Abstract
Neurodegeneration in diseases caused by altered metabolism of mammalian prion protein (PrP) can be averted by reducing PrP expression. To identify novel pathways for PrP down-regulation, we analyzed cells that had adapted to the negative selection pressure of stable overexpression of a disease-causing PrP mutant. A mutant cell line was isolated that selectively and quantitatively routes wild-type and various mutant PrPs for ER retrotranslocation and proteasomal degradation. Biochemical analyses of the mutant cells revealed that a defect in glycosylphosphatidylinositol (GPI) anchor synthesis leads to an unprocessed GPI-anchoring signal sequence that directs both ER retention and efficient retrotranslocation of PrP. An unprocessed GPI signal was sufficient to impart ER retention, but not retrotranslocation, to a heterologous protein, revealing an unexpected role for the mature domain in the metabolism of misprocessed GPI-anchored proteins. Our results provide new insights into the quality control pathways for unprocessed GPI-anchored proteins and identify transamidation of the GPI signal sequence as a step in PrP biosynthesis that is absolutely required for its surface expression. As each GPI signal sequence is unique, these results also identify signal recognition by the GPI-transamidase as a potential step for selective small molecule perturbation of PrP expression.
INTRODUCTION
A wide range of diseases are caused by aberrant folding, processing, trafficking, or degradation of proteins in the secretory pathway (Cohen and Kelly, 2003; Hebert and Molinari, 2007; Otsu and Sitia, 2007). Protein-folding diseases are typically dominant gain-of-function disorders whose pathogenesis is intimately tied to the expression level of the misfolded protein. It is therefore of substantial importance to understand the cellular quality control pathways that discriminate properly folded from misfolded proteins to regulate their maturation. Such studies would provide molecular level insights into the basis of protein-folding diseases and could eventually be exploited to manipulate quality control and influence disease pathogenesis. However, the near ubiquitous use of these quality control pathways makes identifying sufficiently selective points for potential pharmacologic perturbation a daunting challenge.
Several dramatic examples of dominant gain-of-function disorders are caused by misfolding of PrP, a widely expressed cell surface glycoprotein of unknown function (Prusiner, 1998; Aguzzi and Heikenwalder, 2006; Wadsworth and Collinge, 2007). These diseases can be inherited through mutations in Prnp (the gene that codes for PrP) or acquired via a transmissible agent composed of a misfolded isoform of PrP, termed PrPSc. Exogenous PrPSc is capable of converting the normal cellular isoform (PrPC) into additional PrPSc molecules, leading to its accumulation and generation of additional transmissible agent. In the familial diseases, PrP mutations typically cause accumulation of misfolded PrP through poorly understood mechanisms that in some cases also generate PrPSc. Thus, altered PrP folding, metabolism, and accumulation are the proximal causes of both familial and transmissible prion diseases.
Although the downstream pathways leading from misfolded PrP to cellular toxicity are not known, it is clear that ongoing PrP expression is an absolute prerequisite for neuronal cell death and disease progression (Bueler et al., 1993; Prusiner et al., 1993; Brandner et al., 1996). Indeed, not only is there a tight correlation between expression level and the time course of both genetic (Telling et al., 1996; Chiesa et al., 1998; Hegde et al., 1999) and transmissible prion diseases (Prusiner et al., 1990, 1993; Bueler et al., 1993, 1994), but reducing PrP expression even after the onset of symptoms can reverse disease (Mallucci et al., 2003, 2007). Preventing accumulation of more misfolded material by reducing PrP expression allows normal metabolic pathways to restore homeostasis by clearing various disease associated PrP forms (including PrPSc), despite their often aggregated state (Safar et al., 2005). Thus, an attractive and viable strategy for this class of protein folding diseases is to reduce PrP expression (Mallucci and Collinge, 2005). Even a modest reduction in expression can evidently have a substantial impact on disease progression because mice lacking one copy of the Prnp gene exhibit markedly longer incubation times after infection with PrPSc (Bueler et al., 1993; Prusiner et al., 1993; Bueler et al., 1994; Manson et al., 1994). However, the pathways that regulate total cellular levels of PrP, particularly those potentially amenable to selective modulation, are poorly studied and incompletely understood.
As transmissible disease pathogenesis requires that PrPC reach the cell surface (at least transiently; (Caughey and Raymond, 1991; Borchelt et al., 1992), attenuation of PrP expression at any of the earlier biosynthetic steps of transcription, translation, maturation, or trafficking are potentially useful. An especially attractive strategy is to exploit normal quality control (QC) pathways that are designed to recognize and route proteins for degradation. In the case of PrP, the primary site of QC is presumably in the endoplasmic reticulum (ER) lumen, the site of secretory and membrane protein biogenesis and maturation. The ER contains multiple QC pathways composed of different (and sometimes overlapping) subsets of chaperones that operate in parallel to handle different types of substrates. However, the parameters used by any these QC pathways to distinguish normal from abnormal proteins are incompletely understood. The recognition motifs include diverse features such as glycan structure (e.g., the chaperones calnexin and calreticulin; Ellgaard and Helenius, 2001; Hebert and Molinari, 2007), unpaired or inappropriately bonded cysteines (oxido-reductases such as protein disulfide isomerase [PDI]; Hatahet and Ruddock, 2007), and surface-exposed hydrophobic patches (most chaperones, including BiP and PDI; Ni and Lee, 2007). Thus, it is thought that misfolding or misprocessing leads to recognition and prolonged interactions with specific chaperones that can initiate substrate routing into ER-associated degradation (ERAD) pathways (McCracken and Brodsky, 2005; Meusser et al., 2005).
After chaperone-mediated targeting for ERAD, a decisive step in misfolded protein degradation is its export to the cytosol via a putative retrotranslocation channel. Although the identity and nature of the retrotranslocation channel(s) remain contentious (Meusser et al., 2005), it is thought that once substrates are exposed to the cytosol, they are ubiquitinated, extracted from the membrane, deglycosylated (in the case of glycoproteins), and degraded by the proteasome. Thus, the combination of QC and ERAD pathways are designed to recognize, export, and degrade proteins from the ER, and substrate engagement of this pathway is one way to reduce functional expression of a secretory or membrane protein. In fact, examples of protein levels being regulated at the level of QC and ERAD have been described (e.g., HMG-CoA reductase; Hampton, 2002; and apolipoprotein B; Davidson and Shelness, 2000), highlighting the utility of these systems as a means of physiologically and pharmacologically controlling gene expression.
Unfortunately, PrP seems to be especially refractory to recognition, retrotranslocation, and/or degradation by ERAD even when its maturation is impaired in any of several ways. For example, mutations in one or both N-linked glycosylation sites is entirely permissive for PrP trafficking out of the ER, does not substantially reduce steady-state levels of PrP in transgenic and knock-in mice and permits interaction with exogenous PrPSc to support prion replication in cell culture and in mice (Neuendorf et al., 2004; Cancellotti et al., 2005). Mutants that disrupt the single disulfide bond in PrP (Yanai et al., 1999) or PrP made during reducing conditions (Kang et al., 2006), despite being misfolded and incompetent for trafficking to the cell surface, are degraded inefficiently and instead aggregate intracellularly. Deletion of the GPI-anchoring signal sequence (Campana et al., 2007) or conversion to a transmembrane domain (Taraboulos et al., 1995; Kaneko et al., 1997) both result in PrPs that escape QC in the ER and are either secreted or expressed on the cell surface, respectively. Furthermore, various human disease-causing mutants are only partially if at all degraded by ERAD, despite their apparent misfolding or altered topology (Hegde et al., 1998; Ma and Lindquist, 2001; Yedidia et al., 2001; Drisaldi et al., 2003). Finally, a wide range of artificial mutations and deletions are readily tolerated and lead to ER export (Muramoto et al., 1996; Shmerling et al., 1998; Supattapone et al., 2001; Baumann et al., 2007; Li et al., 2007; Hegde et al., 1998) or in some cases, ER storage disease (Muramoto et al., 1997). Thus, in no situation is mutated, misfolded, or misprocessed PrP recognized and efficiently degraded by ERAD. In fact, it has been suggested that retrotranslocation of PrP (even at relatively low levels) could be highly cytotoxic due to the transient generation of cytosolic PrP, perhaps explaining why it is typically such a poor substrate for ERAD. Hence, a major unresolved issue is how or whether PrP can be routed quantitatively for ERAD and whether this would be tolerated, beneficial, or detrimental to cells.
Here, we report the unexpected discovery of a mutant cell line that routes both wild-type and mutant PrPs quantitatively for retrotranslocation and proteasome-dependent degradation without any obvious toxicity. This rerouting was due to an unprocessed GPI-anchoring signal sequence that, in combination with the PrP mature domain, forms a remarkably efficient ERAD substrate. This study has therefore led to the identification of a previously unanticipated site for modulating PrP expression, described a robust model system for the study of PrP QC and retrotranslocation, and more generally, led to new insights into the determinants for QC of GPI-anchored proteins in the ER.
MATERIALS AND METHODS
Cells, Plasmids, and Reagents
Neuro2a (N2a) cells were cultured in DMEM containing 10% FBS in a humidified 37°C incubator at 5% CO2. L-cells, a kind gift from Dr. J. Bonifacino (NIH) and have been previously described (Sugiyama et al., 1991), were cultured in RPMI 1640 medium containing 10% FBS at 5% CO2. Stable cell lines were generated by selection in 200 μg/ml Zeocin (Invitrogen, Carlsbad, CA) for 4 wk, followed by subcloning of individual colonies. PrP expression levels were analyzed in several individual clones by both immunoblotting and immunofluorescence using the 3F4 antibody. One clone expressing wild-type PrP (termed C3) and one that expressed PrP(A117V) at very low steady-state levels (termed A4) were chosen for detailed characterization. All experiments involving transient transfection were transfected with Lipofectamine 2000 (Invitrogen) according to manufacturer's directions and analyzed 18–24 h after transfection. Plasmids encoding wild-type and mutant PrPs in a pCDNA3.1-based vector (Invitrogen) have either been described (Rane et al., 2004) or generated from these constructs using standard site-directed mutagenesis. Hemagglutinin (HA)-tagged PrPs were generated from the nontagged constructs by insertion of synthetic oligonucleotides encoding the epitope at the Bsu36I site (between residues 51 and 52) of PrP. PrPΔGPI was constructed by deletion of the C-terminal 23 amino acids from the PrP coding region. RFP-KDEL, VSVG-GFP, and GFP-GPIFR constructs have been previously described (Presley et al., 1997; Nichols et al., 2001; Snapp et al., 2006) and were kind gifts from J. Lippincott Schwartz (NIH). ETBR-GFP was a kind gift from Dr. R. Schülein (Forschungsinstitut für Molekulare Pharmakologie, Berlin) and has been previously described (Grantcharova et al., 2002). Green fluorescent protein (GFP)-GPI-Chinese hamster ovary (CHO) shown in Figure 7 has a D→N change at position 36 of GFP to introduce a consensus site for glycosylation. Two other constructs were also generated to introduce consensus sites at other positions (L→T at position 137, and DK→YT at positions 211 and 212) and gave identical results to those shown in Figure 7. GFP-GPIPrP was prepared in a pCDNA1.3-based vector and encodes residues 1-40 of bovine preprolactin fused to the complete GFP sequence followed by residues 231-254 of hamster PrP. PrP(S232W) was generated by site-directed mutagenesis of hamster PrP. PrP-Qa was generated by replacing the GPI signal sequence of HA-tagged hamster PrP (see above) with the complete GPI-anchoring sequence of the Qa protein.
