Summary
Computational searches for DNA binding sites often utilize consensus sequences. These search models make assumptions that the frequency of a base pair in an alignment relates to the base pair’s importance in binding and presume that base pairs contribute independently to the overall interaction with the DNA binding protein. These two assumptions have generally been found to be accurate for DNA binding sites. However, these assumptions are often not satisfied for promoters, which are involved in additional steps in transcription initiation after RNA polymerase has bound to the DNA. To test these assumptions for the flagellar regulatory hierarchy, class 2 and class 3 flagellar promoters were randomly mutagenized in Salmonella. Important positions were then saturated for mutagenesis and compared to scores calculated from the consensus sequence. Double mutants were constructed to determine how mutations combined for each promoter type. Mutations in the binding site for FlhD4C2, the activator of class 2 promoters, better satisfied the assumptions for the binding model than did mutations in the class 3 promoter, which is recognized by the σ28 transcription factor. These in vivo results indicate that the activator sites within flagellar promoters can be modeled using simple assumptions but that the DNA sequences recognized by the flagellar sigma factor require more complex models.
Keywords: consensus sequence, gene expression, sigma 28, transcription
Introduction
Flagella provide a competitive advantage to many bacteria by enabling them to swim toward nutrients and away from harmful substances. About sixty genes in Salmonella and E. coli are involved in the construction, function, and regulation of flagella.1 Flagellar genes are activated in a specific order, which may limit the expression of flagellar proteins to when they can be incorporated into the flagellar structure.2,3 This coordination between expression and assembly is in part due to the organization of flagellar promoters into a transcriptional hierarchy of three classes.1 The single class 1 promoter integrates multiple signals concerning the metabolic state of the cell to decide when to transcribe the flhDC genes. Together with the housekeeping sigma factor (σ70), the FlhD4C2 activator complex recognizes and initiates transcription from the class 2 promoters. The class 2 promoters transcribe genes involved in building the flagellar motor, which anchors the flagellum to the membranes and peptidoglycan, and in assembling a flexible, external linker called the hook. One of the class 2 promoters transcribes the gene for the alternative sigma factor σ28, which recognizes flagellar class 3 promoters. Class 3 promoters express the genes for the long external filament, the motor force generators, and the chemosensory system. These late components can then be added onto or interact with the motor and the hook.1
Many class 2 and class 3 flagellar promoters have been characterized through the identification of their transcriptional start sites and by DNA footprint analysis.4–9 This experimental data has facilitated the alignment of flagellar promoters for building consensus sequences. Class 3 promoters have conserved −10 and −35 regions that are recognized by the flagellar sigma factor (σ28).5 At class 2 promoters, FlhD4C2 binds upstream of σ70 to a palindromic DNA site that is not well conserved but has been verified experimentally through footprinting experiments.4,9 FlhD4C2 is unable to activate transcription in strains lacking the C-terminal domain of the alpha subunit of RNA polymerase.10 This suggests that FlhD4C2 contributes to the initial binding of polymerase to class 2 promoters through a direct interaction between FlhD4C2 and the alpha subunit. While a crystal structure of FlhD4C2 has been solved, this structure does not include the DNA to which FlhD4C2 binds.11
Using consensus sequences, several groups have used simple matching5 or more complicated PSSM’s (position specific score matrix)12–14 to search for new flagellar promoters. As has been seen for other promoters and binding sites, these searches detect a large number of false positives.15 This high background is often a result of low information content in a binding site combined with a large DNA search space. However, the large number of false positives may also be due to the assumptions used in building the search models that are intended for simple DNA binding proteins.
A DNA binding site is frequently defined through an alignment of sequences from several closely related genomes, and the consensus sequence derived from this alignment is then used for genome-wide searches.15 Assumptions are usually made about the sequence alignment and the manner in which each nucleotide contributes to a binding site. While the sequences used in an alignment should have arisen independently and be experimentally verified, binding sites for the same operons from closely related species are often used to increase the sample size. Also, the frequency of each nucleotide in an alignment may not be directly related to its importance in the binding site. Stronger binding sites may show greater conservation of bases than weaker binding sites. Finally, it is frequently assumed that each base contributes to the binding site independently from all the others.16 Several groups have provided evidence that most of the contributions to a binding site from individual bases are independent,17–19 but this assumption may not be valid for all binding proteins.
In particular, these assumptions are not generally valid for promoters, which have additional roles to play after binding the DNA. Promoters go through multiple steps during transcription initiation including the binding of RNA polymerase, open complex formation, initiation of RNA synthesis, and promoter clearance. These different steps can be at odds with each other when determining the overall activity of a promoter.20 For example, a promoter that matches the consensus sequence may have low activity because strong binding of RNA polymerase can interfere with promoter clearance.21 Additionally, the strength of a promoter can be affected by poorly conserved DNA outside of the consensus sequences. For σ70-dependent promoters, less conserved sequences like the UP element,22,23 extended −10 element,24,25 discriminator region,26 and the initial transcribed sequence27,28 can affect promoter activity by 10- to 100-fold. Because of the complex kinetics and multiple factors involved in promoter activation, flagellar promoters may not be modeled well using the assumptions for simple DNA binding proteins. Here we test these assumptions for the activator protein of the class 2 flagellar promoters and the sigma factor for the class 3 flagellar promoters through extensive mutational analysis.
In this paper, we used genetics to dissect the consensus sequences for flagellar promoters. Four Salmonella promoters were randomly mutagenized to identify elements important for flagellar transcription. Seventeen positions in a class 3 promoter and 29 positions in a class 2 promoter were then saturated for mutation to all three possible base substitutions. To assess the accuracy of the consensus sequence for flagellar promoters, the activities of these mutants were compared to scores generated from sequence alignments. Additionally, pairs of mutations were constructed to determine how individual mutations combine to affect the overall activity of the promoter. This in vivo study revealed differences between an activator protein and a sigma factor in how transcription was affected by mutations in the promoter sequence. This mutagenesis data illustrates the suitability of these models for flagellar promoters.
Results
Promoter random mutagenesis
To identify bases in the flagellar promoters that are important for transcription, random mutations generated through error-prone PCR were introduced into the promoter regions for four flagellar operons (flgKL, flgMN, fliAZY, and fliDST) in Salmonella typhimurium. These four operons are transcribed from both class 2 and class 3 promoters. While two of the promoter regions that were mutagenized contained both the class 2 and class 3 promoters (fliAZY and fliDST), the two other promoter regions contained only class 3 promoters (flgKL and flgMN). The class 2 promoters for the flgKL and flgMN operons are located one or more genes upstream of the class 3 promoters and were not mutagenized. Fusion of the lac genes to these operons facilitated screening for mutants with altered transcriptional activity (Figure 1).
Figure 1.
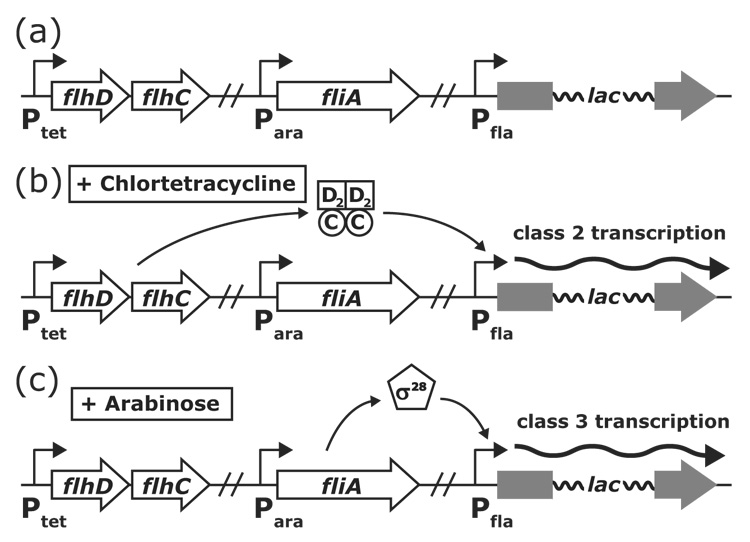
Reporter constructs for the isolation of class 2 and class 3 promoter mutants. (a) Class 2 transcription was assayed separately from class 3 transcription by putting FlhD4C2 and σ28 expression under the control of inducible promoters. (b) When autoclaved chlortetracycline was added, flhDC was expressed from its tetracycline-inducible promoter and resulted in activation of class 2 transcription at 87% of the wildtype level. (c) When arabinose was added, fliA (σ28) was expressed from its arabinose-inducible promoter and activated class 3 transcription at 162% of the wildtype level. Transcription was assayed using a lac-fusion inserted within the first or second genes of the flagellar operon.
For all four promoter regions mutagenized, a total of 151 changes in transcription were detected among the 6,500 colonies that were screened. Seventeen unique flgKL promoter mutants were detected out of 1,500 colonies screened. Twenty unique fliDST promoter mutants were found among 1,300 colonies screened. One flgMN promoter mutant was detected among 1,000 colonies screened. Forty-seven unique fliAZY promoter mutants were found among 2,700 colonies screened. Transitions (A:T <−> G:C) accounted for 76% of the substitution mutants isolated.
Altogether, these recombinants contained 85 unique mutations. Eighty were single base pair substitutions or deletions in the promoter region (Figure 2). Four colonies had multiple mutations, and one colony contained a 123 bp deletion. Most of the single mutations were located in regions of the promoter that matched the consensus sequence for either FlhD4C2 (19 mutations), σ70 (7 mutations), or σ28 (32 mutations). Fourteen mutations were also found near these conserved sequences. This localization of mutations within sequences that match the consensus is most striking in the fliAZY promoter where the class 2 and class 3 promoters overlap (Figure 2c). Moving along the DNA in the direction of transcription, mutations alternate between affecting class 2 transcription and class 3 transcription depending on which consensus sequence is matched in the DNA. Additionally, 10 mutations were in the untranslated region (UTR) of the downstream mRNA, and one mutation was 4 bp after the stop codon for the gene upstream of the flgKL promoter. The mutations in the UTRs and after the stop codon probably affect mRNA stability, introduce pause sites into the mRNA, or interfere with promoter escape. All mutations resulted in decreased transcription except for one mutation in the fliAZY FlhD4C2 binding site and two mutations in the fliDST 5’ UTR. Transcription was decreased by as much as 99% for any single substitution.
Figure 2.
