Scholar]
- Honys D, Twell D. Transcriptome analysis of the haploid male gametophyte development in Arabidopsis. Genome Biology. 2004;5:R85. doi: 10.1186/gb-2004-5-11-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Burgess SM, Hirsch D. β-Glucuronidase from Escherchia coli as a gene-fusion marker. Proceedings of the National Academy of Sciences, USA. 1986;83:8447–8451. doi: 10.1073/pnas.83.22.8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston MO, Schoen DJ. Mutation rates and dominance levels of genes affecting total fitness in two angiosperm species. Science. 1995;267:226–229. doi: 10.1126/science.267.5195.226. [DOI] [PubMed] [Google Scholar]
- Joosen R, Cordewener J, Supena EDJ, et al. Combined transcriptome and proteome analysis identifies pathways and markers associated with the establishment of rapeseed microspore-derived embryo development. Plant Physiology. 2007;144:155–172. doi: 10.1104/pp.107.098723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeppler SM, Kaeppler HF, Rhee Y. Epigenetic aspects of somaclonal variation in plants. Plant Molecular Biology. 2000;43:179–188. doi: 10.1023/a:1006423110134. [DOI] [PubMed] [Google Scholar]
- Kagale S, Uday KD, Krochko JE, Keller WA, Krishna P. Brassinosteroid confers tolerance in Arabidopsis thaliana and Brassica napus to a range of abiotic stresses. Planta. 2007;225:353–364. doi: 10.1007/s00425-006-0361-6. [DOI] [PubMed] [Google Scholar]
- Kagaya Y, Toyoshima R, Okuda R, Usui H, Yamamoto A, Hattori T. LEAFY COTYLEDON1 controls the seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3. Plant Cell Physiology. 2005;46:399–406. doi: 10.1093/pcp/pci048. [DOI] [PubMed] [Google Scholar]
- Kim YW, Moon HK. Enhancement of somatic embryogenesis and plant regeneration in Japanese larch (Larix leptolepis) Plant Cell, Tissue and Organ Culture. 2007;88:241–245. [Google Scholar]
- Komatsuda T, Taguchi-Shiobara F, Oka S, Takaiwa F, Annaka T, Jacobsen HJ. Transfer and mapping of the shoot-differentiation locus Shd1 in barley chromosome 2. Genome. 1995;38:1009–1014. doi: 10.1139/g95-133. [DOI] [PubMed] [Google Scholar]
- Koorneef M, Bade J, Hanhart C, Horsman K, Schel J, Soppe W, Verkerk R, Zabel P. Characterization and mapping of a gene controlling shoot regeneration in tomato. The Plant Journal. 1993;3:131–141. [Google Scholar]
- Kopecky D, Lukaszewski AJ, Gibeault V. Reduction of ploidy level by androgenesis in intergeneric Lolium–Festuca hybrids for turf grass breeding. Crop Science. 2005;45:274–281. [Google Scholar]
- Li H, Devaux P. Enhancement of microspore culture efficiency of recalcitrant barley genotypes. Plant Cell Reports. 2001;20:475–481. [Google Scholar]
- Li H, Rimmer R, Yu M, Sharpe AG, Séguin-Swartz G, Lydiate D, Hegedus DD. Two Brassica napus polygalacturonase inhibitory protein genes are expressed at different levels in response to biotic and abiotic stresses. Planta. 2003;217:299–308. doi: 10.1007/s00425-003-0988-5. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lotan T, Ohto M, Yee KM, West MAL, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell. 1998;93:1195–1205. doi: 10.1016/s0092-8674(00)81463-4. [DOI] [PubMed] [Google Scholar]
- Malik MR, Wang F, Dirpaul JM, Zhou N, Polowick PL, Ferrie AMR, Krochko JE. Transcript profiling and identification of molecular markers for early microspore embryogenesis in Brassica napus. Plant Physiology. 2007;144:134–154. doi: 10.1104/pp.106.092932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa Y, Nakajima N, Abe T, Sakai A, Fujioka S, Kawano S, Kuroiwa T, Yoshida S. Activation of cell proliferation by brassinolide application in tobacco BY-2 cells: effects of brassinolide on cell multiplication, cell-cycle-related gene expression, and organellar DNA contents. Journal of Experimental Botany. 2003;54:2669–2678. doi: 10.1093/jxb/erg312. [DOI] [PubMed] [Google Scholar]
- Mussig C, Altmann T. Genomic brassinosteroid effects. Journal of Plant Growth Regulation. 2003;22:313–324. doi: 10.1007/s00344-003-0061-4. [DOI] [PubMed] [Google Scholar]
- Nair RB, Joy RW, Kurylo E, Shi X, Schnaider J, Datla RSS, Keller WA, Selvaraj G. Identification of a CYP84 family of cytochrome P450-dependent mono-oxygenase genes in Brassica napus and perturbation of their expression for engineering sinapine reduction in the seeds. Plant Physiology. 2000;123:1623–1634. doi: 10.1104/pp.123.4.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan KE, Rose RJ, Gorst JR. Regeneration of Medicago truncatula from tissue culture: increased somatic embryogenesis using explants from regenerated plants. Plant Cell Reports. 1989;8:278–281. doi: 10.1007/BF00274129. [DOI] [PubMed] [Google Scholar]