Figure 7.
The mature domain influences the fate of proteins with unprocessed GPI signals. (A) Pulse-chase analysis in A4 cells of GFP-GPIFR or its glycosylated counterpart (GFP-GPI-CHO) in the absence (Unt.) or presence of 5 μM MG132 (MG.) or 1 μM kifunensine (Kif.). Pulse labeling was for 10 min and chase for either 0 or 6 h. The percent of protein degraded by 6 h was quantified by phosphorimaging and is indicated below each set of lanes. (B) Pulse-chase analysis of PrP turnover in A4 cells in the absence or presence of 10 μg/ml the glycosylation inhibitor tunicamycin. (C) Pulse-chase analysis of GFP-GPIPrP (left panels) and GFP-GPIFR (right panels) in A4 cells. Top panels, PrP immunoprecipitations using the 3F4 antibody; bottom panels, GFP immunoprecipitations. (D) Pulse-chase analysis of transiently transfected WT PrP in A4 and N2a cells in the absence (Unt.) or presence of the reversible reducing agent, dithiothreitol (DTT; 10 mM). +/−DTT indicates samples that were labeled in the presence, but chased in the absence of DTT. The right panel shows the turnover of WT PrP in A4 cells in the absence (Unt.) or presence of ER stress induced by thapsigargin treatment (Tg; 1 μM). (E) Metabolically labeled cell lysates from A4 cells transfected with GFP-GPIFR were immunoprecipitated under nondenaturing conditions using either the PrP-A antibody (left lane) or an anti-GFP antibody (right lane), to detect coassociating proteins. The arrowhead points to the migration of PrP and GFP. Asterisks mark coassociating proteins that are enriched in either the PrP or GPI-GFP immunoprecipitates. (F) Metabolically labeled A4 cell lysates were immunoprecipitated with PrP-A, anti-GFP, or anti-protein disulfide isomerase (PDI) antibodies under nondenaturing conditions (1st IP) followed by denaturation and reimmunoprecipitation with the PrP-A antibody (2nd IP). The arrowhead shows the presence of PrP in the sample that was sequentially immunoprecipitated with anti-PDI and PrP-A antibodies.
Antibodies were described previously or from the following sources: 3F4 mouse monoclonal against PrP (Signet Laboratories, Dedham, MA); PDI (StressGen, San Diego, CA); and GFP (Stefanovic and Hegde, 2007). The PrP-A rabbit antiserum against PrP was generated (Lampire Biological Laboratories, Pipersville, PA) against a synthetic peptide (KKRPKPGGWNTGGSRYC) conjugated to keyhole limpet hemocyanin using standard protocols. Immunoblotting using total brain homogenates from hamster and mouse showed reactivity to only PrP when compared with other PrP antibodies. This antibody reacts against an epitope conserved in all mammalian species and was found to work against human, mouse, and hamster PrP. Dithiothreitol (DTT) and tunicamycin were from Sigma (St. Louis, MO) and were dissolved in water. Thapsigargin, brefeldin A (BFA), MG132, and kifunensine were from Calbiochem (La Jolla, CA) and were dissolved in DMSO, ethanol (BFA) or water (kifunensine). Inhibitor concentrations used are indicated in the corresponding figure legends. Concanavalin A-Sepharose was obtained from Amersham-Pharmacia (Piscataway, NJ. All enzymes for cloning were obtained from New England Biolabs (Beverly, MA) with the exception of Pfu polymerase, which was from Stratagene (La Jolla, CA).
Biochemical Analyses
Metabolic labeling of cells, pulse-chase analysis and immunoprecipitations were performed as previously described (Rane et al., 2004; Kang et al., 2006). In general, cells were preincubated for 30 min in serum-free media lacking methionine and cysteine before addition of 35S-Translabel to initiate the pulse. Chase was initiated by replacing the labeling media with unlabeled complete media. Inhibitors used in pulse-chase experiments were added at the following times before the pulse labeling: 30 min for BFA and kifunensine, 2 h for MG132 and tunicamycin, 10 min for DTT, and 1 min for thapsigargin. Unless otherwise indicated in the figure legends, all inhibitors except thapsigargin (an irreversible inhibitor) were maintained for the duration of the chase. For immunoprecipitation (IP) under denaturing conditions, cells were lysed in 1% SDS, 0.1 M Tris, pH 8, and heated immediately to 100°C in a boiling water bath to fully denature all proteins. After shearing the nucleic acids by repeated vortexing, lysates were diluted 10-fold in ice-cold IP buffer (50 mM HEPES, pH 7.4, 100 mM NaCl, and 1% Triton X-100) before addition of antibodies and protein A-agarose, as previously described (Kang et al., 2006). Nondenaturing IPs were performed on cells lysed directly in cold IP buffer on ice. Lysates were clarified by centrifugation in a microcentrifuge to remove debris and IPed with the relevant antibody and protein A beads (Kang et al., 2006). For sequential IPs, the products of a nondenaturing IP were denatured in 1% SDS, 0.1 M Tris, pH 8, boiled, and diluted 10-fold in IP buffer before adding the second antibody. Preparation of total cell lysates and solubility assays for PrP were performed as previously described (Rane et al., 2004). For separation of hydrophilic and hydrophobic proteins by Triton X-114 (TX-114; Sigma), cells grown in six-well plates were rinsed in PBS and lysed in TX-114 lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 2 mM EDTA, and 1% TX-114) on ice. Cell lysates were homogenized using 25-gauge aluminum hypodermic needles (Monoject; Kendall, Covidien, Mansfield, MA) and placed at 37°C for 5 min to allow phase separation. After centrifugation for 5 min, the hydrophilic (top) and hydrophobic (bottom) phases were carefully transferred to fresh tubes for analysis. PrP glycosylation status was assessed by endoglycosidase H (EndoH) or PNGase F digestion: cell lysates made in 1% SDS, 0.1 M Tris, pH 8, were diluted in buffer containing 1% β-mercaptoethanol/0.05 M Tris, pH 6.8, and heated to 100°C to reduce and denature all proteins. For EndoH digestion, samples were cooled and diluted with G5 buffer (New England Biolabs) to a final concentration of 0.1% SDS, 20 mM Tris, and 50 mM citrate, pH 5.5, before incubation with 5000 U of EndoH for 4 h at 37°C. For PNGase digestion, Triton X-100 (Sigma) was added to a final concentration of 1% to the cooled cell lysates before incubation with 5000 U of PNGase for 4 h at 37°C. After digestion with either enzyme, samples were precipitated using trichloroacetic acid (TCA) according to standard protocols, and recovered protein pellets were dissolved in equal volumes of 1% SDS, and 0.1 M Tris, pH 8. SDS-PAGE separation of proteins and immunoblotting was performed as previously described (Fons et al., 2003). Quantification of pulse-chase experiments utilized a Typhoon Phosphorimager and accompanying software (Molecular Dynamics, Sunnyvale, CA) and autoradiographs on Kodak BioMax film (Eastman Kodak, Rochester, NY) were digitized using Adobe Photoshop for preparation of figures (San Jose, CA).
Immunofluorescence Microscopy
Fluorescence microscopy images were obtained using a Zeiss LSM510 confocal microscope (Thornwood, NY) with accompanying image acquisition software. Imaging conditions and quantitative analyses of localization were as reported previously (Rane et al., 2004) with the following exceptions: a confocal slice corresponding to 2 Airy units and a 63× oil objective were used in the acquisition of all images; for indirect immunofluorescent detection of PrP, an anti-mouse secondary antibody conjugated to Alexa Fluor-488 (Invitrogen) was used at 1:1000 dilution.
Glycolipid Analysis
Labeling and extraction of glycolipids was performed as previously (Lemansky et al., 1991) with some modifications. Briefly, 60–90% confluent cells in 10-cm plates were rinsed twice in glucose-free medium and incubated in RPMI 1640 medium without glucose containing 10% dialyzed FBS and 10 μg/ml tunicamycin for 1 h at 37°C. Each dish of cells was labeled with 0.25 mCi/ml [3H]mannose (Perkin Elmer-Cetus, Norwalk, CT) for 2 h at 37°C, rinsed once in PBS, scraped into a polypropylene tube, sedimented, and extracted twice with chloroform-methanol-water (CMW; 10:10:3 ratio). The pooled extracts were dried under vacuum, dissolved in water-saturated 1-butanol and extracted with butanol-saturated water. The aqueous phase was then re-extracted in butanol, and the pooled butanol phases were dried under vacuum to a volume of ∼50 μl and spotted onto TLC plates (Merck, Whitehouse Station, NJ). Chromatograms were developed in CMW, dried in air, sprayed with Enhance fluorography reagent (Dupont, Wilmington, DE) and exposed to film.
RESULTS
Isolation of a Mutant Cell Line with Altered PrP Expression
On the application of long-term negative selection pressure, somatic cell lines often acquire mutations that partially or completely alleviate the impediment to normal growth. Understanding the genetic or biochemical basis of specific adaptations to a selection pressure can reveal new and unanticipated parameters that influence a specific cellular process. Indeed, biochemical and genetic analysis of somatic cell mutants isolated by imposed selection pressure have provided critical insight into such diverse areas as cholesterol metabolism (Goldstein et al., 2002) and protein trafficking (Hyman, 1988; Lemansky et al., 1991; Hirose et al., 1992; Ohtsuka et al., 1993; Ghaedi et al., 1999). A comparable approach should also be useful in identifying posttranscriptional pathways that regulate cellular PrP levels. We therefore analyzed PrP expression in clonal lines generated by stable transfection of a mouse neuroblastoma cell line (N2a) with disease-causing PrP mutants expressed behind the strong and constitutive CMV promoter. Although most clones that show poor expression are presumably due to low transgene copy number, recombination, or site of integration, we reasoned that at least some may be due to somatic mutations induced by selection pressure to down-regulate mutant PrP expression.