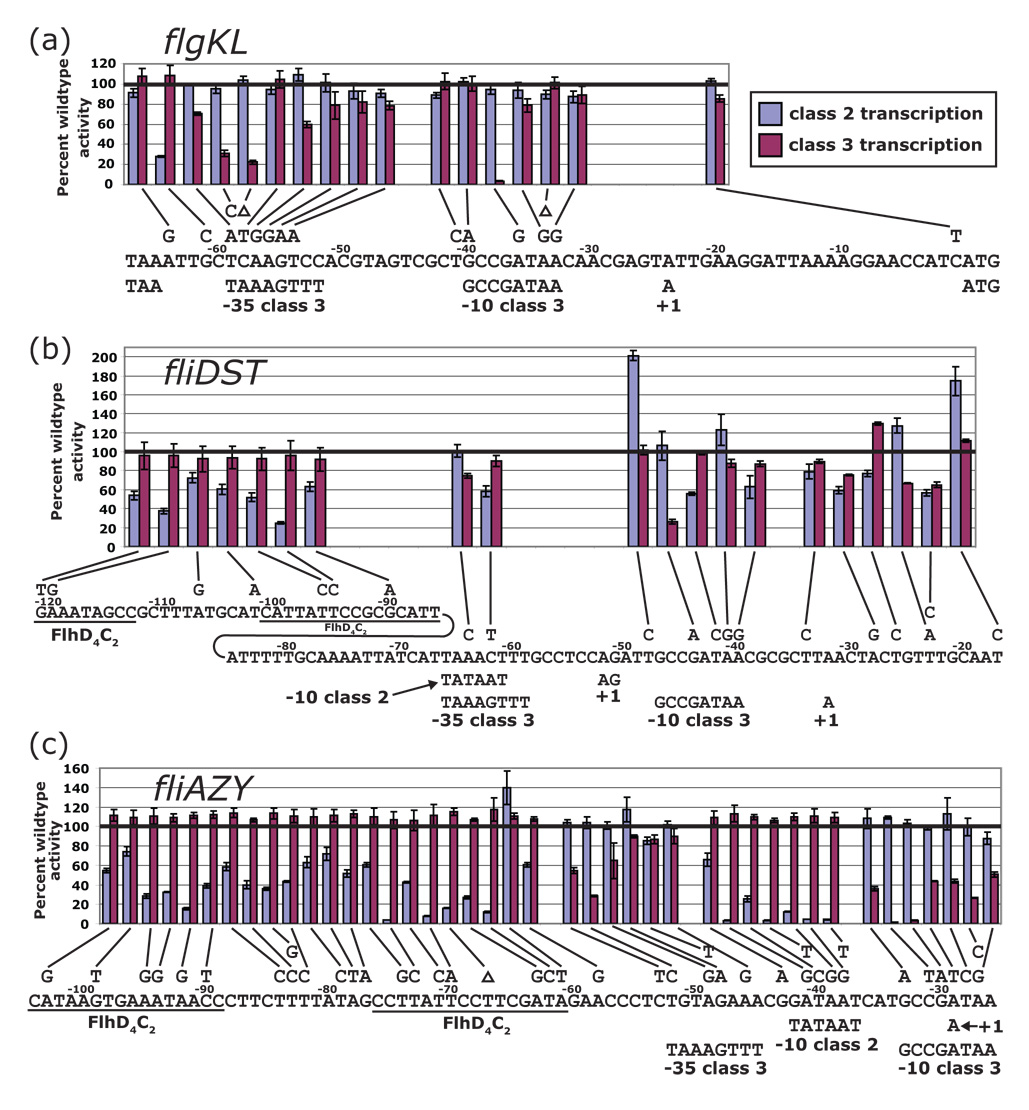
Locations of random mutations and their affects on class 2 and class 3 flagellar transcription. The flgKL (a), fliDST (b), and fliAZY (c) promoters were screened for mutations affecting class 2 or class 3 transcription. The single base pair mutants that were isolated are displayed above the promoter sequences, and transcriptional start sites47,48 and consensus sequences are below. FlhD4C2 binding sites are underlined. When either class 2 transcription (FlhD4C2) or class 3 transcription (σ28) was induced, the activities of lac-fusions to these mutant promoters were quantified and normalized to the wildtype promoter (100%). These β-galactosidase activities are connected by a line to their respective mutations. The numbers above the DNA indicate the distance from the start codon.
Of 1,000 colonies screened for mutations in the flgMN class 3 promoter, only one colony (a −65 C:T mutant, in the −10 region) showed decreased transcription. The rarity of mutations isolated at this promoter is most likely the result of the lac reporter being inserted in the gene for the anti-sigma factor FlgM. Without FlgM, class 3 transcription levels are very high, and small decreases in activity probably would be missed on the indicator plates used. Also, high levels of free σ28 could compensate for binding defects in the flgMN promoter to reduce the number of mutants affecting transcription.
Saturation mutagenesis of the flgKL class 3 promoter
The random mutagenesis demonstrated that most of the point mutations affecting transcription from the class 3 promoter are located in sequences matching the consensus. Therefore, primers were designed to construct all three possible nucleotide changes at each conserved position in the flgKL class 3 promoter. For eight conserved positions in the −35 region and nine conserved positions in the −10 region, 39 additional mutations were generated that primarily decreased class 3 transcription (Figure 3b).
Figure 3.
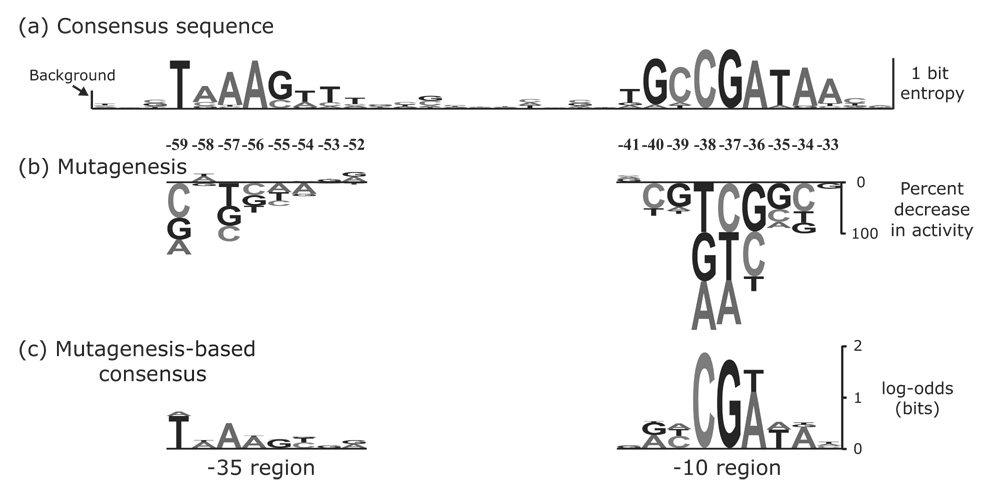
Comparison of the class 3 consensus sequence to the activities of flgKL promoter mutants.(a) The class 3 consensus sequence was constructed from the alignment of 17 flagellar promoters in Table 2 of Ide, et. al.5 The height of each WebLogo-like nucleotide stack49 corresponds to that position’s entropy,16 which is a measure of the conservation of nucleotides at that position. The frequency of each nucleotide in the alignment is reflected by the height of that nucleotide within the stack. On the left side of the figure, the height of the bar labeled background is twice the small sample correction as calculated by WebLogo.49 The small sample correction gives the average entropy value for a random alignment of sequences.50 (b) All single base pair mutations for eight positions in the −35 region and nine positions in the −10 region of the flgKL class 3 promoter were constructed. The height of each letter corresponds to the percent change in promoter activity as quantified by β-galactosidase assays. Letters above or below the line indicate an increase or decrease, respectively, in promoter activity. The numbers above the DNA indicate the distance from the start codon. (c) The measured activities in (b) were used as counts in a new “alignment” in order to generate a new consensus sequence. The log-odds scores displayed were calculated using these counts and are a measure of the conservation of nucleotides in the alignment.
The mutagenesis data reveals that some positions in the promoter are more important for activity than would be expected from the consensus sequence. The ‘CGA’ in the center of the −10 region (gcCGAtaa) seems to be most important for activity, even though other bases in the −10 and −35 regions are nearly as conserved (−59T, −56A, −40G, −39C, −35T, −34A). Mutations at −59T and −57A result in the largest decreases in transcription in the −35 hexamer (TaAagttt), despite similarly well-conserved bases at −56A and −55G. Several bases show some conservation but contribute very little to the activity of the promoter (−58A, −54T, −53T, −52T, −41T, and −33A).
Similar results were observed in Yu, et. al., where a chlamydial class 3 promoter was saturated for mutagenesis and assayed for transcriptional activity in vitro.29 While both their in vitro and our in vivo data assigned similar importance to positions within the class 3 promoter (e.g., the ‘CGA’ in the middle of the −10 hexamer), we were unable to directly correlate the data sets (data not shown). Changes in activity in our in vivo data were generally reflected by much larger changes in their in vitro data. These discrepancies probably resulted from a combination of studying different class 3 promoters that have spacers of different lengths under different conditions (in vitro vs. in vivo).
One explanation for this pattern is that σ28 is overexpressed in our reporter system. As demonstrated later, these high σ28 levels probably compensate for binding defects in the promoter. Bases involved in initial binding of σ28 would not be as important for activity with these high levels of σ28. Therefore, those bases that are more important for activity than expected from the consensus sequence (e.g., the ‘CGA’ in the middle of the −10 hexamer) are likely to be important for steps in transcription initiation after initial binding.
Insertions and deletions in the spacer for the flgKL class 3 promoter
In E. coli and Salmonella, the well-conserved octamers of the −10 and −35 regions in class 3 promoters are separated by an 11 bp spacer.5 However, in the in vitro study of Yu, et. al.,29 spacers of 11 bp or 12 bp worked equally well for class 3 transcription using E. coli σ28 and RNA polymerase. Also, a 10 bp spacer showed only a 40% decrease in promoter activity. To determine whether transcription in vivo shows the same tolerance for different spacer lengths, insertions or deletions were introduced into the spacer for the flgKL class 3 promoter (Figure 4). The pattern of transcription in vivo was nearly identical to the Yu, et. al. in vitro data.29 The only major difference was that the promoter with the 10 bp spacer was not transcribed well in vivo but exhibited about 60% of wildtype activity in vitro. Given the nearly equal activities for class 3 promoters with 11 bp and 12 bp spacers, it is unclear why only 11 bp spacers are observed in Salmonella and E. coli.
Figure 4.

Insertions and deletions were constructed in the spacer of the flgKL class 3 promoter.β-galactosidase activities for these mutants were measured and normalized to the activity of the wildtype promoter (100%). These mutants were assayed in a background containing nearly wildtype levels of class 3 transcription (110% of wildtype).
Construction of a class 2 promoter consensus sequence
A consensus sequence for the palindromic FlhD4C2 binding site in the class 2 promoter was constructed to help determine which positions were likely to be important for transcription. Using the FlhD4C2 consensus sequence from Stafford, et. al. as an initial search pattern,13 promoter regions from Salmonella, E. coli, and Proteus that have experimental evidence for containing a class 2 promoter were searched. To increase the sample size, promoter regions for flagellar genes in related organisms (Erwinia, Photorhabdus, Shigella, and Yersinia) were also searched. Both halves of the palindromic binding site for each of the 43 matches were aligned to construct a consensus sequence. As has been observed before,6 the FlhD4C2 binding site is not as well conserved as the σ28 binding site (Figure 3a and Figure 5a). This conservation can be quantified by determining how much information or nonrandomness is present in the alignment. Based upon this entropy measurement, the σ28 site (12.4 bits entropy) contains 15 times more information than the FlhD4C2 half-site (8.5 bits entropy).
Figure 5.