- Osborn TC, Butrulle DV, Sharpe AG, Pickering KJ, Parkin IAP, Parker JS, Lydiate DJ. Detection and effects of a homeologous reciprocal transposition in Brassica napus. Genetics. 2003;165:1569–1577. doi: 10.1093/genetics/165.3.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauls KP, Chan J, Woronuk G, Schulze D, Brazolot J. When microspores decide to become embryos—cellular and molecular changes. Canadian Journal of Botany. 2006;84:668–678. [Google Scholar]
- Pullman GS, Zhang Y, Phan BH. Brassinolide improves embryogenic tissue initiation in conifers and rice. Plant Cell Reports. 2003;22:96–104. doi: 10.1007/s00299-003-0674-x. [DOI] [PubMed] [Google Scholar]
- Saal LH, Troein C, Vallon-Christersson J, Gruvberger S, Borg A, Peterson C. BioArray Software Environment (BASE): a platform for comprehensive management and analysis of microarray data. Genome Biology. 2002;3:software0003.1–0003.6. doi: 10.1186/gb-2002-3-8-software0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schianteralli E, De la Pena A, Candela M. Use of recombinant inbred lines (RIL) to identify, locate and map major genes and quantitative trait loci involved with in vitro regeneration ability in Arabidopsis thaliana. Theoretical and Applied Genetics. 2001;102:335–341. [Google Scholar]
- Schmidt ED, Guzzo F, Toonen MA, de Vries SC. A leucine-rich repeat containing receptor-like kinase marks somatic plant cells competent to form embryos. Development. 1997;124:2049–2062. doi: 10.1242/dev.124.10.2049. [DOI] [PubMed] [Google Scholar]
- Schultz ST, Lynch M, Willis JH. Spontaneous deleterious mutation in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA. 1999;96:13393–11398. doi: 10.1073/pnas.96.20.11393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proceedings of the National Academy of Sciences, USA. 2001;98:11806–11811. doi: 10.1073/pnas.201413498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi-Shiobara F, Lin SY, Tanno K, Komatsuda T, Yano M, Sasaki T, Oka S. Mapping quantitative trait loci associated with regeneration ability of seed callus in rice, Oryza sativa L. Theoretical and Applied Genetics. 1997;95:828–833. [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Thomas WTB, Forster BP, Gertsson B. Doubled haploids in breeding. In: Maluszynski M, Kasha KJ, Forster BP, Szarejko I, editors. Doubled haploid production in crop plants. A manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2003. pp. 337–349. [Google Scholar]
- To A, Valon C, Savino G, Guilleminot J, Devic M, Giraudat J, Parcy F. A network of local and redundant gene regulation governs Arabidopsis seed maturation. The Plant Cell. 2006;18:1642–1651. doi: 10.1105/tpc.105.039925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuwamoto R, Fukuoka H, Takahata Y. Identification and characterization of genes expressed in early embryogenesis from microspores of Brassica napus. Planta. 2007;225:641–652. doi: 10.1007/s00425-006-0388-8. [DOI] [PubMed] [Google Scholar]
- Tusher V-G, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences, USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udall JA, Quijada PA, Osborn TC. Detection of chromosomal rearrangements derived from homeologous recombination in four mapping populations of Brassica napus L. Genetics. 2005;169:967–979. doi: 10.1534/genetics.104.033209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Guo J, Lambert KN, Lin Y. Developmental control of Arabidopsis seed oil biosynthesis. Planta. 2007;226:773–783. doi: 10.1007/s00425-007-0524-0. [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLOS ONE. 2007;2 doi: 10.1371/journal.pone.0000718. e718. doi:10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang D, Datla R, Li F, et al. Development of a Brassica seed cDNA microarray. Genome. 2008;51:236–242. doi: 10.1139/G07-115. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhang FL, Aoki S, Takahata Y. RAPD markers linked to microspore embryogenic ability in Brassica crops. Euphytica. 2003;131:207–213. [Google Scholar]
- Zhang FL, Takahata Y. Inheritance of microspore embryogenic ability in Brassica crops. Theoretical and Applied Genetics. 2001;103:254–258. [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiology. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Katavic V, Giblin ME, Barton DL, MacKenzie SL, Keller WA, Hu X, Taylor DC. Modification of seed oil content and acyl composition in the Brassicaceae by expression of a yeast sn-2 acyltransferase gene. The Plant Cell. 1997;9:909–923. doi: 10.1105/tpc.9.6.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.