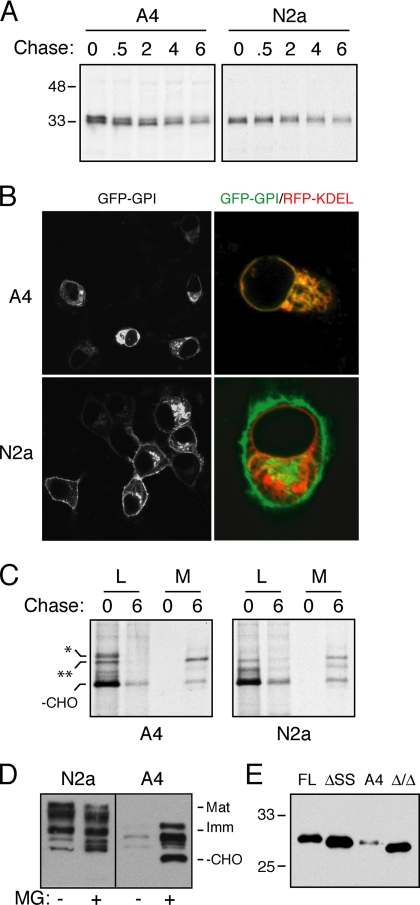
The disease-causing PrP(A117V) mutant can be expressed in cultured cells upon transient transfection without any obvious differences from wild-type PrP in expression level, localization, solubility, glycosylation pattern, or metabolism (Figure 1A). However, during the isolation of stable cell lines expressing PrP(A117V), a significant number of clones had poor or no expression. Our interest was drawn to one such clone, termed A4, that not only showed very low steady-state levels, but exclusively immature and intracellular PrP glycoforms (Figure 1, B and C). Surprisingly, mature forms of wild-type mouse PrP (MoPrP) endogenously expressed by the parental N2a cell line were no longer detectable in A4 cells (Figure 1B). Hence, expression and processing of both the transgene-expressed PrP(A117V) and endogenous MoPrP were altered in A4 cells, suggesting a cellular, rather than a transgene mutation. This was confirmed by demonstrating altered expression patterns in A4 cells, but not in parental N2a cells, of transiently transfected plasmids encoding either wild-type PrP or PrP(A117V; Figure 1D). Not only are the exogenously transfected PrPs expressed at lower steady-state levels in A4 cells, but they show only immature glycoforms by SDS-PAGE (Figure 1D), complete sensitivity to EndoH (Figure 1E), and an exclusively ER localization (Figure 1F). These data together indicate that both endogenously and exogenously derived wild-type or mutant PrPs fail to be expressed as mature, glycosylated, cell surface molecules in A4 cells. Instead, the low level, exclusively immature, ER-restricted PrP species was suggestive of altered PrP biosynthesis, processing, or trafficking in the early secretory pathway of A4 cells.
Figure 1.
Altered PrP expression in the A4 mutant cell line. (A) Total cell lysates from N2a cells transiently transfected with either wild type (WT) or the A117V PrP mutant were separated into detergent soluble (S) and insoluble (P) fractions, resolved by SDS-PAGE, and immunoblotted using an antibody (3F4) that specifically detects transfected but not endogenous PrP. The migration of different PrP species are indicated: Mat, mature PrP containing fully modified glycans; Imm, Immature PrP containing core or incompletely modified glycans; −CHO, unglycosylated PrP. Bottom panels, the corresponding indirect immunofluorescence images using the 3F4 antibody. (B) Immunoblot analysis of the parental N2a cells and two derived cell lines (C3 and A4) that stably express WT or PrP(A117V), respectively. The left panel was probed with 3F4 to detect expression of the stably transfected product, and the right panel with PrP-A (a pan-PrP antibody) to detect both endogenous and transfected products. (C) Indirect immunofluorescent localization of PrP in C3 and A4 cells using the 3F4 antibody. Identical detector settings were used to allow comparison of relative expression levels. The lower panels show single cells to illustrate the different PrP localization patterns in C3 versus A4 cells, the latter of which colocalized with RFP-KDEL, an ER marker introduced by transient transfection. (D) A4 and parental N2a cells were transiently transfected with plasmids encoding either WT or A117V PrP and analyzed by fractionation and 3F4 immunoblotting as in A. (E) Total cell lysates from A4 and N2a cells transiently transfected with WT PrP were digested with EndoH (E), with PNGase F (P) or left untreated (−) before immuoblotting with 3F4. The lower panel shows this immunoblot stripped and reprobed with an antibody against TRAPα, an ER resident glycoprotein. (F) A4 and parental N2a cells were transiently transfected with WT PrP and analyzed by indirect immunofluorescence using 3F4. Note the lack of surface expression in A4 cells, in which all of the PrP colocalized with RFP-KDEL (bottom panels).
A4 Cells Constitutively and Selectively Degrade PrP from the ER
To identify the altered step(s) in PrP expression in A4 cells, we examined the biosynthesis and trafficking of the stably expressed PrP(A117V) by pulse-chase labeling experiments. For comparison, we used the previously characterized C3 cell line (derived from the same parental N2a cells at the same time as A4 cells) that stably expresses wild-type PrP. Ordinarily, PrP [and PrP(A117V)] expressed in N2a cells is core glycosylated, folded within a few minutes in the ER lumen, trafficked via the Golgi to the cell surface within ∼30 min, and degraded with a t1/2 of ∼6 h in the endo-lysosomal system. Transit through the Golgi is accompanied by trimming and modification of glycans that is readily assayed by slower migration of PrP on SDS-PAGE and acquisition of resistance to EndoH. In contrast to this normal PrP metabolism (illustrated by C3 cells), PrP(A117V) synthesized in A4 cells during 10 min of labeling disappeared rapidly (t1/2 = 1- 2 h) without the appearance of mature glycoforms characteristic of trafficking through the Golgi (Figure 2A). Similar results were obtained for transiently expressed human PrP (HuPrP), HA-tagged HuPrP, or two disease-associated C-terminal PrP mutants (H187R and E200K) transfected into A4 cells: in each case, PrP was degraded in A4 cells without detectable generation of mature glycosylated species (Figure 2, B and C). Yet, these constructs were metabolized by a distinctly different pathway in the parental N2a cells, and mature PrPs were readily generated as the primary product during the chase period.
Figure 2.
Constitutive ERAD of PrP in A4 cells. (A) Pulse-chase analysis of the stably transfected PrP products (immunoprecipitated using 3F4) in either A4 or C3 cells. Pulse labeling with [35S]methionine was for 10 min, followed by chase for the indicated times (in hours). (B and C) A4 and parental N2a cells were transiently transfected with the indicated constructs and analyzed by pulse chase as in A. The samples in B were immunoprecipitated with 3F4, whereas the HA-tagged proteins in C were recovered using anti-HA antibody. (D) Pulse-chase analysis of A4 cells performed in the absence or presence of a proteasome inhibitor (5 μM MG132) or mannosidase I inhibitor (1 μM kifunensine). (E) Steady-state levels of PrP in A4 cells (detected using 3F4) after treatments with MG132 or kifunensine for the indicated times (in hours). (F) Pulse-chase analysis of A4 cells transiently transfected with the indicated mutant PrPs performed in the absence or presence of proteasome inhibitor (5 μM MG132). (G) Indirect immunofluorescence (using 3F4) of A4 cells before and after treatment with 5 μM MG132 for 4 h.
As the initial biosynthesis of PrP (judged by the glycosylation pattern and amount synthesized at the pulse-point) was the same in A4 and N2a cells, we inferred that the defect in A4 cells was at a step after successful PrP translocation into the ER lumen. Furthermore, the reduced half-life and lack of Golgi-specific glycan modifications suggested that PrP may be retained in the ER and degraded by ERAD in A4 cells. Typically, glycoproteins subjected to ERAD are processed by mannosidase I, retrotranslocated into the cytosol, deglycosylated by N-glycanse, and degraded by the ubiquitin-proteasome system. To test the involvement of this pathway in PrP degradation by A4 cells, we analyzed PrP metabolism upon inhibition of ERAD at two distinct steps with either a proteasome inhibitor (MG132) or mannosidase I inhibitor (kifunensine). Both treatments led to the stabilization of PrP (in both steady-state and pulse-chase experiments), with accumulation of different forms: an unglycosylated species with MG132, and a core-glycosylated species with kifunensine (Figure 2, D and E). Importantly, pulse-chase analysis in MG132-treated cells showed a clear conversion of core-glycosylated PrP into unglycosylated PrP over time, definitively illustrating PrP retrotranslocation from the lumen (where it is glycosylated) to the cytosol (where it is deglycosylated).
Similar results were seen for exogenously transfected HuPrP and mutants (H187R and E200K), suggesting that all PrPs are routed to ERAD in A4 cells (Figure 2F). Importantly, even upon prolonged inhibition of PrP degradation in A4 cells overexpressing any of these PrPs, mature forms did not appear. Rather, the accumulated PrP was exclusively in immature forms, suggesting that the lack of mature PrP expression was not simply a consequence of rapid routing into ERAD before exit from the ER. Congruent with these biochemical data, immunofluorescent detection of PrP in MG132-treated A4 cells showed no detectable surface expression despite both an increase in the number and brightness of cells detectably expressing PrP (Figure 2G). Thus, wild-type and mutant PrPs are efficiently recognized and routed into the classical ERAD pathway in A4 cells, despite the fact that these same mutants are poorly degraded by ERAD upon misfolding in N2a cells. This routing into ERAD is apparently via a high capacity pathway because it was observed (albeit with a lower rate) even in the transient transfection experiments that grossly overexpress PrP. Furthermore, the normal cell morphology and growth kinetics of A4 cells relative to the parental cells (data not shown) suggested a relatively specific effect on a subset of total proteins (including PrP), because constitutive degradation of all glycoproteins or defects in ER-to-Golgi trafficking would be incompatible with viability.