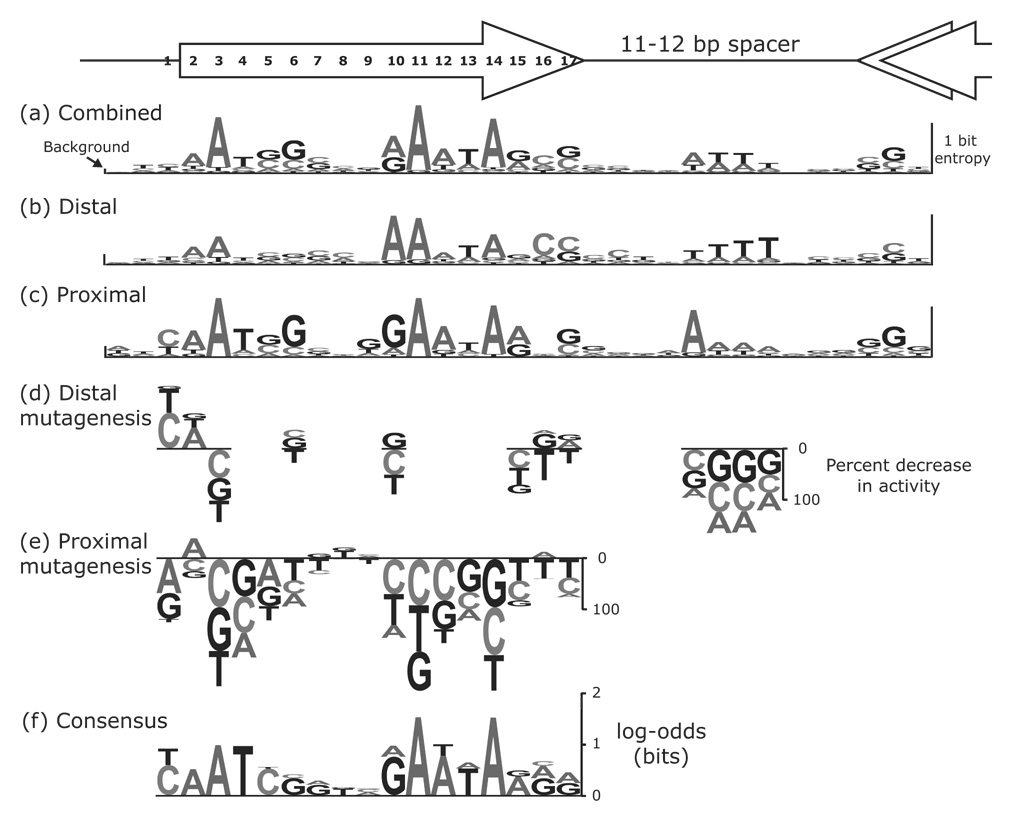
Comparison of the class 2 consensus sequence to the activities of fliAZY promoter mutants.Eighty-six proximal and distal FlhD4C2 sites were aligned to give the entropies in (a). The entropies for distal sites (b) or proximal sites (c) from an alignment of 30 “unidirectional” promoters are also displayed. The height of each WebLogo-like nucleotide stack49 corresponds to that position’s entropy,16 which is a measure of the conservation of nucleotides at that position. The frequency of each nucleotide in the alignment is reflected by the height of that nucleotide within the stack. On the left side of the figure, the height of the bar labeled background is twice the small sample correction as calculated by WebLogo.49 The small sample correction gives the average entropy value for a random alignment of sequences.50 All single base pair mutations at eight positions in the distal site (d), at four positions in the spacer (d), and at 17 positions in the proximal site (e) of the fliAZY promoter were constructed. The height of each letter corresponds to the percent change in promoter activity as quantified by β-galactosidase assays. Letters above or below the line indicate an increase or decrease, respectively, in promoter activity. (f) The measured activities for 17 positions in the distal and proximal sites were used as counts in order to generate a new consensus sequence. The activities for equivalent positions in the distal and proximal sites were combined, and the log-odds scores that are displayed were calculated using these counts. The log-odds scores are a measure of the conservation of nucleotides in the alignment.
In reality, the class 3 promoter likely contains much more entropy than reported here. Because each of the four nucleotides might not be represented at each position in a sequence alignment, “pseudocounts” are added into the regular counts of nucleotides at each position. Pseudocounts introduce flexibility into the model to correct for small sample sizes. Pseudocounts allow potential binding site matches to contain nucleotides that were not observed in the alignment. Because 17 experimentally verified promoters were used in the class 3 promoter alignment, the four pseudocounts that were added greatly reduced the entropy of the overall site. These four pseudocounts had a much smaller effect on the 86 half-sites in the FlhD4C2 alignment. If only one pseudocount had been added, the σ28 site (19 bits entropy) would contain twice the entropy or over 700 times more information than the FlhD4C2 half-site (9.5 bits entropy). FlhD4C2 likely recognizes class 2 promoters specifically by interacting with two of these less conserved DNA sites.
Since some of the 43 binding sites in the alignment are shared between divergent promoters (flgB-L and flgAMN, and fliE and fliF-K), an alignment including these binding sites might obscure directional information in the promoter. Therefore, the consensus sequence for the alignment of the “unidirectional” binding sites that activate a single promoter (30 sites) was constructed (Figure 5b,c). We refer to the half of the palindromic binding site closest to the −10 hexamer as the “proximal” site and the half-site farther upstream as the “distal” site. While the proximal and distal sites have much in common, several positions have different conserved bases (positions 1, 4, 6, 9, 10, 15, 16, and 17). Finally, several T’s are conserved in the center of the spacer when oriented to the distal site, whereas A’s are conserved at those same positions when looking at the proximal site.
Saturation mutagenesis of the fliAZY class 2 promoter
Most point mutations that affected class 2 transcription were either in the −10 hexamer, the sequences matching the FlhD4C2 binding site, or in the spacer between the FlhD4C2 proximal and distal sites. Since the −10 hexamer for σ70 has already been well characterized, we further investigated the class 2 promoter by saturating mutagenesis for the proximal FlhD4C2 site. The mutagenesis data for the proximal site tended to reflect the conservation of bases in the consensus sequence (Figure 5e). The most important base pairs in the proximal site were grouped into two segments (YaATCg---GAATAarr; where Y = C/T and R = A/G) that were separated by three base pairs that had little effect on activity when mutated. Although the structure of the FlhD4C2 complex was recently solved in the absence of its DNA binding site,11 the authors proposed a model where each DNA half-site made two major contacts with an FlhD dimer. These two contacts were separated by about 4 bp of noninteracting DNA, which is very similar to the 3 bp of nonessential DNA that we see here.
Five of the positions in the distal site that were predicted by the sequence alignments to be different from the corresponding positions in the proximal site were also saturated for mutagenesis. Three of these positions (10, 15, and 17) showed similar nucleotide preferences for both the proximal and distal sites when mutated (Figure 5d,e). At position 6, the distal site preferred a G/C and the proximal site preferred a G as predicted by the consensus sequences. At position 16, the distal site preferred a G and the proximal site preferred an A, which was not predicted by the consensus sequences. These differences could reflect a different requirement for binding where FlhD4C2 contacts RNA polymerase. This mutagenesis data also suggests that most of the differences in the consensus sequences were not significant, and it is not surprising that these poorly conserved consensus sequences were not entirely reliable.
Additionally, another group has suggested that the −35 hexamer for σ70 (TTGACA) overlaps the outermost three bases in FlhD4C2’s proximal site (TTG and TTA on the nontemplate strand).13 Therefore, we saturated mutagenesis for these positions in both the proximal and distal sites. If these three positions in the proximal site are important for interacting with σ70, then mutating these same bases in the upstream distal site should not affect promoter activity. The mutagenesis revealed that both sites prefer TTG and TTA (Figure 5d,e), suggesting that these nucleotides are conserved to allow binding of FlhD4C2. However, the fliAZY promoter that was mutagenized might not utilize this potential −35 hexamer due to a short 14 bp spacer for σ70.
Finally, the sequence alignments indicated that the center of the spacer between the FlhD4C2 proximal and distal sites is enriched for T’s. Individual mutations to A, C, or G at these positions reduced the activity of the promoter, which suggests that there is a preference for T’s (Figure 5d). Since a run of A’s or T’s can put a bend in the DNA,30 these four T’s may be needed for DNA curvature and not for protein interactions. To test this, these four T’s were replaced with four A’s, and this new fliAZY promoter initiated transcription at near wildtype levels (Figure 6). In the model for FlhD4C2 interaction with its binding site,11 a bend is located in this spacer, which agrees with our data suggesting a preference for a run of A’s or T’s.
Figure 6.

Insertions, deletions, and multiple substitutions were constructed in the FlhD4C2 spacer of the fliAZY promoter. β-galactosidase activities for these mutants were measured and normalized to the activity of the wildtype promoter (100%).
Insertions and deletions in FlhD4C2’s spacer
To determine which spacers are compatible with a functional FlhD4C2 binding site, insertions and deletions were constructed in the 12 bp spacer of the fliAZY promoter (Figure 6). The promoter does well with an 11 bp spacer and tolerates some 10 bp spacers. A 13 bp spacer, however, does not function well. This mutagenesis data somewhat agrees with our sequence alignments, where the FlhD4C2 spacer is predicted to be 11 or 12 bp in length. Additionally, DNA encompassing one turn of the DNA helix (10 or 11 bp) from either the flgB or fliE spacer was inserted into the fliAZY spacer. These mutants showed low transcriptional activity (Figure 6). This suggests that the FlhD4C2 sites need to be close to each other and not merely positioned on the proper face of the DNA.
Comparison of log-odds scores to the activities of mutated promoters
The changes in promoter activity due to mutations were compared to the corresponding changes in log-odds scores calculated from the sequence alignments (Figure 7). Log-odds scores assigned to individual nucleotides are derived from the frequency of a given nucleotide within an alignment. The odds that a nucleotide at a certain position is not there by chance is the “odds” in the log-odds score. For example, there are 2:1 odds if the nucleotide is present at twice the normal frequency. The unit for a log-odds score is the bit since the logarithm (base 2) of the “odds” is used. A change in a log-odds score for a binding site is often assumed to directly correspond to the change in binding energy or the change in promoter activity.15
Figure 7.
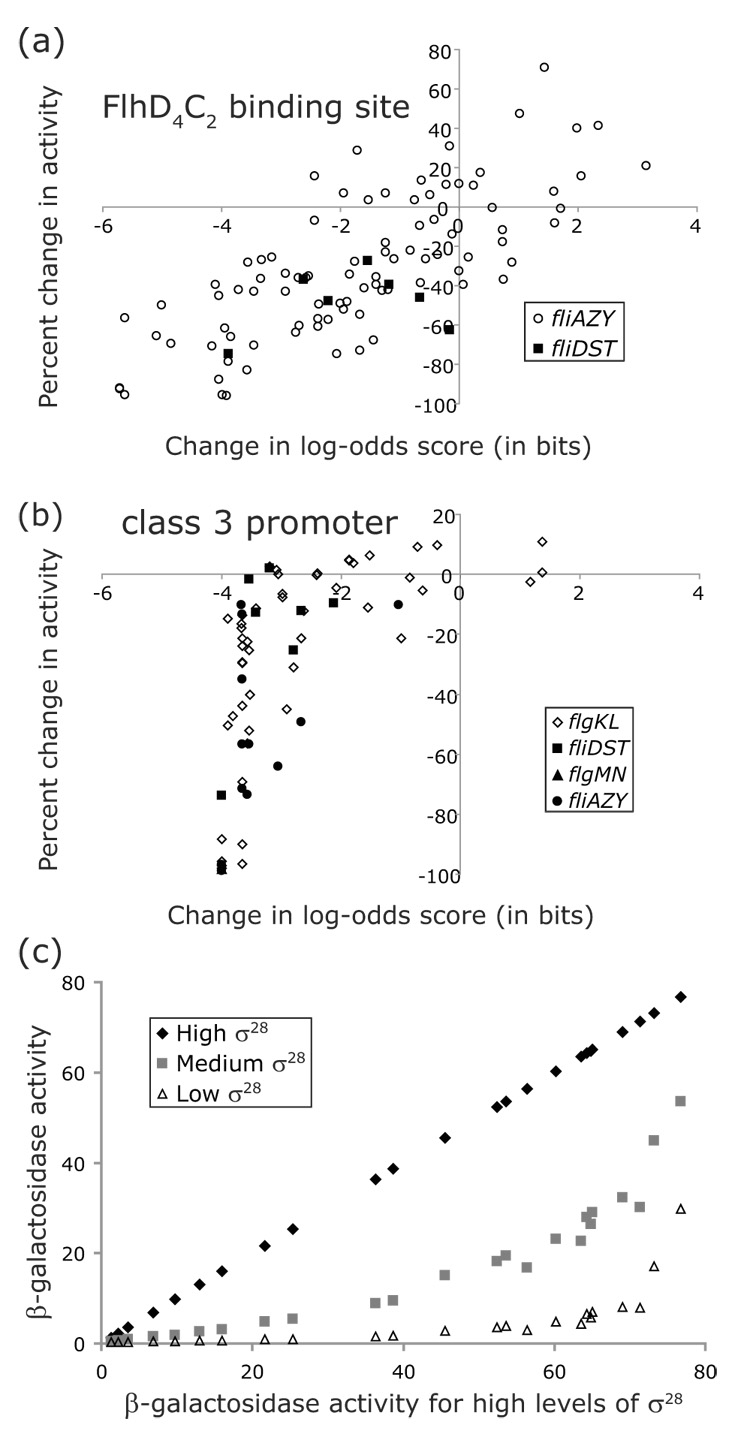
Comparison of log-odds scores to the activities of mutated promoters. For mutations in the FlhD4C2 binding sites (a) or the class 3 promoters (b), the change in log-odds scores calculated from the consensus sequence was plotted against the observed change in promoter activity. (c) The activities of a subset of the class 3 flgKL promoter mutants were assayed in backgrounds expressing high (162% of wildtype), medium (110% of wildtype), or low (64% of wildtype) levels of class 3 transcription. The activities of the mutants in these three backgrounds were plotted against their activity in the high class 3 transcription background. The β-galactosidase activities in (c) are expressed in units of nmol/min/OD650/ml.