The GPI Anchoring Signal Sequence Is Required for ERAD of PrP in A4 Cells
To gain insight into the basis of PrP degradation in A4 cells, we examined the relative substrate specificity of constitutive ERAD. Pulse-chase analysis of total glycoproteins (captured using concanavalin A) in A4 cells revealed no major differences in the profile of proteins, their relative amounts of synthesis, or half-lives compared with the parental N2a cells (Figure 3A). Analysis of specific model membrane proteins including GFP-tagged vesicular stomatitis virus G-protein (VSVG-GFP) and GFP-tagged endothelin-B receptor (ETBR-GFP) showed apparently normal maturation in A4 cells compared with the control N2A cells. Both glycoproteins had similar kinetics of maturation (as judged by glycan modifications), half-lives of degradation, and steady-state localization at the plasma membrane and endomembrane system (Figure 3, B and C).
Figure 3.
Normal glycoprotein metabolism in A4 cells. (A) The synthesis and turnover of total glycoproteins (captured using immobilized concanavalin-A) was assessed in A4 and N2a cells using pulse-chase analysis. Pulse labeling with [35S]methionine was for 15 min, followed by chase for the indicated times (in hours). The positions of molecular weight markers (in kDa) are indicated on the left. (B) Synthesis and turnover of VSVG-GFP and ETBR-GFP in A4 and parental N2a cells was followed by pulse-chase analysis as in Figure 2A. Immunoprecipitation was with anti-GFP antibody. (C) Representative fluorescence images of A4 and N2a cells transfected with VSVG-GFP and ETBR-GFP.
Unlike these (and most) glycoproteins, PrP contains a GPI anchor. We therefore asked if GPI-anchored proteins in general are targeted for ERAD in A4 cells. Pulse-chase analysis of a minimal GPI-anchored protein, GFP-GPIFR (GFP with the folate receptor GPI signal), in A4 and N2A cells showed comparable expression levels and turnover kinetics (Figure 4A). However, a subtle difference in the migration of the GFP-GPI FR species between the two cell types suggested some difference in modification, prompting us to examine its localization by confocal microscopy. GFP-GPI FR was predominantly localized to the plasma membrane and Golgi of N2a cells, but remained entirely in the ER of A4 cells (Figure 4B). Thus, A4 cells are altered in the trafficking of a heterologous GPI-anchored protein.
Figure 4.
The GPI-anchoring signal sequence is necessary for ERAD of PrP in A4 cells. (A) The turnover of GFP-GPIFR was analyzed in transiently transfected A4 and N2a cells by pulse-chase labeling and immunoprecipitation (with anti-GFP). Pulse labeling was for 15 min, followed by chase for the indicated times (in hours). (B) Left, fluorescence images of A4 and N2a cells transfected with GFP-GPIFR; right, enlarged views of A4 and N2a cells cotransfected with GFP-GPIFR (in green) and RFP-KDEL (in red). Yellow indicates colocalization of the two proteins. (C) The metabolism of PrP lacking the GPI-anchoring signal sequence (PrP-ΔGPI) was determined in A4 and parental N2a cells by pulse-chase analysis followed by immunoprecipitation of cell lysates (L) and culture media (M) with 3F4. Labeling was for 10 min, followed by chase for either 0 or 6 h. The positions of stably expressed PrP(A117V) and transiently transfected PrP-ΔGPI are indicated by the single and double asterisks, respectively. (D) Steady-state levels of PrP-Qa in N2a and A4 cells (detected using 3F4) in the presence (+) or absence (−) of MG132 for 6 h. (E) Unglycosylated PrP from tunicamycin-treated A4 cells was compared in its migration to in vitro–synthesized full-length PrP (FL), PrP lacking the N-terminal signal sequence (ΔSS), or PrP lacking both the N- and C-terminal signals (Δ/Δ). PrP was detected by immunoblotting using the 3F4 antibody.
Converse to the observations with GFP-GPI FR, conversion of PrP to a secretory protein by deletion of its GPI-anchoring signal sequence (PrP-ΔGPI) normalized its metabolism in A4 cells. Anchorless PrP was synthesized, core glycosylated (albeit inefficiently as described previously; Campana et al., 2007), and secreted into the medium in both A4 and parental N2a cells with equal efficiency (Figure 4C). PrP(A117V) expressed in the same A4 cells as PrP-ΔGPI was completely degraded without any secretion into the media. Reintroducing an unrelated GPI anchor (from the Qa protein) onto PrP (PrP-Qa) restored the differences between the N2a and A4 cells (Figure 4D). These findings suggested that A4 cells are selectively deficient in the proper trafficking and/or metabolism of proteins that contain a GPI-anchoring signal sequence. For PrP, ER retention and ERAD in A4 cells can be bypassed by deleting the GPI-anchoring signal sequence, whereas the addition of a GPI signal to GFP was sufficient to alter its trafficking.
Degradation of PrP in A4 Cells Is Due to Lack of Addition of a GPI Anchor
Requirement of the GPI-anchoring sequence for altered PrP metabolism in A4 cells can be explained in two possible ways. One possibility is that the A4 cells are defective in the trafficking of GPI-anchored proteins out of the ER. In this view, GPI-anchored proteins are retained in the ER despite their proper maturation and, depending on features of the mature protein, are routed for ERAD. Alternatively, A4 cells could be defective in the correct processing of the GPI signal sequence. Hence, substrates would either remain unprocessed or modified with an immature GPI anchor, resulting in a feature that could be recognized for ER retention and degradation.
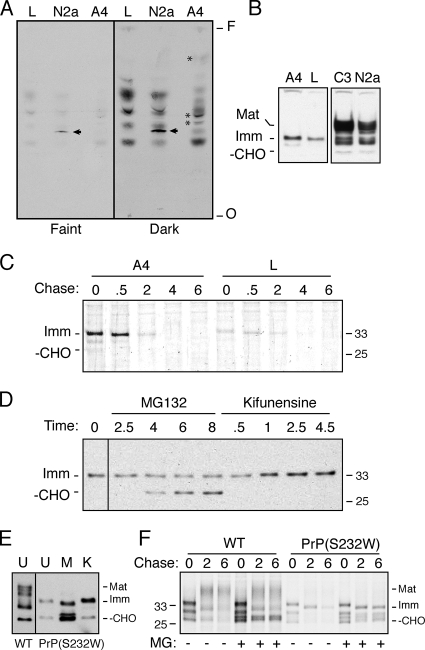
Several observations argued for a processing rather than trafficking defect. First, prolonged retention of PrP in the ER with BFA was insufficient to route PrP for ERAD in N2a cells (Figure 5A). Second, PrP synthesized in A4 cells (but not in N2a cells) fractionated predominantly into the aqueous phase of a TX-114 phase partitioning experiment (Figure 5B), arguing against an attached lipid. Third, PrP(A117V) synthesized in A4 cells migrated slightly slower than PrP-ΔGPI expressed in the same cells (Figure 4C). And finally, PrP(A117V) from A4 cells comigrates precisely with a form of PrP whose N-terminal signal has been removed, but whose C-terminal GPI anchor signal had not be processed (Figure 4E). These data indicate that PrP made in A4 cells does not contain the hydrophobic GPI lipid anchor and based on its migration, retains an uncleaved GPI-anchoring signal sequence. Retention of the GPI signal would also explain the difference in migration seen with GFP-GPI FR expressed in A4 versus N2a cells. Thus, A4 cells are defective in GPI processing such that an unprocessed GPI signal sequence may be retained on the substrate, leading to its altered trafficking and metabolism.
Figure 5.
A4 cells are defective in GPI signal processing rather than post-ER trafficking. (A) Pulse-chase analysis of transiently transfected WT PrP in N2a cells in the absence (−) or presence (+) of 10 μg/ml brefeldin A (BFA), a drug that prevents ER-to-Golgi trafficking. (B) Total lysates (T) from untransfected A4 and N2a cells were prepared in TX-114 containing buffer and separated into aqueous (A) and hydrophobic (H) phases. These samples were immunoblotted with the PrP-A antibody. Note that recovery of material from the hydrophobic phase is less efficient under our assay conditions. The asterisk denotes a nonspecific soluble protein detected by the PrP-A antibody that serves as a control for complete and comparable recovery of proteins from the aqueous phase of both cell types.
Processing of the GPI-anchoring signal is mediated by the ER resident GPI-transamidase enzyme complex that recognizes and cleaves the signal on a substrate and replaces it with a preassembled GPI anchor. Hence, the presence of an uncleaved GPI signal sequence in A4 cells could be due to either a defective transamidase enzyme or the inability to synthesize a fully mature GPI lipid anchor. To distinguish between these possibilities, we performed TLC of glycolipids from A4 cells labeled with [3H]mannose. As controls, we used the glycolipid profile of the parental N2a cells (as a marker for mature GPIs) and lipids from a cell line known to be defective in the first step of GPI anchor biosynthesis (LM-TK− cells or L-cells) as a control for non-GPI mannose-containing lipids. Neither L-cells nor A4 cells contained the mature GPI lipid anchor seen in N2a cells (Figure 6A, left panel). Even upon overexposure of the TLC (right panel), mature GPI lipid could not be observed in the A4 cells, although putative immature species could now be seen (asterisks, Figure 6A). This argues that the efficient routing of PrP (and various mutants) to ERAD in A4 cells can be traced to a deficiency in mature GPI anchors, leading to deficient GPI transamidation and an uncleaved GPI-anchoring signal sequence that routes PrP for ERAD.
Figure 6.
Defects in GPI anchor biosynthesis in A4 and L-cells routes PrP for ERAD. (A) A4, N2a, and GPI anchor-deficient L-cells were labeled with [3H]mannose for 2 h, extracted with organic solvent to isolate the glycolipid fraction, and analyzed by TLC and autoradiography. The arrowhead points to the fully mature GPI lipid anchor, the major mannose-containing lipid in N2a cells that is lacking in both A4 and L-cells. The darker exposure on the right revealed several presumably immature mannolipid species (asterisks) present in A4 cells that are not found in L-cells. O, origin; F, solvent front. (B) Total cell lysates from untransfected A4, N2a, C3 and L-cells were immunoblotted using the PrP-A antibody to detect steady-state levels of endogenously expressed PrP. (C) Biosynthesis and maturation of endogenous PrP in A4 and L-cells were assessed by pulse-chase analysis followed by immunoprecipitation using the PrP-A antibody. Pulse labeling with [35S]methionine was for 10 min, followed by chase for the indicated times (in hours). (D) Steady-state levels of PrP in L-cells (detected using PrP-A antibody) after treatments with 5 μM MG132 or 1 μM kifunensine for the indicated times (in hours). (E) Steady-state levels of PrP(S232W) in N2a cells (detected using 3F4) after treatments with 5 μM MG132 (M) or 1 μM kifunensine (K) for 4 h. U, untreated cell lysates. Untreated lysates from WT PrP expressing cells are shown for comparison (WT). (F) Pulse-chase analysis of WT and PrP(S232W) in N2a cells performed in the absence (−) or presence (+) of 5 μM MG132.