To test this assumption for the FlhD4C2 binding site and the class 3 promoter, the changes in promoter activities due to mutations were plotted against the changes in log-odds scores (Figure 7a,b). While there was an overall linear relationship between activity and log-odds score for the mutations in the FlhD4C2 binding site, there was a wide range of observed activities for each score (correlation coefficient of r = 0.74). Therefore, the log-odds score for the FlhD4C2 binding site would likely be an adequate predictor for transcription from class 2 promoters. In contrast, there was a nonlinear relationship between activities and log-odds scores for the class 3 promoter. Almost half the mutations resulted in activities that were within 15% of the wildtype promoter activity but had a wide range of log-odds scores. Most of the remaining mutations had log-odds scores between −3 and −4 but had a wide range of activities.
An explanation for this pattern is that σ28 was overexpressed in our strains and these high σ28 levels compensated for small binding defects in the promoter. These same σ28 levels may not be high enough to compensate for larger binding defects. To determine whether overexpression of σ28 was responsible for this pattern, we reduced σ28 levels by mutating the arabinose promoter that transcribes σ28 in our reporter system. As judged by a transcriptional fusion to the class 3 motAB promoter, the base changes introduced in the arabinose promoter were able to reduce class 3 transcription from 162% of wildtype levels in the overexpression strain to either 110% or 64% of the wildtype level. A subset of our class 3 flgKL promoter mutants were transferred to these backgrounds that expressed reduced levels of σ28 (Figure 7c). If the initial binding of σ28 at the flgKL promoter was not a rate-limiting step in transcription initiation, the activities of all the mutants should have been reduced by the same percentage when σ28 levels were reduced. Since mutants with intermediate levels of class 3 transcription (e.g., 50% of wildtype) decreased their activity the most when σ28 levels were reduced, the pattern in Figure 6b is likely an artifact of overexpressing σ28. The correlation coefficient for the medium level of class 3 transcription (r = 0.77 for 15 mutants) revealed a somewhat more linear relationship between activity and log-odds score than under the original high levels of class 3 transcription (r = 0.60 for 71 mutants). These results underscore the importance of using physiologically relevant levels of transcription factors when examining promoters.
Interestingly, only a 62% increase in class 3 transcription levels was enough to compensate for binding defects in the flgKL promoter. σ28 may normally be expressed inside the cell at levels where it is sensitive to small changes in binding affinity in potential class 3 promoters. The binding affinity of the class 3 promoters may be optimized so that the level of σ28 in the cell increases and decreases past the steepest part of a Michaelis-Menton curve to produce the largest changes in promoter activity.
Comparison of predicted and observed activities for double mutants
Another assumption frequently made in computational models is that individual base pairs exert independent effects on the overall ability of a protein to bind DNA.15 When all activities are expressed as a fraction of the activity of the wildtype promoter and two mutations are combined into one binding site, the activity of the double mutant is expected to equal the activity of the first mutant multiplied by the activity of the second mutant. To test this assumption for flagellar promoters, pairs of mutations were combined for certain positions within the promoters. To minimize the work and cost involved, double mutants were constructed for selected pairs of positions within the binding sites. These pairs were chosen in order to sample many positions and a large range of predicted activities. For the FlhD4C2 binding site in the fliAZY promoter, mutations for positions within the proximal site and for positions between the distal and proximal sites were combined (Figure 8a). The activities of the double mutants match well with the predicted activities. For the σ28-dependent promoter for the flgKL operon, mutations for positions within the −10 region, within the −35 region, and between the −10 and −35 regions were combined (Figure 8b). The activities of these double mutants do not directly match with the predicted activities. In most cases for the class 3 promoter, when two mutations of decreased activity were combined, the activity of the resulting double mutant was lower than what was predicted.
Figure 8.
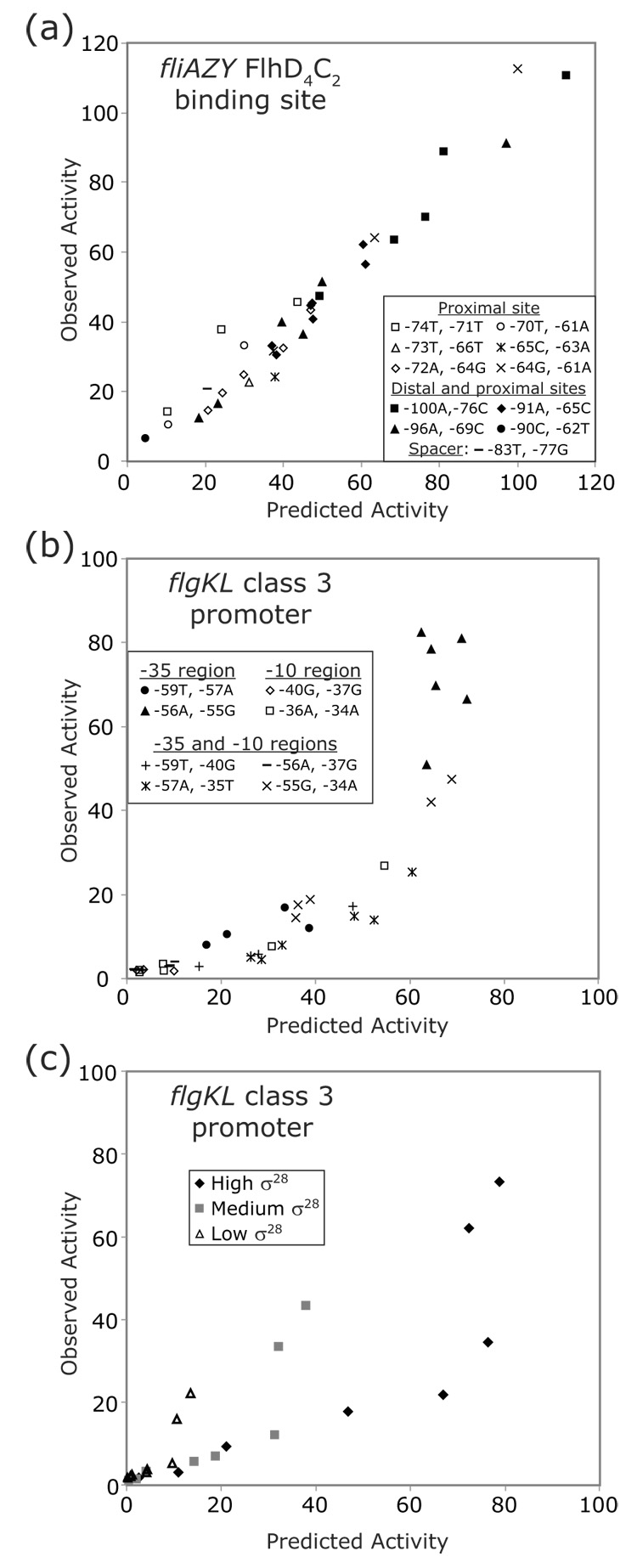
Comparison of the predicted and observed activities for double mutants. The predicted activity for a double mutant is equal to the activity of the first mutant multiplied by the activity of the second mutant. The observed activity is the quantified β-galactosidase activity of the double mutant. All these activities are normalized to the wildtype promoter. The wildtype nucleotides for the mutated positions are listed in the keys. (a) Double mutants within the proximal site, in both the distal and proximal sites, and within the spacer for the fliAZY FlhD4C2 binding site were constructed. (b) Double mutants within the −35 region, within the −10 region, and in both the −35 and −10 regions for the flgKL class 3 promoter were constructed. (c) Eight of the class 3 double mutants were assayed in backgrounds expressing high (162% of wildtype), medium (110% of wildtype), or low (64% of wildtype) levels of class 3 transcription.
Once again, this nonlinear pattern could be the result of high σ28 levels compensating for small defects in binding to the promoter but not compensating for large defects in binding (i.e., the double mutant). Therefore, eight of the double mutants were moved into the backgrounds with reduced σ28 levels (Figure 8c). Since the defects in transcription for the single mutants became more pronounced when σ28 levels were reduced, it is not surprising that the predicted activities of the double mutants (lower activity #1 × lower activity #2) decreased when σ28 levels were reduced. However, the same pattern of expression for the double mutants under high σ28 levels is repeated when there are medium or low levels of σ28. For all three σ28 levels, the predicted activity matches the observed activity for the two most highly expressed double mutants and perhaps a few of the lowest expressed mutants. For the other double mutants, the observed activity is about 40% of the predicted activity for all three σ28 levels. Therefore, the pattern for the activity of the double mutants is not an artifact of overexpressing σ28. The repetition of the pattern for a range of σ28 levels indicates that steps in transcription initiation after binding are being affected.
Introduction of elements of other class 3 promoters into the flgKL promoter
To determine how well alignment data could predict the activity of real class 3 promoters, elements of the tar, motA, cheV, and aer promoters were introduced into the flgKL class 3 promoter. Since the alignment data showed significant conservation in only the −10 and −35 regions, these elements were first substituted into the flgKL promoter (Figure 9, gray circles). As the modified promoters increasingly matched the consensus sequence, the activities generally increased. The exception to this trend is that the introduced −10 and −35 regions of the motA promoter increased activity more than the similarly scored wildtype flgKL promoter and the higher scoring tar promoter. To determine how sequences that are not conserved affect class 3 transcription, the intergenic region upstream of the transcription start site of the flgKL promoter was replaced by sequences from other class 3 promoters comprising the DNA upstream of the transcription start site through 25 bp upstream of the −35 region (Figure 9, black circles).The additional sequences increased the activity from promoters containing the less-conserved aer and cheV −10 and −35 regions and reduced activity from promoters containing the better-conserved motA and tar −10 and −35 regions. For the four promoters examined, these nonconserved sequences had a significant moderating effect on overall promoter activity. While the data for these modified promoters did not reveal any strong relationship between conservation and activity, the two less-conserved promoters did exhibit lower activities than the two better-conserved promoters.
Figure 9.
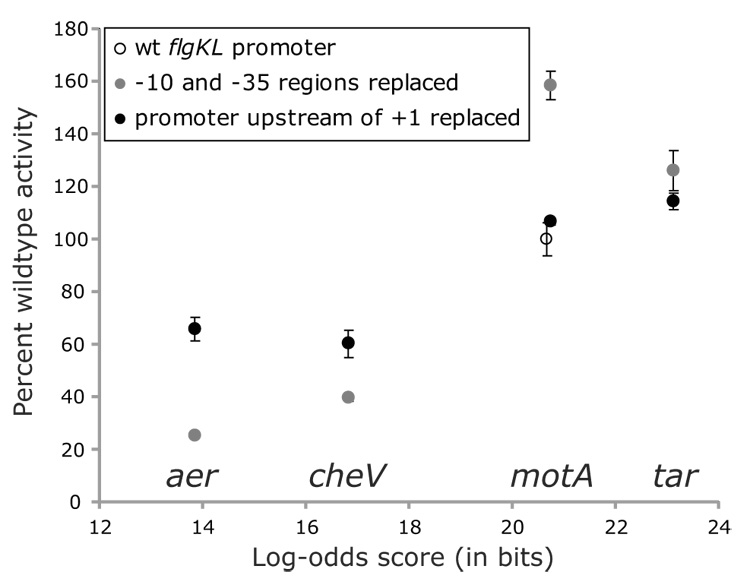
Effect of replacing elements of the flgKL class 3 promoter with elements of other class 3 promoters. For each of four class 3 promoters, the −10 and −35 regions alone were substituted into the flgKL class 3 promoter (gray circles). Also, the sequence upstream of the transcription start site through 25 bp upstream of the −35 region for each of the four promoters was used to replace the flgKL promoter upstream of the transcription start site (black circles). The activities of these hybrid promoters were plotted against the total log-odds score for their −10 and −35 regions. A high log-odds score indicates a close match to the consensus sequence. All activities are normalized to the level of transcription from the wildtype flgKL promoter (empty circle).