To further validate and generalize this conclusion, we performed two additional experiments. First, we analyzed PrP metabolism in the GPI anchor defective L-cells in which PrP is predicted to be routed for constitutive ERAD exactly as in A4 cells. Steady-state analysis of the endogenous PrP expressed in these mouse fibroblasts revealed only immature glycoforms that comigrated with PrP made in A4 cells (Figure 6B). Pulse-chase analysis revealed that L-cells synthesize and target PrP for degradation with approximately the same kinetics as in A4 cells (Figure 6C). In both steady-state and pulse-chase experiments, treatment with MG132 and kifunensine confirmed that L-cells employ the classical ERAD pathway (Figure 6D). Second, we generated a PrP mutant that cannot be transamidated because of the introduction of a bulky residue at the +1 position of the GPI-anchoring sequence [PrP(S232W); Kodukula et al., 1993]. On expression in normal N2a cells, PrP(S232W) was synthesized exclusively in immature forms and was expressed at very low steady-state levels. Treatment with MG132 or kifunensine resulted in stabilization of immature PrP forms, suggesting that PrP(S232W) was subject to constitutive ERAD in normal cells (Figure 6E). Pulse-chase analysis of PrP(S232W) in the absence and presence of MG132 confirmed its constitutive degradation by a proteasome-dependent pathway (Figure 6F). Taken together, these data demonstrate that wild-type and mutant PrPs can be routed extremely efficiently into the retrotranslocation and ERAD pathway by preventing normal processing of the GPI-anchoring signal. Consistent with this conclusion, a genetic disease causing PrP mutant (Q217R) that partially interferes with GPI signal sequence processing leads to ER retention of the unprocessed PrP (Singh et al., 1997).
A Role for the Mature Domain in the Fate of Misprocessed GPI-anchored Proteins
GFP-GPI FR is not substantially shortened in its half-life in A4 cells (relative to N2a cells) and has a much slower turnover rate than PrP (compare Figures 2A and 4A). Yet, both proteins are efficiently retained in the ER and prevented from trafficking to the cell surface in A4 cells. These observations indicated that a GPI-anchoring signal is not in itself sufficient to direct a substrate efficiently to ERAD, suggesting a role for the mature domain in determining the fate of unprocessed GPI-anchored proteins. Indeed, GFP-GPIFR turnover in A4 cells, despite its retention in the ER, does not occur via a pathway sensitive to proteasome inhibition (Figure 7A). Surprisingly, even the addition of a glycan at any of three different positions on GFP-GPIFR (GFP-GPI-CHO) was insufficient to increase the degradation rate of GFP-GPI FR or route any of it into a pathway that is inhibitable by either kifunensine or MG132 (Figure 7A). Similar results were seen with two other GFP-GPI-CHO constructs with glycans at different positions (data not shown). Conversely, preventing the glycosylation of PrP by tunicamycin treatment of A4 cells had no effect on its rate of degradation (Figure 7B). Thus, although glycosylated PrP follows (at least partially) an ERAD pathway that is sensitive to inhibitors of glycan trimming, a glycan is not obligatory for ERAD of PrP with an unprocessed GPI-anchoring signal sequence.
We also considered the possibility that the PrP GPI sequence contained a unique feature not shared by other GPI sequences, which could be used to route the protein for ERAD. To test this idea, we generated a GFP-GPI construct using the GPI signal sequence from PrP (GFP-GPIPrP). Pulse-chase analysis showed that GFP-GPIPrP, similar to GFP-GPIFR and in contrast to PrP, is refractory to rapid degradation in A4 cells (Figure 7C). This demonstrates that the PrP GPI signal sequence is not the sole determinant of ERAD for PrP because a heterologous protein cannot be routed for degradation in A4 cells with just this sequence. Instead, highly efficient ERAD in A4 cells requires not only an unprocessed GPI-anchoring signal sequence, but also as yet unidentified features of the mature domain that are found in PrP, but not in GFP. Neither the PrP mature domain (e.g., PrP-ΔGPI) nor a GPI-anchoring sequence (e.g., GFP-GPIFR and GFP-GPIPrP) alone was sufficient for efficient ERAD in A4 cells. Only when PrP contains an unprocessed GPI-anchoring signal (either its native sequence or the one from Qa) is it routed for efficient ERAD.
As targeting of misfolded or misprocessed proteins for ERAD is thought to be mediated by coassociating chaperones (Nakatsukasa and Brodsky, 2008), it seemed plausible that the different fates of PrP and GFP-GPI FR involved differences in their associations. Nondenaturing immunoprecipitations of PrP and GFP-GPIFR from radiolabeled A4 cells (Figure 7E) did in fact reveal differences in coprecipitating proteins (the identities of which remain unknown). One potential difference could be an association with protein disulfide isomerase (PDI), an oxido-reductase and chaperone known to make contact with PrP during its initial biosynthesis at the ER (Kim and Hegde, 2002). By contrast, GFP does not contain any disulfide bonds, and, based on sequential immunoprecipitation experiments, does not seem to coassociate with PDI (data not shown). This raised the possibility that PDI (and/or related oxido-reductases) might play a role in the differential routing of PrP and GFP-GPIFR to ERAD in A4 cells.
To test this idea, we analyzed ERAD of PrP in pulse-chase experiments performed on A4 cells under reducing conditions (with 10 mM DTT) that inhibit the chaperone activity of PDI (Tsai et al., 2001). PrP degradation was completely inhibited under these conditions, but was largely restored upon removal of the reducing agent (Figure 7D). This effect was not simply a consequence of ER stress caused by reducing conditions, because stress induced by a different method (ER Ca2+ depletion by thapsigargin) did not preclude PrP degradation (Figure 7D). Furthermore, we could demonstrate by sequential IP analyses that at least a proportion of PrP is coassociated with PDI in pulse-labeled A4 cells (Figure 7F). These results are consistent with the involvement of redox-sensitive factor(s) in the routing of PrP for ERAD. As PrP, known to interact with PDI during its biosynthesis, can be coprecipitated with PDI from A4 cells, the PDI chaperone cycle is inhibited by reducing conditions (Tsai et al., 2001) and PDI cysteine-mutants have been shown to be retained in the ER by PDI (Capellari et al., 1999), PDI is a plausible candidate for facilitating ERAD of PrP. Furthermore, this could also explain the lack of ERAD for GFP-GPIFR because this protein does not contain disulfide bonds in its folded state and does not seem to interact with PDI. Although the mechanistic details of the pathway(s) used for PrP degradation in A4 cells remain to be elucidated, the comparative analyses of PrP and GFP-GPIFR have revealed an unanticipated heterogeneity in the fate of misprocessed GPI-anchored proteins that depends directly on features of the mature domain, perhaps as a consequence of its different chaperone interactions.
DISCUSSION
In this study, our analysis of the A4 mutant cell line that arose spontaneously as a consequence of stable overexpression of disease-causing mutant PrP led to the identification of a pathway for highly efficient posttranslational attenuation of PrP expression. The defect in A4 cells proved to be in the synthesis of mature GPI lipid anchors with consequent misprocessing of GPI-anchored proteins. The fate of a GPI-anchored protein in A4 cells was found to be surprisingly dependent on the mature domain, indicating a previously unanticipated heterogeneity in the QC pathways that dispose of proteins containing unprocessed GPI-anchoring signal sequences. In the case of PrP and disease-causing PrP mutants, failure to process the GPI-anchoring signal sequence leads to its quantitative retrotranslocation and proteasome-dependent degradation by a redox-sensitive pathway possibly involving PDI family members. By contrast, a heterologous GPI-anchored protein was retained in the ER and turned over with slow kinetics by an as yet uncharacterized nonproteasomal pathway. Our results illustrate the utility of somatic cell mutant generation and analyses as a means for uncovering adaptive pathways of toxic protein down-regulation. The implications of these findings for PrP-associated diseases, as well as general insights into the QC of GPI-anchored proteins are discussed in subsequent sections below.
Implications for PrP Biology
Combined with previous data on PrP metabolism, GPI anchor addition, and ERAD pathways, our results suggest a working model for the regulation of PrP degradation and trafficking by GPI signal sequence transamidation (Figure 8). During its cotranslational translocation into the ER lumen (step 1), PrP makes contacts with chaperones including PDI (Kim and Hegde, 2002). On completion of synthesis, the GPI-anchoring signal sequence (which is typically not hydrophobic or long enough to form a transmembrane domain) is transported through the translocon into the ER lumen (Dalley and Bulleid, 2003) before interacting with the GPI8 subunit of the GPI transamidase complex (Spurway et al., 2001; Vidugiriene et al., 2001; Chen et al., 2003). When fully assembled GPI anchors are available, the signal-transamidase-GPI anchor complex undergoes a concerted cleavage and transamidation reaction that replaces the GPI signal sequence with the GPI anchor (steps 2 and 3; Orlean and Menon, 2007). In the absence of GPI anchors (e.g., in A4 or L-cells), PrP retains the GPI signal sequence and, based on coimmunoprecipitation analysis (Figure 7F), may remain in association with chaperones including PDI. We postulate that an inability of PrP to fold correctly in the presence of an unprocessed GPI signal sequence allows its prolonged association with the PDI-containing chaperone complex. This association presumably maintains PrP molecules (including various mutants) in a largely unfolded state that contemporary models posit is needed for retrotranslocation. Subsequently, an interaction between PDI and Derlin (Bernardi et al., 2007; Schelhaas et al., 2007) would deliver PrP substrates to the retrotranslocation machinery for export out of the ER, ubiquitination, deglycosylation, and proteasomal degradation (steps 4 and 5).
Figure 8.
Working model for the regulation of PrP degradation and trafficking by GPI signal sequence transamidation. (1) Interaction of PrP with PDI (green) during cotranslational translocation into the ER. (2) Recognition of and interaction with the GPI-anchoring signal sequence (red) by the GPI transamidase enzyme complex (blue) leads to the replacement of the GPI signal sequence with a preassembled GPI lipid anchor in cells (e.g., N2a) where GPI anchor biosynthesis is intact (3). This lipid anchored PrP species is now competent for ER exit. PrP species that contain a GPI signal sequence that is inserted into the ER membrane as a transmembrane segment and anchorless PrP generated by transamidation of the signal by a nucleophile such as water are also species that remain competent for trafficking out of the ER. (4) In cells defective in GPI anchor synthesis (e.g., A4 and L-cells) the GPI signal-containing form of PrP may maintain prolonged interactions with chaperones such as PDI. (5) These interactions eventually deliver misprocessed PrP to the ERAD pathway, perhaps through interaction of PDI with the Derlin associated retrotranslocation machinery.
For some substrates or under some conditions, a failure in transamidation may lead the GPI signal sequence to insert into the membrane as a transmembrane segment (Waneck et al., 1988). For PrP, such insertion might shield the hydrophobic GPI anchor signal in the lipid bilayer, thereby allowing the mature domain to fold correctly and traffic to the cell surface (Kaneko et al., 1997). Hence, the key event in routing unprocessed PrP for degradation could be an uncleaved GPI signal that remains unshielded by the membrane. Another potential route for PrP to escape the ER in GPI-deficient cells is if the transamidation reaction proceeds with another nucleophile (such as water), as can occur for model GPI proteins in vitro (Maxwell et al., 1995) and in some cases, in vivo (Lisanti et al., 1991). In this instance, PrP would essentially be anchorless, resulting in its secretion (Campana et al., 2007; dotted arrows, step 3).
This model indicates that preventing processing of the GPI signal sequence from PrP can be used to posttranslationally attenuate its expression. Although inhibiting all or most GPI signal processing (as occurs in L-cells or A4 cells) is presumably not a viable therapeutic strategy in vivo, our results point to the interaction between GPI transamidase and the GPI-anchoring signal sequence of PrP (step 2 in Figure 8) as a potentially selective step for perturbation. The transamidase-signal interaction is poorly understood at present, but appears to depend critically on a central hydrophobic core common to all GPI-anchoring signal sequences. Despite this shared feature, GPI signals are highly variable from substrate to substrate and typically show little or no sequence homology to each other. This means that although general biophysical parameter(s) such as overall hydrophobicity are being recognized, each signal–transamidase interaction is unique in subtle ways. Hence, these substrate-specific differences may be amenable to selective small molecule perturbation. Precedent for this concept was recently provided by studies of N-terminal ER targeting signals which, like GPI signals, share only general features such as hydrophobicity (von Heijne, 1985) that nonetheless suffice for recognition by both SRP and the Sec61 translocon (Rapoport, 2007). Remarkably, this signal-Sec61 interaction was not only shown to differ subtly from substrate to substrate (Kim and Hegde, 2002), but also amenable to substrate-selective perturbation by small molecules (Besemer et al., 2005; Garrison et al., 2005) that inhibit the activity of only a small subset of signal sequences. The target of these translocation inhibitors proved to be Sec61 (MacKinnon et al., 2007), illustrating that a rather generic recognition event mediated by a “housekeeping” protein can nonetheless be exploited for selective inhibition.
Whether down-regulation of PrP via retrotranslocation and ERAD would be beneficial or detrimental remains a matter of considerable debate because of cytosolic PrP (cyPrP) has been suggested to be protective (Roucou et al., 2003), toxic (Ma et al., 2002; Wang et al., 2005), or inert (Mironov et al., 2003). The toxicity of cyPrP was further used to suggest that presumptive low-level retrotranslocation of disease-causing PrP mutants might be the basis of their neurodegenerative phenotype. Yet, we found no evidence of toxicity even when PrP or three independent mutants (A117V, H187R, E200K) were overexpressed and quantitatively routed for retrotranslocation and ERAD. These apparently contradictory results can potentially be explained by the fact that enforced expression of PrP directly in the cytosol (by deletion of its N- and C-terminal signal sequences) is qualitatively different from retrotranslocation of PrP from the ER lumen. The former never enters the ER and is handled by different QC pathways (e.g., cytosolic chaperones) than proteins retrotranslocated from the ER. Thus, a very tight coupling of retrotranslocation with ubiquitination and degradation may essentially never fully expose PrP to the cytosol during ERAD, whereas signal-deleted PrP would undergo rounds of attempted folding in the cytosol before being routed for degradation. Our description of a cleavage-deficient PrP that undergoes constitutive retrotranslocation (Figure 6, E and F) could be used to definitively assess in transgenic mice the controversial pathological (Ma et al., 2002) versus protective (Roucou et al., 2003) roles for PrP retrotranslocation.
Implications for ER Quality Control
An unexpected finding from this study is that an uncleaved GPI signal sequence is not sufficient to mediate QC and ERAD of all proteins by the same pathway. Although an unprocessed terminal hydrophobic domain would seem to be an ideal degradation motif, this proved overly simplistic. A well-folded mature domain such as GFP, while being retained in the ER, was not retrotranslocated to an appreciable extent. Even the addition of glycans, which presumably alters the chaperones that bind to GFP (Molinari and Helenius, 2000), did not influence its degradation. Yet, GFP is not intrinsically resistant to retrotranslocation because it has been appended to the lumenal side of transmembrane proteins that remain competent for ERAD. Thus, the ER retention mediated by an unprocessed GPI signal sequence can be uncoupled from its routing into ERAD. This conclusion may help explain why several previous studies on the fate of unprocessed GPI-anchored proteins have sometimes arrived at conflicting conclusions. For example, an especially thorough early study of GPI-anchored proteins in anchor-deficient L-cells demonstrated degradation by a pre-Golgi pathway (which presumably is the currently known ERAD pathway) for both native and heterologous proteins (Delahunty et al., 1993). On the basis of these results, our finding of PrP degradation by ERAD in anchor-deficient cells would seem predictable. Yet, several studies manipulating the GPI anchor of PrP have failed to demonstrate efficient ERAD (Taraboulos et al., 1995; Kaneko et al., 1997; Winklhofer et al., 2003; Kiachopoulos et al., 2005), and studies of other unprocessed GPI-anchored proteins have suggested the involvement of a non-ER, possibly autophagic, pathway (Field et al., 1994). Similarly, although we find that PrP degradation is inhibited by reducing conditions, other studies have found these same conditions stimulatory for degradation of unprocessed GPI-anchored proteins (Wainwright and Field, 1997). One reason for these different conclusions probably lies in the different substrates being analyzed in the different studies. Thus, the GPI signal, while specifying ER retention and degradation, does not do so by a single mechanism.
A second parameter that may be important in interpreting these studies involves the means of generating the unprocessed GPI-anchored substrate for analysis. Several studies have analyzed GPI-anchored proteins with a mutated (and uncleavable) signal as a model. However, it is not always clear that a mutated anchoring signal would be equivalent in its metabolism to a native anchoring sequence that fails to be processed. For example, certain mutants that inhibit cleavage do so by essentially converting the GPI signal to a transmembrane domain (TMD), which for PrP allows its efficient trafficking to the cell surface (Kaneko et al., 1997). Similarly, other mutants designed to prevent cleavage may simultaneously alter the properties of the GPI signal (or result in cleavage at alternative sites) such that the protein's behavior is not directly analogous to an uncleaved native GPI signal. This may explain why a presumably uncleavable mutant of the PrP GPI signal led to secretion rather than ERAD in an earlier study (Winklhofer et al., 2003). Because at present, the parameters that influence ER retention and degradation of unprocessed GPI-anchored proteins remain poorly defined, the use of GPI signal mutants should be evaluated with caution.
At present, it is not clear how GFP-GPIFR is turned over in A4 cells and why it is incompetent for ERAD. Typically, excessive aggregation or saturation of ERAD results in rerouting to other yet uncharacterized pathways for degradation of misfolded ER proteins. In this case, neither seems plausible because the GFP fluorescence was homogeneously distributed throughout the ER and its expression is comparable to PrP. We have observed that a small proportion of GFP is secreted into the media after cleavage at a furin-like protease site near the C-terminus of GFP (data not shown), but this minor population is insufficient to explain its turnover. In stark contrast to GFP-GPIFR, PrP is a robust retrotranslocation substrate regardless of its glycosylation state or disease-causing mutations. This strong dependence on the mature domain for the pathway of degradation, together with the different fates for a GPI-anchoring signal in the absence of transamidation (step 3 in Figure 8), may explain otherwise conflicting reports regarding the metabolism of unprocessed GPI-anchored proteins ranging from ERAD, ER retention, post-ER accumulation, secretion, or cell surface expression. It will therefore be important to now dissect the parameters of the mature domain that influence the pathways of degradation. An important clue in this respect may be the different chaperone(s) that differentially associate with proteins of different characteristics. The differential metabolism of PrP and GFP-GPIFR in A4 cells may provide an important model system to study the distinct pathways available to unprocessed GPI-anchored proteins for degradation.
ACKNOWLEDGMENTS
We are grateful to N. Rane for generating and first noticing the behavior of A4 cells; to C. Wunder and A. Menon for advice on GPI and TLC analysis; J. Bonifacino (NIH, Bethesda, MD) and J. Lippincott-Schwartz (NIH) for reagents and useful discussions; G. Patterson for help with confocal microscopy; R. Schülein (FMP, Berlin, Germany) for constructs; M. Lehrman for advice on tunicamycin use; and our lab members for useful discussions. This work was supported by the intramural research program of the National Institute of Child Health and Human Development at the National Institutes of Health.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-01-0087) on May 28, 2008.