Discussion
The mutagenesis data in this paper illustrates some of the difficulties involved in predicting flagellar promoters within a genome sequence. While the location of most of the point mutations affecting transcription from the class 2 and class 3 promoters could be predicted from the consensus sequences, a few mutations were isolated near these conserved regions or in the UTR. These nonconserved regions played a significant role in determining the overall activity when substituted into a class 3 promoter. Additionally, sites for the activator FlhD4C2 (the class 2 promoter) better satisfied assumptions about protein binding than did σ28 sites (the class 3 promoter). The activities for mutations in an FlhD4C2 binding site correlated with the frequency of nucleotides in an alignment and combined independently with other mutations. The σ28 site did not satisfy either assumption as well. This may reflect the sensitivity of class 3 promoters to σ28 levels as well as the multiple roles that σ28 plays in transcription initiation (e.g., initial binding, open complex formation, and promoter clearance). This genetic data suggests that the binding sites for flagellar transcription factors and sigma factors should not be modeled the same way.
The saturation mutagenesis for the class 2 promoter revealed the nucleotide preferences for the poorly conserved FlhD4C2 binding site. This study systematically mutated each position in one half of the palindromic FlhD4C2 binding site. This saturation mutagenesis highlighted two segments separated by three nonessential base pairs in the half-site that were important for activity. In a model for FlhD4C2 binding, two contacts were formed between an FlhD dimer and each half-site in the DNA.11 Our data provides some support for this model since these two contacts formed with the DNA is similar to the two important segments identified by the mutagenesis data. Additionally, this model proposed a 110° bend in the spacer DNA that separates the two half-sites. Our mutagenesis data demonstrated that a run of A’s or T’s is preferred in the spacer. Since a run of A’s or T’s can put a bend in the DNA, this data provides some further support for the model. To definitively test this model, crosslinking experiments or mutagenesis of FlhD and FlhC would need to be performed.
The saturation mutagenesis for the class 3 promoter revealed that some base pairs were far more important for activity than would be expected from the consensus sequence. In particular, the ‘CGA’ in the middle of the −10 region was as well conserved as other bases in the alignments but had much larger effects on transcription when mutated. Similar results were obtained by in vitro transcription for a different class 3 promoter.29 However, when the amount of σ28 in our reporter system was decreased to better approximate the wildtype level, mutations that previously had little effect on class 3 transcription now exhibited greater defects in transcription. This indicated that overexpressing σ28 could compensate for small binding defects in the promoter. Therefore, the consensus sequence is likely to be a better approximation for activity than was revealed by our class 3 promoter mutagenesis data.
Surprisingly, this compensation for binding defects occurred when overexpressed σ28 increased transcription from wildtype class 3 promoters by only 62%. This suggests that σ28 is normally present in the cell at levels that are responsive to small changes in binding affinity at class 3 promoters. Conversely, when σ28 levels increase after the flagellar regulon is induced, initially low σ28 levels may turn on strong class 3 promoters first as proposed by Kalir, et. al.3 These strong promoters could express genes for adaptor proteins (FlgK, FlgL, and FliD) needed earlier in flagellar assembly than other class 3 expressed genes (e.g., the flagellin subunits).
The lack of normal flagellar regulation in our reporter strain might also contribute to the nonlinear relationship between activity and log-odds score for the class 3 promoter. By expressing σ28 from the arabinose promoter in the absence of class 1 and class 2 transcription, σ28 was always expressed despite the lack of both the motor and hook. Class 3 transcription normally occurs only after completion of these two structures. As a result, σ28 expression was not coordinated with flagellar assembly and lacked the normal negative feedback through FlgM that moderates its activity. The activity of flagellar promoters in the context of normal flagellar regulation might exhibit a better correlation to the log-odds score.
An assumption in binding models is that each base pair affects binding independently from other base pairs in the binding site. Double mutations in the FlhD4C2 binding site satisfied this assumption. For the class 3 promoter, most mutations did not combine independently. Most mutants had an observed activity that was 40% of what was predicted. This pattern held even when σ28 levels were reduced. The unusual behavior of σ28 could be due to the additional complexity of a sigma factor. Besides recognizing its binding site, sigma factors help RNA polymerase to melt the DNA strands, initiate RNA synthesis, and aid in promoter clearance.31 While initial binding of σ28 was revealed to be a critical stage for class 3 transcription in this study, the double mutant analysis demonstrated that other steps in transcription initiation contribute to the strength of a class 3 promoter. The ‘CGA’ in the −10 region that was so important for activity most likely plays a role in these later steps. Since mutating the ‘CGA’ nucleotides resulted in severe transcription defects even under high σ28 levels, these mutations probably affect steps after binding. In contrast, FlhD4C2 most likely activates transcription simply by binding upstream of RNA polymerase and contacting the alpha subunit.10 The extra roles that σ28 plays may alter the manner in which bases in the promoter contribute to transcription initiation.
Genome-wide searches for DNA binding sites tend to be difficult due to large numbers of false positives.15 This high background is often a result of low information content in a binding site combined with a large DNA search space. Also, not all proteins may satisfy assumptions that relate binding ability to the frequency of a base pair in an alignment and presume that base pairs contribute independently to the overall site. In this study, we demonstrated in vivo that an activator for the class 2 flagellar promoters satisfied these assumptions and that the alternate sigma factor for the class 3 promoters perhaps satisfied only one assumption. These results suggest that simple models for DNA binding can be applied to the activator proteins for promoters but are not as appropriate for the sigma factors. Allowing for this alternate behavior presents additional challenges in designing computational models for promoters. However, these insights may enable better recognition of DNA binding sites and promoters.
Materials and Methods
Bacterial strains
The strains used in this study were derived from Salmonella typhimurium strain LT2 and are listed in Table 1. Promoter mutants are listed in Supplemental Table S9.
Table 1.
Salmonella strains used in this study.
| Strain | Genotype | Referencea | |
|---|---|---|---|
| TH437 | LT2 | J. Roth | |
| TH2090 | fliG5101::MudJ | lab collection | |
| TH2142 | hisD9953::MudJ his-9944::MudI | Hughes and Roth29 | |
| TH2534 | flgA5211::MudJ | Gillen and Hughes33 | |
| TH2570 | flgC5215::MudJ | Gillen and Hughes33 | |
| TH2779 | flgM5222::MudJ | Gillen and Hughes33 | |
| TH2856 | fliA5059::Tn10dCm | Karlinsey, et. al.34 | |
| TH2945 | ataA::[P22 Kn9 PfliA(−600)-lacZYA-'9] | M. Chadsey and G. Chilcott | |
| TH3730 | PflhDC5451::Tn10dTc[Δ25] | Karlinsey, et. al.35 | |
| TH4212 | fliS5480::MudK | H. Bonifield | |
| TH4702 | pKD46 (λ-red recombinase plasmid, AmpR, temperature sensitive replication) | Datsenko and Wanner26 | |
| TH4721 | flgK5396::MudJ | Aldridge, et. al.36 | |
| TH5504 | ΔfliA5647::FRT | Aldridge, et. al.27 | |
| TH5794 | PfliD5744::Tn10dTc ΔflgHI958 fljB e,n,x vh2 | lab collection | |
| TH6701 | ΔaraBAD925::tetRA | P. Aldridge | |
| TH7023 | ΔIS200IV::cat(fli-5583) | lab collection | |
| TH7270 | flgJ5964::tetRA(inserted after stop codon) n | ||
| TH7395 | ΔflgM5628::FRT motA5461::MudJ ΔaraBAD923::flgM-FKF ParaB935 | Aldridge, et. al.27 | |
| TH7396 | ΔflgM5628::FRT motA5461::MudJ ΔaraBAD923::flgM-FKF ParaB936 | Aldridge, et. al.27 | |
| TH8006 | pKD46 / ΔaraBAD925::tetRA ΔfliA5647::FRT | ||
| TH8082 | PflhDC5451::Tn10dTc[Δ25] ΔaraBAD943::fliA ΔfliA5647::FRT | ||
| TH8083 | CRR4108[PflhDC5451::Tn10dTc[Δ25](TcS)] ΔaraBAD943::fliA ΔfliA5647::FRT | ||
| TH8091 | flgA6093::tetRA (inserted after stop codon) | ||
| TH8239 | ΔaraBAD943::fliA ΔfliA5647::FRT | ||
| TH8315 | pKD46 / CRR4108[PflhDC5451::Tn10dTc[ Δ25](TcS)] ΔaraBAD943::fliA ΔfliA5647::FRT flgK5396::MudJ flgJ5964::tetRA | ||
| TH8321 | pKD46 / CRR4108[PflhDC5451::Tn10dTc[Δ25](TcS)] ΔaraBAD943::fliA ΔfliA5647::FRT fliS5480::MudK fliD5744::Tn10dTc | ||
| TH8922 | fliA5890:Tn10dTc[ Δ25, IS10R(Δ2808–2894)] Δtar-flhD2039 fljBe,n,x vh2 | ||
| TH8925 | ΔaraBAD956::fliA ΔfliA5647::FRT | ||
| TH8926 | ΔaraBAD956::fliA ΔfliA5647::FRT PflhDC5451::Tn10dTc[Δ25] | ||
| TH8927 | ΔaraBAD956::fliA ΔfliA5647::FRT CRR4107[PflhDC5451::Tn10dTc[Δ25](TcS)] | ||
| TH8928 | ΔaraBAD956::fliA ΔfliA5647::FRT CRR4107[PflhDC5451::Tn10dTc[Δ25](TcS)] ataA::P22[sieA'-Kn6-PfliA(−583 to +1) - lacZYA'-'9] | ||
| TH8929 | ΔaraBAD956::fliA ΔfliA5647::FRT CRR4107[PflhDC5451::Tn10dTc[Δ25](TcS)] flgK5396::MudJ | ||
| TH8931 | ΔaraBAD956::fliA ΔfliA5647::FRT CRR4107[PflhDC5451::Tn10dTc[Δ25](TcS)] fliS5480::MudK | ||
| TH8933 | ΔaraBAD956::fliA ΔfliA5647::FRT CRR4107[PflhDC5451::Tn10dTc[Δ25](TcS)] flgM5222::MudJ | ||
| TH8936 | pKD46 / CRR4107[PflhDC5451::Tn10dTc[ Δ25](TcS)] ΔaraBAD956::fliA ΔfliA5647::FRT flgK5396::MudJ flgJ5964::tetRA | ||