REFERENCES
- Aguzzi A., Heikenwalder M. Pathogenesis of prion diseases: current status and future outlook. Nat. Rev. Microbiol. 2006;4:765–775. doi: 10.1038/nrmicro1492. [DOI] [PubMed] [Google Scholar]
- Baumann F., Tolnay M., Brabeck C., Pahnke J., Kloz U., Niemann H. H., Heikenwalder M., Rulicke T., Burkle A., Aguzzi A. Lethal recessive myelin toxicity of prion protein lacking its central domain. EMBO J. 2007;26:538–547. doi: 10.1038/sj.emboj.7601510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi K. M., Forster M. L., Lencer W. I., Tsai B. Derlin-1 facilitates the retro-translocation of cholera toxin. Mol. Biol. Cell. 2008;19:877–884. doi: 10.1091/mbc.E07-08-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besemer J., Harant H., Wang S., Oberhauser B., Marquardt K., Foster C. A., Schreiner E. P., de Vries J. E., Dascher-Nadel C., Lindley I. J. Selective inhibition of cotranslational translocation of vascular cell adhesion molecule 1. Nature. 2005;436:290–293. doi: 10.1038/nature03670. [DOI] [PubMed] [Google Scholar]
- Borchelt D. R., Taraboulos A., Prusiner S. B. Evidence for synthesis of scrapie prion proteins in the endocytic pathway. J. Biol. Chem. 1992;267:16188–16199. [PubMed] [Google Scholar]
- Brandner S., Isenmann S., Raeber A., Fischer M., Sailer A., Kobayashi Y., Marino S., Weissmann C., Aguzzi A. Normal host prion protein necessary for scrapie-induced neurotoxicity. Nature. 1996;379:339–343. doi: 10.1038/379339a0. [DOI] [PubMed] [Google Scholar]
- Bueler H., Aguzzi A., Sailer A., Greiner R. A., Autenried P., Aguet M., Weissmann C. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- Bueler H., Raeber A., Sailer A., Fischer M., Aguzzi A., Weissmann C. High prion and PrPSc levels but delayed onset of disease in scrapie-inoculated mice heterozygous for a disrupted PrP gene. Mol. Med. 1994;1:19–30. [PMC free article] [PubMed] [Google Scholar]
- Campana V., Caputo A., Sarnataro D., Paladino S., Tivodar S., Zurzolo C. Characterization of the properties and trafficking of an anchorless form of the prion protein. J. Biol. Chem. 2007;282:22747–22756. doi: 10.1074/jbc.M701468200. [DOI] [PubMed] [Google Scholar]
- Cancellotti E., Wiseman F., Tuzi N. L., Baybutt H., Monaghan P., Aitchison L., Simpson J., Manson J. C. Altered glycosylated PrP proteins can have different neuronal trafficking in brain but do not acquire scrapie-like properties. J. Biol. Chem. 2005;280:42909–42918. doi: 10.1074/jbc.M509557200. [DOI] [PubMed] [Google Scholar]
- Capellari S., Zaidi S. I., Urig C. B., Perry G., Smith M. A., Petersen R. B. Prion protein glycosylation is sensitive to redox change. J. Biol. Chem. 1999;274:34846–34850. doi: 10.1074/jbc.274.49.34846. [DOI] [PubMed] [Google Scholar]
- Caughey B., Raymond G. J. The scrapie-associated form of PrP is made from a cell surface precursor that is both protease- and phospholipase-sensitive. J. Biol. Chem. 1991;266:18217–18223. [PubMed] [Google Scholar]
- Chen R., Anderson V., Hiroi Y., Medof M. E. Proprotein interaction with the GPI transamidase. J. Cell. Biochem. 2003;88:1025–1037. doi: 10.1002/jcb.10439. [DOI] [PubMed] [Google Scholar]
- Chiesa R., Piccardo P., Ghetti B., Harris D. A. Neurological illness in transgenic mice expressing a prion protein with an insertional mutation. Neuron. 1998;21:1339–1351. doi: 10.1016/s0896-6273(00)80653-4. [DOI] [PubMed] [Google Scholar]
- Cohen F. E., Kelly J. W. Therapeutic approaches to protein-misfolding diseases. Nature. 2003;426:905–909. doi: 10.1038/nature02265. [DOI] [PubMed] [Google Scholar]
- Dalley J. A., Bulleid N. J. The endoplasmic reticulum (ER) translocon can differentiate between hydrophobic sequences allowing signals for glycosylphosphatidylinositol anchor addition to be fully translocated into the ER lumen. J. Biol. Chem. 2003;278:51749–51757. doi: 10.1074/jbc.M303978200. [DOI] [PubMed] [Google Scholar]
- Davidson N. O., Shelness G. S. APOLIPOPROTEIN B: mRNA editing, lipoprotein assembly, and presecretory degradation. Annu. Rev. Nutr. 2000;20:169–193. doi: 10.1146/annurev.nutr.20.1.169. [DOI] [PubMed] [Google Scholar]
- Delahunty M. D., Stafford F. J., Yuan L. C., Shaz D., Bonifacino J. S. Uncleaved signals for glycosylphosphatidylinositol anchoring cause retention of precursor proteins in the endoplasmic reticulum. J. Biol. Chem. 1993;268:12017–12027. [PubMed] [Google Scholar]
- Drisaldi B., Stewart R. S., Adles C., Stewart L. R., Quaglio E., Biasini E., Fioriti L., Chiesa R., Harris D. A. Mutant PrP is delayed in its exit from the endoplasmic reticulum, but neither wild-type nor mutant PrP undergoes retrotranslocation prior to proteasomal degradation. J. Biol. Chem. 2003;278:21732–21743. doi: 10.1074/jbc.M213247200. [DOI] [PubMed] [Google Scholar]
- Ellgaard L., Helenius A. ER quality control: towards an understanding at the molecular level. Curr. Opin. Cell Biol. 2001;13:431–437. doi: 10.1016/s0955-0674(00)00233-7. [DOI] [PubMed] [Google Scholar]
- Field M. C., Moran P., Li W., Keller G. A., Caras I. W. Retention and degradation of proteins containing an uncleaved glycosylphosphatidylinositol signal. J. Biol. Chem. 1994;269:10830–10837. [PubMed] [Google Scholar]
- Fons R. D., Bogert B. A., Hegde R. S. Substrate-specific function of the translocon-associated protein complex during translocation across the ER membrane. J. Cell Biol. 2003;160:529–539. doi: 10.1083/jcb.200210095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison J. L., Kunkel E. J., Hegde R. S., Taunton J. A substrate-specific inhibitor of protein translocation into the endoplasmic reticulum. Nature. 2005;436:285–289. doi: 10.1038/nature03821. [DOI] [PubMed] [Google Scholar]
- Ghaedi K., Kawai A., Okumoto K., Tamura S., Shimozawa N., Suzuki Y., Kondo N., Fujiki Y. Isolation and characterization of novel peroxisome biogenesis-defective Chinese hamster ovary cell mutants using green fluorescent protein. Exp. Cell Res. 1999;248:489–497. doi: 10.1006/excr.1999.4413. [DOI] [PubMed] [Google Scholar]
- Goldstein J. L., Rawson R. B., Brown M. S. Mutant mammalian cells as tools to delineate the sterol regulatory element-binding protein pathway for feedback regulation of lipid synthesis. Arch. Biochem. Biophys. 2002;397:139–148. doi: 10.1006/abbi.2001.2615. [DOI] [PubMed] [Google Scholar]
- Grantcharova E., Furkert J., Reusch H. P., Krell H. W., Papsdorf G., Beyermann M., Schulein R., Rosenthal W., Oksche A. The extracellular N terminus of the endothelin B (ETB) receptor is cleaved by a metalloprotease in an agonist-dependent process. J. Biol. Chem. 2002;277:43933–43941. doi: 10.1074/jbc.M208407200. [DOI] [PubMed] [Google Scholar]
- Hampton R. Y. ER-associated degradation in protein quality control and cellular regulation. Curr. Opin. Cell Biol. 2002;14:476–482. doi: 10.1016/s0955-0674(02)00358-7. [DOI] [PubMed] [Google Scholar]
- Hatahet F., Ruddock L. W. Substrate recognition by the protein disulfide isomerases. FEBS J. 2007;274:5223–5234. doi: 10.1111/j.1742-4658.2007.06058.x. [DOI] [PubMed] [Google Scholar]
- Hebert D. N., Molinari M. In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiol. Rev. 2007;87:1377–1408. doi: 10.1152/physrev.00050.2006. [DOI] [PubMed] [Google Scholar]
- Hegde R. S., Mastrianni J. A., Scott M. R., DeFea K. A., Tremblay P., Torchia M., DeArmond S. J., Prusiner S. B., Lingappa V. R. A transmembrane form of the prion protein in neurodegenerative disease. Science. 1998;279:827–834. doi: 10.1126/science.279.5352.827. [DOI] [PubMed] [Google Scholar]
- Hegde R. S., Tremblay P., Groth D., DeArmond S. J., Prusiner S. B., Lingappa V. R. Transmissible and genetic prion diseases share a common pathway of neurodegeneration. Nature. 1999;402:822–826. doi: 10.1038/45574. [DOI] [PubMed] [Google Scholar]
- Hirose S., Mohney R. P., Mutka S. C., Ravi L., Singleton D. R., Perry G., Tartakoff A. M., Medof M. E. Derivation and characterization of glycoinositol-phospholipid anchor-defective human K562 cell clones. J. Biol. Chem. 1992;267:5272–5278. [PubMed] [Google Scholar]
- Hyman R. Somatic genetic analysis of the expression of cell surface molecules. Trends Genet. 1988;4:5–8. doi: 10.1016/0168-9525(88)90120-5. [DOI] [PubMed] [Google Scholar]
- Kaneko K., Vey M., Scott M., Pilkuhn S., Cohen F. E., Prusiner S. B. COOH-terminal sequence of the cellular prion protein directs subcellular trafficking and controls conversion into the scrapie isoform. Proc. Natl. Acad. Sci. USA. 1997;94:2333–2338. doi: 10.1073/pnas.94.6.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S. W., Rane N. S., Kim S. J., Garrison J. L., Taunton J., Hegde R. S. Substrate-specific translocational attenuation during ER stress defines a pre-emptive quality control pathway. Cell. 2006;127:999–1013. doi: 10.1016/j.cell.2006.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiachopoulos S., Bracher A., Winklhofer K. F., Tatzelt J. Pathogenic mutations located in the hydrophobic core of the prion protein interfere with folding and attachment of the glycosylphosphatidylinositol anchor. J. Biol. Chem. 2005;280:9320–9329. doi: 10.1074/jbc.M412525200. [DOI] [PubMed] [Google Scholar]