| TH8937 | pKD46 / CRR4107[PflhDC5451::Tn10dTc[Δ25](TcS)] ΔaraBAD956::fliA ΔfliA5647::FRT fliS5480::MudK fliD5744::Tn10dTc | ||
| TH8938 | pKD46 / CRR4107[PflhDC5451::Tn10dTc[Δ25](TcS)] ΔaraBAD956::fliA ΔfliA5647::FRT flgM5222::MudJ flgA6093::tetRA | ||
| TH8939 | pKD46 / CRR4107[PflhDC5451::Tn10dTc[ Δ25](TcS)] ΔaraBAD956::fliA ΔfliA5647::FRT ataA::P22[sieA'-Kn6-PfliA(−583 to +1)-lacZYA'-'9] | ||
| TH9250 | fliA6399::tetRA (replaces −79 to −44 bp from GTG with tetRA) | ||
| TH9252 | pKD46 / CRR4107[PflhDC5451::Tn10dTc[ Δ25](TcS)] ΔaraBAD956::fliA ΔfliA5647::FRT ataA::P22[sieA'-Km6-PfliA(−583 to +1) 6400::tetRA-lacZYA'-'9] | ||
| TH10022 | Δfli-5583::cat(ΔIS200IV) fliA5890::Tn10dTc[ Δ25, IS10R(Δ2808–2894)] Δtar-flhD2039 fljBe,n,xvh2 | ||
| TH10032 | ΔaraBAD956::fliA fliZ6591::MudJ fliA5890::Tn10dTc[ Δ25, IS10R(Δ2808–2894)] | ||
| TH10049 | fliZ6591::MudJ | ||
| TH10132 | CRR4107[PflhDC5451::Tn10dTc[Δ25](TcS)] ΔaraBAD956::fliA fliZ6591::MudJ ΔfliA5647::FRT | ||
| TH10151 | pKD46 / CRR4107[PflhDC5451::Tn10dTc[Δ25](TcS)] ΔaraBAD956::fliA fliZ6591::MudJ ΔfliA5647::FRT fliA6399::tetRA | ||
| TH10826 | fliA6785::tetRA (inserted −43 bp from GTG) | ||
| TH10896 | pKD46 / CRR4107[PflhDC5451::Tn10dTc[Δ25](TcS)] ΔaraBAD956::fliA fliZ6591::MudJ ΔfliA5647::FRT fliA6785::tetRA | ||
| TH11808 | fliA7226::tetRA (inserted −79 bp from GTG) | ||
| TH12065 | pKD46 / CRR4107[PflhDC5451::Tn10dTc[Δ25](TcS)] ΔaraBAD956::fliA fliZ6591::MudJ ΔfliA5647::FRT fliA7226::tetRA | ||
| TH12359 | CRR4107[PflhDC5451::Tn10dTc[Δ25](TcS)] ΔaraBAD956::fliA ΔfliA5647::FRT motA5461::MudJ | ||
| TH12906 | Para998::tetRA ΔaraBAD956::fliA ΔfliA5647::FRT | ||
| TH12927 | CRR4107[PflhDC5451::Tn10dTc[Δ25](TcS)] ΔaraBAD956::fliA fliA5059::Tn10dCm flgC5215::MudJ | ||
| TH12930 | CRR4107[PflhDC5451::Tn10dTc[Δ25](TcS)] ΔaraBAD956::fliA fliA5059::Tn10dCm flgA5211::MudJ | ||
| TH12933 | CRR4107[PflhDC5451::Tn10dTc[Δ25](TcS)] ΔaraBAD956::fliA fliA5059::Tn10dCm fliG5101::MudJ | ||
| TH12990 | pKD46 / CRR4107[PflhDC5451::Tn10dTc[Δ25](TcS)] ΔaraBAD956::fliA Para998::tetRA ΔfliA5647::FRT motA5461::MudJ | ||
| TH13035 | CRR4107[PflhDC5451::Tn10dTc[Δ25](TcS)] ΔaraBAD956::fliA ParaB935(−85 A:T) ΔfliA5647::FRT motA5461::MudJ | ||
| TH13036 | CRR4107[PflhDC5451::Tn10dTc[Δ25](TcS)] ΔaraBAD956::fliA ParaB936(−41 T:C) ΔfliA5647::FRT motA5461::MudJ | ||
| TH13127 | CRR4107[PflhDC5451::Tn10dTc[Δ25](TcS)] ΔaraBAD956::fliA ParaB935 ΔfliA5647::FRT flgJ6310::tetRA | ||
| TH13128 | CRR4107[PflhDC5451::Tn10dTc[Δ25](TcS)] ΔaraBAD956::fliA ParaB936 ΔfliA5647::FRT flgJ6310::tetRA | ||
| TH13161 | CRR4107[PflhDC5451::Tn10dTc[Δ25](TcS)] ParaB936 ΔaraBAD956::fliA ΔfliA5647::FRT flgK5396::MudJ | ||
| TH13237 | CRR4107[PflhDC5451::Tn10dTc[Δ25](TcS)] ParaB935 ΔaraBAD956::fliA ΔfliA5647::FRT flgK5396::MudJ | ||
Strains given no reference were constructed for this study.
Media and general techniques
Cultures of bacterial strains and phage P22 lysates were prepared as described,32 except that X-gal was added to plates at a concentration of 40 µg/ml and LB (per liter: 10 g Bacto tryptone, 5 g Bacto yeast extract, 5 g NaCl) was used as a rich medium for growing bacteria. Chlortetracycline (50 µg/ml, autoclaved) or 0.2% arabinose (Ara) were used to induce transcription from the tetA and araBAD promoters, respectively. Tetracycline-sensitive (TetS) selections and transductions were performed as described,33,34 except that LB was used instead of NB. Primers were synthesized by Integrated DNA Technologies (Coralville, IA). PCR products were sequenced at the DNA Sequencing Facility (Department of Biochemistry) at the University of Washington or at the DNA Sequencing Core Facility at the University of Utah.
Using the λ-red recombination proteins expressed from plasmid pKD46, PCR products were recombined into the chromosome.35 Each 25 ml LB-Ampicillin-Ara culture was inoculated with a single colony and grown with shaking at 30°C until the culture reached an OD600 of 0.6. The cells were washed twice in cold, sterile water and resuspended to a final volume of 100 µl to 500 µl with water. An ethanol-precipitated PCR product was electroporated into 50 µl of cells, incubated at room temperature for 1 hour, and spread on selective media. Recombinants were selected at 37°C or above to ensure loss of the temperature-sensitive pKD46 plasmid.35
Overview of random mutagenesis of flagellar promoters
Random mutations were introduced into the promoter regions for the four flagellar operons of Salmonella that are transcribed from both class 2 and class 3 promoters. Random mutations in the promoter DNA sites were generated by amplifying the promoter regions using high concentrations of Taq polymerase (5U Promega Taq polymerase per 20 µl reaction)36. These PCR products were moved into the chromosome by using λ-red recombination to replace a TetR cassette in the promoter region. TetS recombinants were isolated on Tet-sensitive selective media. Individual colonies were purified by single colony isolation on LB-plates before being screened on lactose indicator media (LB-X-gal, MacConkey-lactose, and Triphenyl Tetrazolium Chloride (TTC)-lactose plates) for changes in transcription from lac fusion reporters inserted downstream of the mutagenized promoters.
To assay class 2 transcription independent of class 3 transcription, a strain that expresses the flhDC operon from a tetracycline-inducible promoter and fliA (σ28) from an arabinose-inducible promoter was constructed (Figure 1). When this strain (TH8927) is grown in the presence of chlortetracycline, flhDC is transcribed and class 2 promoters are activated. When the strain is grown in media containing arabinose, fliA (σ28) is transcribed and class 3 (σ28-dependent) promoters are activated. Mutants could immediately be screened for changes in class 2 transcription independent of changes in class 3 transcription by checking the Lac phenotype on indicator plates containing chlortetracycline or arabinose, respectively.
Construction of strains used in random mutagenesis
The ΔaraBAD956::fliA+ allele in TH8927 was constructed by amplifying the fliA coding sequence with primers araBfliAF and araDfliAR (primers are listed in Table 2) and then using λ-red recombination to replace the ΔaraBAD::tetRA in TH8006 with the amplified fliA+ DNA. The tetRA cassette encodes the tetracycline resistance determinants from transposons Tn10 and Tn10dTc. This ΔaraBAD956::fliA+ allele replaces the araB start codon through the araD stop codon with the start codon (ATG) through the stop codon of fliA. The PflhDC5451::Tn10dTc[ Δ25] allele from TH3730 was transduced into TH8925 to give TH8926. The PflhDC5451::Tn10dTc[Δ25] allele is an operon fusion of tetA to flhDC that is expressed from the tetA promoter. A spontaneous mutant of TH8926 that was no longer TetR but could induce class 2 transcription was selected for on Tet-sensitive plates (TH8927). This TetS mutant is presumably defective in the tetracycline resistance gene (tetA) but still transcribes the flhDC operon from PtetA.
Table 2.
Primers used in this study.
| Primer name | Sequence |
|---|---|
| ara5’ | cgtcttactccatccagaaaaacagg |
| araC+5R | ccatgatttctctacccc |
| flgA +417/+436 | cgtcagtttgcgcgatctcg |
| flgJ 5’ RT-PCR | gatcaccaccactgaatacg |
| flgK +100/+81 | aacccgcaacgttataattg |
| flgK +42/+23 R | ggcgttaagtccgctcatgg |
| flgK −105/−86 F | tcagcaaaacctacagcgcg |
| flgM +24/+5 R | aggtgaggtacggtcaatgc |
| flgM +61/+45 | tggtttcgcgcgtctgg |
| flgM5′-UP | gaaccgtcgattctgatg |
| fliA +27/+8 | accttcagcggtatacagtg |
| fliA +5/−15 | ttcacgataaacagccctgc |
| fliA +720/+697 | ctataacttacccagtttggtgcg |
| fliA −156/−137 | tctggctgattttattctgc |
| fliA −225/−209 | gcggctggtaagagagc |
| fliA#4 | tagtctatacgttgtgcggc |
| fliC+39R | caacagcgacaggctgtttg |
| fliC13 | gttctttgtcaggtctgtc |
| fliD +4/−16 | ccatgccttcttcctttttg |
| fliD −196/−177 F | gtaacccttgtatcggcacc |
| fliD+190R | aacgcggtatttgccgtc |
| araBfliA F | actgtttctccatacctgtttttctggatggagtaagacgatgaattcactgtataccgc |
| araDfliA R | ttcatcaacgcgccccccatgggacgcgtttttagaggcactataacttacccagtttgg |
| flgAtetR | aaccgtcgattctgatgggaatattcttattaacctataattaagacccactttcacatt |
| flgJtetR | cagcaaaacctacagcgcgaatctcgacaatctcttttaattaagacccactttcacatt |
| flgKtetA | cgttaagtccgctcatggcgtgattaatcaagctggacatctaagcacttgtctcctg |
| flgMtetA | tgaatatctcatcggcagccgcgacaaaatctttacacaactaagcacttgtctcctg |
| fliAPtetA | taaacagccctgcgttaaatgagttatcggcatgattatcctaagcacttgtctcctg |
| fliAPtetA2 | atccgtttctacagagggttctatcgaaggaataaggctactaagcacttgtctcctg |
| fliAPtetR | aaaaggcgctacaggttacataagtgaaataacccttcttttaagacccactttcacatt |
| fliAtetR2 | ttatagccttattccttcgatagaaccctctgtagaaacgttaagacccactttcacatt |
| ParaBtetR | gtcttactccatccagaaaaacaggtatggagaaacagtattaagacccactttcaca |
| ParaBtetA | gtccatatcgaccaggacgacagagcttccgtctccgcaactaagcacttgtctcctg |
MudJ and MudK vectors were used for the construction of lac transcriptional and translational fusions, respectively, to operons of interest.37 The lac reporter fusions used in this study were flgM5222::MudJ (TH2779), fliS5480::MudK (TH4212), flgK5396::MudJ (TH4721), and fliZ6591::MudJ (TH10049). The fliZ6591::MudJ insertion allele was isolated in this study and is described in a later section. The TetR markers used in the Tet-sensitive selections were PfliD5744::Tn10dTc (TH5794), flgJ5964::tetRA (TH7270), flgA6093::tetRA (TH8091), fliA6399::tetRA (TH9250), fliA6785::tetRA (TH10826), and fliA7226::tetRA (TH11808). In order to construct the tetRA cassette insertion alleles, primers flgJtetR and flgKtetA (TH7270), flgAtetR and flgMtetA (TH8091), fliAPtetR and fliAPtetA (TH9250), fliAtetR2 and fliAPtetA (TH10826), and fliAPtetR and fliAPtetA2 (TH11808) were used to amplify tetRA cassettes from TH5794. The λ-red system was used to recombine these PCR products into the chromosome, and TetR colonies were selected. These lac reporters and TetR markers were moved by transduction into TH8927.
Random mutagenesis of promoters
The flgMN class 3 promoter was mutagenized (replacing −132 to +24 bp relative to the flgM start codon) by recombining an error prone PCR product (primers flgM5’UP and flgM+24/+5R, 45°C annealing) into TH8938. The flgMN promoter regions of recombinants were sequenced using primer flgA+417/+436 and the amplified promoter (primers flgA+417/+436 and flgM+61/+45, 45°C annealing).
The flgKL class 3 promoter was mutagenized (replacing −105 to +42 bp relative to the flgK start codon) by recombining an error prone PCR product (primers flgK−105/−86F and flgK+42/+23R, 60°C annealing) into TH8315 or TH8936. The flgKL promoter region of recombinants was sequenced using primer flgJ5’RTPCR and the amplified promoter (primers flgJ5’RTPCR and flgK+100/+81, 45°C annealing).
The fliDST class 2 and class 3 promoter region was mutagenized (replacing −196 to +75 bp relative to the fliD start codon) by recombining an error prone PCR product (primers fliD−196/−177F and fliC13, 50°C annealing) into TH8321. Alternatively, the fliDST promoter was mutagenized (replacing −196 to +4 bp relative to the fliD start codon) by recombining an error prone PCR product (primers fliD−196/−177F and fliD+4/−16, 60°C annealing) into TH8937. The fliDST promoter regions of recombinants were sequenced using primer fliC+39R and the amplified promoter (primers fliD+190R and fliC+39R, 60°C annealing).
The fliAZY class 2 and class 3 promoter region was mutagenized (replacing −156 to +5 bp relative to the fliA start codon) by recombining an error prone PCR product (primers fliA+5/−15 and fliA−156/−137, 55°C annealing) into TH9252 or TH10151. The fliAZY promoter region of recombinants was sequenced using primer fliA+720/+697 and the amplified promoter (primers fliA#4 and fliA−225/−209, 60°C annealing).
Twelve flgKL and 12 fliDST promoter mutants were isolated using TH8315 and TH8321. It was then discovered that the ΔaraBAD943::fliA allele in these strains contained a mutation (P190S). These promoter mutants were then transduced to backgrounds containing wildtype fliA+ (ΔaraBAD956::fliA+). All transcriptional activities were quantified in this wildtype fliA+ background. No significant difference in Lac activity for these strains was observed between wildtype fliA and the fliA(P190S) allele.
Eleven fliAZY promoter mutants were isolated using TH9252, which contains a lac fusion to the fliAZY promoter on a P22 prophage. When β-galactosidase assays were attempted on these strains, the β-galactosidase activities disappeared quickly after the cells were lysed (~2 minute half-life). Therefore, the fliZ6591::MudJ was isolated (see below) and used for all additional screens of fliAZY promoter mutants. The 11 previously isolated mutations were amplified, moved into TH10151, and quantified.
Isolation of fliZ::MudJ
In order to detect changes in transcription from the fliAZY promoter, an insertion of a MudJ transcriptional reporter in this operon was isolated. Random MudJ insertions were generated38 in a strain containing the fliAZY operon transcribed from PtetA (TH10022). Eighty thousand colonies were pooled, and a phage stock was prepared on the pooled colonies. This pool of random insertions was transduced into TH8925 (ΔaraBAD::fliA ΔfliA::FRT) and spread onto LB-Tet-Kan-EGTA plates to select for transductions that had brought in the fliAZY region (selecting TetR) and nearby MudJ insertions (selecting for MudJ-encoded kanamycin resistance). Four hundred transductants were patched to X-gal, MacConkey-lactose, and TTC-lactose plates containing either arabinose, chlortetracycline, or no inducer. Five colonies exhibited a Lac+ phenotype on the chlortetracycline plates and were Lac− on the arabinose and no inducer plates. This pattern would be expected for an insertion in the PtetA-fliAZY operon but not in other class 3 transcribed genes. One out of the five insertions was located in fliZ by PCR. By sequencing into the right end, the MudJ was confirmed to be inserted after V38 in fliZ. In TH10049, this fliZ6591::MudJ was transduced into a wildtype background (TH437).
Construction of specific promoter mutations
Mutations in the fliAZY FlhD4C2 spacer (the DNA between the distal and proximal halves of the FlhD4C2 binding site) and for the saturation mutagenesis were generated using primers containing the desired mutations. These primers were designed with about 40 bp of homology to the promoter (for recombination), then the mutation, and about 20 bp of homology to the promoter (for amplification; ~55°C Tm). These primers were then used with an appropriate primer from the random mutagenesis experiment to amplify the promoter. λ-red recombination was used to move these mutations into the chromosome. When two or three mutations were desired at a certain position, a mixed base was used in the primer. After recombination, 16 or 32 colonies, respectively, were checked for Lac activity on lactose indicator plates, and representative colonies were sequenced. Double mutants were constructed by designing both mutations into a primer or by amplifying a mutant promoter with a primer containing a second mutation (Supplementary Tables S10 and S11).
Measuring the levels of class 2 and class 3 transcription in the reporter strains
Transcriptional fusions to class 2 and class 3 promoters were utilized to determine whether FlhD4C2 and σ28 were being overexpressed in the reporter strains. When FlhD4C2 was expressed from the tet promoter in the absence of σ28, lac fusions inserted in the class 2 expressed genes flgA flgC, and fliG exhibited transcription levels that were 71%, 92%, and 97% of the wildtype level, respectively. Averaging these three measurements, class 2 transcription was 87% of the wildtype level in the reporter strains. When σ28 was expressed from the arabinose promoter in the absence of flhDC transcription, a lac fusion inserted in the class 3 gene motA was transcribed at 162% of the wildtype level.
Construction of mutations in the arabinose promoter
To reduce σ28 levels, mutations were constructed in the arabinose promoter region that expresses σ28 in the reporter strains. First, a tetRA cassette was amplified from TH5794 using primers ParaBtetR and ParaBtetA (49°C annealing temperature) and inserted into the arabinose promoter using λ-red recombination (TH12906). This tetRA was transferred to a strain containing a transcriptional fusion to the motA class 3 promoter (TH12990). Next, two promoter mutations that were initially isolated in Aldridge, et. al.36 to lower FlgM levels in a Para-flgM strain were tested. Primers araC+5R and ara5’ were used to amplify these mutations from strains TH7395 (−85 A:T) and TH7396 (−41 T:C). λ-red recombination was used to move these PCR products into the chromosome to replace the tetRA in the arabinose promoter. TH13035 expressed the motA class 3 transcriptional fusion at 110% of wildtype levels, and TH13036 expressed the fusion at 64% of wildtype levels. These mutated arabinose promoter alleles were transferred to a strain containing a tetRA insertion in the flgKL promoter and a wildtype motA gene (TH13127 and TH13128). flgKL promoter mutants were transduced into these backgrounds by selecting for the KanR flgK::MudJ allele and screening for loss of the tetRA insertion.
Formation of consensus sequences
In order to construct consensus sequences, a position specific score matrix algorithm (PSSM)16 was implemented in Java. The PSSM uses the frequency of nucleotides at each position in an alignment to assign log-odds scores. The log-odds score is equal to the logarithm (base 2) of the frequency of a nucleotide at a position in an alignment divided by the background frequency of that nucleotide. The unit for a log-odds score is the bit since the logarithm (base 2) of the “odds” are used. The PSSM evaluates potential binding sites by adding up log-odds scores for each position in the binding site.
This “log-odds” method was utilized instead of the closely related information theory approach. The difference between these two techniques arises from the choice of background frequencies for calculating the significance of each nucleotide in a sequence alignment. If a nucleotide appears more frequently in the alignment than observed in the background distribution of nucleotides, a positive score is calculated. For the log-odds method that was used, the frequency of nucleotides in the intergenic regions were used for the background frequencies.16 In information theory, each nucleotide is given the same background frequency (i.e., 0.25).39
The argument for using information theory is that there is no “external observer” of the genome composition involved in the binding process. The information gained by the system would only depend on the change in entropy from the regulatory protein being unbound in the before state to being bound to the DNA site in the after state. Since the regulatory protein “does not make physical contact with the nucleic acid bases in the before state the composition of the genome should not matter”.39 We argue that the log-odds method is a valid technique because the evolutionary process is an observer of the system. In order for a regulatory protein to specifically recognize its DNA binding sites in the genome, the DNA binding sites must contain enough information to be distinguished from random sequences. If the genome is GC rich and the sites the regulatory protein binds to are GC rich, there are more random sequences to which the regulatory protein can bind than if the genome were AT rich. The regulatory protein will be selected against if it improperly regulates essential genes or important pathways through binding of too many of these random sequences. Therefore, the evolutionary process likely takes into account the background frequencies of nucleotides in the genome when adapting a regulatory protein for that genome. Since the DNA binding sites for the regulator are then used in sequence alignments, we argue that it is valid for the log-odds method to take into account these background frequencies.
The nucleotide frequencies in the intergenic regions of a genome were therefore used as the background frequencies for calculating log-odds. Moreover, essentially the same results were obtained when the figures were prepared using information theory (data not shown). Few differences were observed because the frequency of A’s or T’s in the intergenic regions of the organisms we studied (0.28 in Salmonella, for example) are close to the 0.25 frequency used in information theory.
Sequences used in alignments were weighted according to similarity using a neighbor-joining algorithm.16 Four pseudocounts (multiplied by the background frequencies) were added into the observed counts for each nucleotide at each position.
The consensus sequence for the FlhD4C2 binding sites was constructed through multiple rounds of PSSM searching. Only the upstream regions for genes (flgA, flgB, flhB, fliA, fliB, fliD, fliE, fliF, fliL, fepE, narK, rpiA, serA, srfA, yecR, yffO, and yqfE) that have some experimental evidence for an FlhD4C2 binding site4,6,9,13,40,41 were searched. The search was limited to 20 bp upstream of the start codon through 100 bp of the gene immediately upstream. After each round of searching, the top matches were used to form a new PSSM. Proximal and distal sites were combined so as not to bias the PSSM toward either site. Matches were not used in the new PSSM if they were nowhere near where the biological data suggested they should be.
The simple consensus sequence identified in Stafford, et. al.13 was used for an initial search. Since simple consensus sequences are not very effective PSSM’s, matches with scores above 4 bits were considered. All later searches used a more restrictive cutoff of 10 bits. Salmonella, E. coli, and Proteus mirablis (gi numbers 16763390, 49175990, 6959881, and 1857436; Proteus flhB sequence in Claret and Hughes42), for which biological data on flagellar promoters is available, were iteratively searched until no new matches were detected. To increase the sample size, we then searched upstream of flagellar genes in related organisms (Erwinia, Photorhabdus, Shigella, and Yersinia; gi numbers 50118965, 37524032, 30061571, and 51594359) until no new matches were detected. Spacers of 10, 11, and 12 bp were tried in each search. However, the final iterations removed all but one of the promoters that matched best with the 10 bp spacer.
β-galactosidase assays
10 µl of an overnight culture (LB) was subcultured into 3 ml of LB + arabinose or LB + chlortetracycline. Tubes were incubated with shaking at 37°C until they reached a mid-log density of 70 Klett units, which corresponds to an OD600 of about 0.7. Cultures were put on ice, spun down, and resuspended in 3 ml of cold, buffered saline. Between 50 µl and 0.5 ml of culture was added to 0.55 ml of complete Z-buffer34 (Z-buffer plus 5 µl 10% SDS and 100 µl chloroform) and buffered saline to give a total aqueous volume of 1.05 ml. The assay was continued as described.34,43–46 For each strain, assays were performed for three independent biological replicates.
Supplementary Material
Acknowledgements
This study was supported by Public Health Service Grants GM56141 and GM54501308 from the National Institutes of Health. We thank the members of the Hughes lab for helpful discussions and critical reading of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chilcott GS, Hughes KT. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol Mol Biol Rev. 2000;64:694–708. doi: 10.1128/mmbr.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hughes KT, Gillen KL, Semon MJ, Karlinsey JE. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science. 1993;262:1277–1280. doi: 10.1126/science.8235660. [DOI] [PubMed] [Google Scholar]
- 3.Kalir S, McClure J, Pabbaraju K, Southward C, Ronen M, Leibler S, Surette MG, Alon U. Ordering genes in a flagella pathway by analysis of expression kinetics from living bacteria. Science. 2001;292:2080–2083. doi: 10.1126/science.1058758. [DOI] [PubMed] [Google Scholar]
- 4.Liu X, Matsumura P. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J Bacteriol. 1994;176:7345–7351. doi: 10.1128/jb.176.23.7345-7351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ide N, Ikebe T, Kutsukake K. Reevaluation of the promoter structure of the class 3 flagellar operons of Escherichia coli and Salmonella. Genes Genet Syst. 1999;74:113–116. doi: 10.1266/ggs.74.113. [DOI] [PubMed] [Google Scholar]
- 6.Ikebe T, Iyoda S, Kutsukake K. Promoter analysis of the class 2 flagellar operons of Salmonella. Genes Genet Syst. 1999;74:179–183. doi: 10.1266/ggs.74.179. [DOI] [PubMed] [Google Scholar]
- 7.Schaubach OL, Dombroski AJ. Transcription initiation at the flagellin promoter by RNA polymerase carrying sigma28 from Salmonella typhimurium. J Biol Chem. 1999;274:8757–8763. doi: 10.1074/jbc.274.13.8757. [DOI] [PubMed] [Google Scholar]
- 8.Givens JR, McGovern CL, Dombroski AJ. Formation of intermediate transcription initiation complexes at pfliD and pflgM by sigma(28) RNA polymerase. J Bacteriol. 2001;183:6244–6252. doi: 10.1128/JB.183.21.6244-6252.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claret L, Hughes C. Interaction of the atypical prokaryotic transcription activator FlhD2C2 with early promoters of the flagellar gene hierarchy. J Mol Biol. 2002;321:185–199. doi: 10.1016/s0022-2836(02)00600-9. [DOI] [PubMed] [Google Scholar]
- 10.Liu X, Fujita N, Ishihama A, Matsumura P. The C-terminal region of the alpha subunit of Escherichia coli RNA polymerase is required for transcriptional activation of the flagellar level II operons by the FlhD/FlhC complex. J Bacteriol. 1995;177:5186–5188. doi: 10.1128/jb.177.17.5186-5188.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang S, Fleming RT, Westbrook EM, Matsumura P, McKay DB. Structure of the Escherichia coli FlhDC complex, a prokaryotic heteromeric regulator of transcription. J Mol Biol. 2006;355:798–808. doi: 10.1016/j.jmb.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 12.Park K, Choi S, Ko M, Park C. Novel sigmaF-dependent genes of Escherichia coli found using a specified promoter consensus. FEMS Microbiol Lett. 2001;202:243–250. doi: 10.1111/j.1574-6968.2001.tb10811.x. [DOI] [PubMed] [Google Scholar]
- 13.Stafford GP, Ogi T, Hughes C. Binding and transcriptional activation of non-flagellar genes by the Escherichia coli flagellar master regulator FlhD2C2. Microbiology. 2005;151:1779–1788. doi: 10.1099/mic.0.27879-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu HH, Kibler D, Tan M. In silico prediction and functional validation of sigma28-regulated genes in Chlamydia and Escherichia coli. J Bacteriol. 2006;188:8206–8212. doi: 10.1128/JB.01082-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stormo GD. DNA binding sites: representation and discovery. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- 16.Durbin R, Eddy S, Krogh A, Michison G. Biological Sequence Analysis. Cambridge: Cambridge University Press; 1998. [Google Scholar]
- 17.Roulet E, Busso S, Camargo AA, Simpson AJ, Mermod N, Bucher P. High-throughput SELEX SAGE method for quantitative modeling of transcription-factor binding sites. Nat Biotechnol. 2002;20:831–835. doi: 10.1038/nbt718. [DOI] [PubMed] [Google Scholar]
- 18.Benos PV, Bulyk ML, Stormo GD. Additivity in protein-DNA interactions: how good an approximation is it? Nucleic Acids Res. 2002;30:4442–4451. doi: 10.1093/nar/gkf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bulyk ML, Johnson PL, Church GM. Nucleotides of transcription factor binding sites exert interdependent effects on the binding affinities of transcription factors. Nucleic Acids Res. 2002;30:1255–1261. doi: 10.1093/nar/30.5.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brunner M, Bujard H. Promoter recognition and promoter strength in the Escherichia coli system. Embo J. 1987;6:3139–3144. doi: 10.1002/j.1460-2075.1987.tb02624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strainic MG, Jr, Sullivan JJ, Velevis A, deHaseth PL. Promoter recognition by Escherichia coli RNA polymerase: effects of the UP element on open complex formation and promoter clearance. Biochemistry. 1998;37:18074–18080. doi: 10.1021/bi9813431. [DOI] [PubMed] [Google Scholar]
- 22.Rao L, Ross W, Appleman JA, Gaal T, Leirmo S, Schlax PJ, Record MT, Jr, Gourse RL. Factor independent activation of rrnB P1. An "extended" promoter with an upstream element that dramatically increases promoter strength. J Mol Biol. 1994;235:1421–1435. doi: 10.1006/jmbi.1994.1098. [DOI] [PubMed] [Google Scholar]
- 23.Ross W, Aiyar SE, Salomon J, Gourse RL. Escherichia coli promoters with UP elements of different strengths: modular structure of bacterial promoters. J Bacteriol. 1998;180:5375–5383. doi: 10.1128/jb.180.20.5375-5383.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keilty S, Rosenberg M. Constitutive function of a positively regulated promoter reveals new sequences essential for activity. J Biol Chem. 1987;262:6389–6395. [PubMed] [Google Scholar]
- 25.Burr T, Mitchell J, Kolb A, Minchin S, Busby S. DNA sequence elements located immediately upstream of the −10 hexamer in Escherichia coli promoters: a systematic study. Nucleic Acids Res. 2000;28:1864–1870. doi: 10.1093/nar/28.9.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Josaitis CA, Gaal T, Gourse RL. Stringent control and growth-rate-dependent control have nonidentical promoter sequence requirements. Proc Natl Acad Sci U S A. 1995;92:1117–1121. doi: 10.1073/pnas.92.4.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kammerer W, Deuschle U, Gentz R, Bujard H. Functional dissection of Escherichia coli promoters: information in the transcribed region is involved in late steps of the overall process. Embo J. 1986;5:2995–3000. doi: 10.1002/j.1460-2075.1986.tb04597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan CL, Gross CA. The anti-initial transcribed sequence, a portable sequence that impedes promoter escape, requires sigma70 for function. J Biol Chem. 2001;276:38201–38209. doi: 10.1074/jbc.M104764200. [DOI] [PubMed] [Google Scholar]
- 29.Yu HH, Di Russo EG, Rounds MA, Tan M. Mutational analysis of the promoter recognized by Chlamydia and Escherichia coli sigma(28) RNA polymerase. J Bacteriol. 2006;188:5524–5531. doi: 10.1128/JB.00480-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hagerman PJ. Sequence-directed curvature of DNA. Annu Rev Biochem. 1990;59:755–781. doi: 10.1146/annurev.bi.59.070190.003543. [DOI] [PubMed] [Google Scholar]
- 31.deHaseth PL, Zupancic ML, Record MT., Jr RNA polymerase-promoter interactions: the comings and goings of RNA polymerase. J Bacteriol. 1998;180:3019–3025. doi: 10.1128/jb.180.12.3019-3025.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gillen KL, Hughes KT. Molecular characterization of flgM, a gene encoding a negative regulator of flagellin synthesis in Salmonella typhimurium. J Bacteriol. 1991;173:6453–6459. doi: 10.1128/jb.173.20.6453-6459.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maloy SR, Nunn WD. Selection for loss of tetracycline resistance by Escherichia coli. J Bacteriol. 1981;145:1110–1111. doi: 10.1128/jb.145.2.1110-1111.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maloy SR. Experimental Techniques in Bacterial Genetics. Boston: Jones and Bartlett; 1990. [Google Scholar]
- 35.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aldridge PD, Karlinsey JE, Aldridge C, Birchall C, Thompson D, Yagasaki J, Hughes KT. The flagellar-specific transcription factor, sigma28, is the Type III secretion chaperone for the flagellar-specific anti-sigma28 factor FlgM. Genes Dev. 2006;20:2315–2326. doi: 10.1101/gad.380406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Groisman EA. In vivo genetic engineering with bacteriophage Mu. Methods Enzymol. 1991;204:180–212. doi: 10.1016/0076-6879(91)04010-l. [DOI] [PubMed] [Google Scholar]
- 38.Hughes KT, Roth JR. Transitory cis complementation: a method for providing transposition functions to defective transposons. Genetics. 1988;119:9–12. doi: 10.1093/genetics/119.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider TD. Information content of individual genetic sequences. J Theor Biol. 1997;189:427–441. doi: 10.1006/jtbi.1997.0540. [DOI] [PubMed] [Google Scholar]
- 40.Prüß BM, Campbell JW, Van Dyk TK, Zhu C, Kogan Y, Matsumura P. FlhD/FlhC is a regulator of anaerobic respiration and the Entner-Doudoroff pathway through induction of the methyl-accepting chemotaxis protein Aer. J Bacteriol. 2003;185:534–543. doi: 10.1128/JB.185.2.534-543.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frye J, Karlinsey JE, Felise HR, Marzolf B, Dowidar N, McClelland M, Hughes KT. Identification of new flagellar genes of Salmonella enterica serovar Typhimurium. J Bacteriol. 2006;188:2233–2243. doi: 10.1128/JB.188.6.2233-2243.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Claret L, Hughes C. Functions of the subunits in the FlhD(2)C(2) transcriptional master regulator of bacterial flagellum biogenesis and swarming. J Mol Biol. 2000;303:467–478. doi: 10.1006/jmbi.2000.4149. [DOI] [PubMed] [Google Scholar]
- 43.Gillen KL, Hughes KT. Transcription from two promoters and autoregulation contribute to the control of expression of the Salmonella typhimurium flagellar regulatory gene flgM. J Bacteriol. 1993;175:7006–7015. doi: 10.1128/jb.175.21.7006-7015.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karlinsey JE, Tsui HC, Winkler ME, Hughes KT. Flk couples flgM translation to flagellar ring assembly in Salmonella typhimurium. J Bacteriol. 1998;180:5384–5397. doi: 10.1128/jb.180.20.5384-5397.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karlinsey JE, Tanaka S, Bettenworth V, Yamaguchi S, Boos W, Aizawa SI, Hughes KT. Completion of the hook-basal body complex of the Salmonella typhimurium flagellum is coupled to FlgM secretion and fliC transcription. Mol Microbiol. 2000;37:1220–1231. doi: 10.1046/j.1365-2958.2000.02081.x. [DOI] [PubMed] [Google Scholar]
- 46.Aldridge P, Karlinsey J, Hughes KT. The type III secretion chaperone FlgN regulates flagellar assembly via a negative feedback loop containing its chaperone substrates FlgK and FlgL. Mol Microbiol. 2003;49:1333–1345. doi: 10.1046/j.1365-2958.2003.03637.x. [DOI] [PubMed] [Google Scholar]
- 47.Kutsukake K, Ide N. Transcriptional analysis of the flgK and fliD operons of Salmonella typhimurium which encode flagellar hook-associated proteins. Mol Gen Genet. 1995;247:275–281. doi: 10.1007/BF00293195. [DOI] [PubMed] [Google Scholar]
- 48.Ikebe T, Iyoda S, Kutsukake K. Structure and expression of the fliA operon of Salmonella typhimurium. Microbiology. 1999;145(Pt 6):1389–1396. doi: 10.1099/13500872-145-6-1389. [DOI] [PubMed] [Google Scholar]
- 49.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneider TD, Stormo GD, Gold L, Ehrenfeucht A. Information content of binding sites on nucleotide sequences. J Mol Biol. 1986;188:415–431. doi: 10.1016/0022-2836(86)90165-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