- Kim S. J., Hegde R. S. Cotranslational partitioning of nascent prion protein into multiple populations at the translocation channel. Mol. Biol. Cell. 2002;13:3775–3786. doi: 10.1091/mbc.E02-05-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodukula K., Gerber L. D., Amthauer R., Brink L., Udenfriend S. Biosynthesis of glycosylphosphatidylinositol (GPI)-anchored membrane proteins in intact cells: specific amino acid requirements adjacent to the site of cleavage and GPI attachment. J. Cell Biol. 1993;120:657–664. doi: 10.1083/jcb.120.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemansky P., Gupta D. K., Meyale S., Tucker G., Tartakoff A. M. Atypical mannolipids characterize Thy-1-negative lymphoma mutants. Mol. Cell. Biol. 1991;11:3879–3885. doi: 10.1128/mcb.11.8.3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A., Christensen H. M., Stewart L. R., Roth K. A., Chiesa R., Harris D. A. Neonatal lethality in transgenic mice expressing prion protein with a deletion of residues 105–125. EMBO J. 2007;26:548–558. doi: 10.1038/sj.emboj.7601507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisanti M. P., Field M. C., Caras I. W., Menon A. K., Rodriguez-Boulan E. Mannosamine, a novel inhibitor of glycosylphosphatidylinositol incorporation into proteins. EMBO J. 1991;10:1969–1977. doi: 10.1002/j.1460-2075.1991.tb07726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Lindquist S. Wild-type PrP and a mutant associated with prion disease are subject to retrograde transport and proteasome degradation. Proc. Natl. Acad. Sci. USA. 2001;98:14955–14960. doi: 10.1073/pnas.011578098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Wollmann R., Lindquist S. Neurotoxicity and neurodegeneration when PrP accumulates in the cytosol. Science. 2002;298:1781–1785. doi: 10.1126/science.1073725. [DOI] [PubMed] [Google Scholar]
- MacKinnon A. L., Garrison J. L., Hegde R. S., Taunton J. Photo-leucine incorporation reveals the target of a cyclodepsipeptide inhibitor of cotranslational translocation. J. Am. Chem. Soc. 2007;129:14560–14561. doi: 10.1021/ja076250y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallucci G., Collinge J. Rational targeting for prion therapeutics. Nat. Rev. Neurosci. 2005;6:23–34. doi: 10.1038/nrn1584. [DOI] [PubMed] [Google Scholar]
- Mallucci G., Dickinson A., Linehan J., Klohn P. C., Brandner S., Collinge J. Depleting neuronal PrP in prion infection prevents disease and reverses spongiosis. Science. 2003;302:871–874. doi: 10.1126/science.1090187. [DOI] [PubMed] [Google Scholar]
- Mallucci G. R., White M. D., Farmer M., Dickinson A., Khatun H., Powell A. D., Brandner S., Jefferys J. G., Collinge J. Targeting cellular prion protein reverses early cognitive deficits and neurophysiological dysfunction in prion-infected mice. Neuron. 2007;53:325–335. doi: 10.1016/j.neuron.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Manson J. C., Clarke A. R., McBride P. A., McConnell I., Hope J. PrP gene dosage determines the timing but not the final intensity or distribution of lesions in scrapie pathology. Neurodegeneration. 1994;3:331–340. [PubMed] [Google Scholar]
- Maxwell S. E., Ramalingam S., Gerber L. D., Udenfriend S. Cleavage without anchor addition accompanies the processing of a nascent protein to its glycosylphosphatidylinositol-anchored form. Proc. Natl. Acad. Sci. USA. 1995;92:1550–1554. doi: 10.1073/pnas.92.5.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken A. A., Brodsky J. L. Recognition and delivery of ERAD substrates to the proteasome and alternative paths for cell survival. Curr. Top. Microbiol. Immunol. 2005;300:17–40. doi: 10.1007/3-540-28007-3_2. [DOI] [PubMed] [Google Scholar]
- Meusser B., Hirsch C., Jarosch E., Sommer T. ERAD: the long road to destruction. Nat. Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- Mironov A., Jr., Latawiec D., Wille H., Bouzamondo-Bernstein E., Legname G., Williamson R. A., Burton D., DeArmond S. J., Prusiner S. B., Peters P. J. Cytosolic prion protein in neurons. J. Neurosci. 2003;23:7183–7193. doi: 10.1523/JNEUROSCI.23-18-07183.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari M., Helenius A. Chaperone selection during glycoprotein translocation into the endoplasmic reticulum. Science. 2000;288:331–333. doi: 10.1126/science.288.5464.331. [DOI] [PubMed] [Google Scholar]
- Muramoto T., DeArmond S. J., Scott M., Telling G. C., Cohen F. E., Prusiner S. B. Heritable disorder resembling neuronal storage disease in mice expressing prion protein with deletion of an alpha-helix. Nat. Med. 1997;3:750–755. doi: 10.1038/nm0797-750. [DOI] [PubMed] [Google Scholar]
- Muramoto T., Scott M., Cohen F. E., Prusiner S. B. Recombinant scrapie-like prion protein of 106 amino acids is soluble. Proc. Natl. Acad. Sci. USA. 1996;93:15457–15462. doi: 10.1073/pnas.93.26.15457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsukasa K., Brodsky J. L. The Recognition and Retrotranslocation of Misfolded Proteins from the Endoplasmic Reticulum. Traffic. 2008 doi: 10.1111/j.1600-0854.2008.00729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuendorf E., Weber A., Saalmueller A., Schatzl H., Reifenberg K., Pfaff E., Groschup M. H. Glycosylation deficiency at either one of the two glycan attachment sites of cellular prion protein preserves susceptibility to bovine spongiform encephalopathy and scrapie infections. J. Biol. Chem. 2004;279:53306–53316. doi: 10.1074/jbc.M410796200. [DOI] [PubMed] [Google Scholar]
- Ni M., Lee A. S. ER chaperones in mammalian development and human diseases. FEBS Lett. 2007;581:3641–3651. doi: 10.1016/j.febslet.2007.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols B. J., Kenworthy A. K., Polishchuk R. S., Lodge R., Roberts T. H., Hirschberg K., Phair R. D., Lippincott-Schwartz J. Rapid cycling of lipid raft markers between the cell surface and Golgi complex. J. Cell Biol. 2001;153:529–541. doi: 10.1083/jcb.153.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka T., Nishijima M., Akamatsu Y. A somatic cell mutant defective in phosphatidylglycerophosphate synthase, with impaired phosphatidylglycerol and cardiolipin biosynthesis. J. Biol. Chem. 1993;268:22908–22913. [PubMed] [Google Scholar]
- Orlean P., Menon A. K. Thematic review series: lipid posttranslational modifications. GPI anchoring of protein in yeast and mammalian cells, or: how we learned to stop worrying and love glycophospholipids. J. Lipid Res. 2007;48:993–1011. doi: 10.1194/jlr.R700002-JLR200. [DOI] [PubMed] [Google Scholar]
- Otsu M., Sitia R. Diseases originating from altered protein quality control in the endoplasmic reticulum. Curr. Med. Chem. 2007;14:1639–1652. doi: 10.2174/092986707780830952. [DOI] [PubMed] [Google Scholar]
- Presley J. F., Cole N. B., Schroer T. A., Hirschberg K., Zaal K. J., Lippincott-Schwartz J. ER-to-Golgi transport visualized in living cells. Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- Prusiner S. B. The prion diseases. Brain Pathol. 1998;8:499–513. doi: 10.1111/j.1750-3639.1998.tb00171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner S. B., Groth D., Serban A., Koehler R., Foster D., Torchia M., Burton D., Yang S. L., DeArmond S. J. Ablation of the prion protein (PrP) gene in mice prevents scrapie and facilitates production of anti-PrP antibodies. Proc. Natl. Acad. Sci. USA. 1993;90:10608–10612. doi: 10.1073/pnas.90.22.10608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner S. B., Scott M., Foster D., Pan K. M., Groth D., Mirenda C., Torchia M., Yang S. L., Serban D., Carlson G. A., et al. Transgenetic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell. 1990;63:673–686. doi: 10.1016/0092-8674(90)90134-z. [DOI] [PubMed] [Google Scholar]
- Rane N. S., Yonkovich J. L., Hegde R. S. Protection from cytosolic prion protein toxicity by modulation of protein translocation. EMBO J. 2004;23:4550–4559. doi: 10.1038/sj.emboj.7600462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport T. A. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- Roucou X., Guo Q., Zhang Y., Goodyer C. G., LeBlanc A. C. Cytosolic prion protein is not toxic and protects against Bax-mediated cell death in human primary neurons. J. Biol. Chem. 2003;278:40877–40881. doi: 10.1074/jbc.M306177200. [DOI] [PubMed] [Google Scholar]
- Safar J. G., DeArmond S. J., Kociuba K., Deering C., Didorenko S., Bouzamondo-Bernstein E., Prusiner S. B., Tremblay P. Prion clearance in bigenic mice. J. Gen. Virol. 2005;86:2913–2923. doi: 10.1099/vir.0.80947-0. [DOI] [PubMed] [Google Scholar]
- Schelhaas M., Malmstrom J., Pelkmans L., Haugstetter J., Ellgaard L., Grunewald K., Helenius A. Simian Virus 40 depends on ER protein folding and quality control factors for entry into host cells. Cell. 2007;131:516–529. doi: 10.1016/j.cell.2007.09.038. [DOI] [PubMed] [Google Scholar]
- Shmerling D., Hegyi I., Fischer M., Blattler T., Brandner S., Gotz J., Rulicke T., Flechsig E., Cozzio A., von Mering C., Hangartner C., Aguzzi A., Weissmann C. Expression of amino-terminally truncated PrP in the mouse leading to ataxia and specific cerebellar lesions. Cell. 1998;93:203–214. doi: 10.1016/s0092-8674(00)81572-x. [DOI] [PubMed] [Google Scholar]
- Singh N., Zanusso G., Chen S. G., Fujioka H., Richardson S., Gambetti P., Petersen R. B. Prion protein aggregation reverted by low temperature in transfected cells carrying a prion protein gene mutation. J. Biol. Chem. 1997;272:28461–28470. doi: 10.1074/jbc.272.45.28461. [DOI] [PubMed] [Google Scholar]
- Snapp E. L., Sharma A., Lippincott-Schwartz J., Hegde R. S. Monitoring chaperone engagement of substrates in the endoplasmic reticulum of live cells. Proc. Natl. Acad. Sci. USA. 2006;103:6536–6541. doi: 10.1073/pnas.0510657103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurway T. D., Dalley J. A., High S., Bulleid N. J. Early events in glycosylphosphatidylinositol anchor addition. substrate proteins associate with the transamidase subunit gpi8p. J. Biol. Chem. 2001;276:15975–15982. doi: 10.1074/jbc.M010128200. [DOI] [PubMed] [Google Scholar]
- Stefanovic S., Hegde R. S. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell. 2007;128:1147–1159. doi: 10.1016/j.cell.2007.01.036. [DOI] [PubMed] [Google Scholar]
- Sugiyama E., DeGasperi R., Urakaze M., Chang H. M., Thomas L. J., Hyman R., Warren C. D., Yeh E. T. Identification of defects in glycosylphosphatidylinositol anchor biosynthesis in the Thy-1 expression mutants. J. Biol. Chem. 1991;266:12119–12122. [PubMed] [Google Scholar]
- Supattapone S., Bouzamondo E., Ball H. L., Wille H., Nguyen H. O., Cohen F. E., DeArmond S. J., Prusiner S. B., Scott M. A protease-resistant 61-residue prion peptide causes neurodegeneration in transgenic mice. Mol. Cell. Biol. 2001;21:2608–2616. doi: 10.1128/MCB.21.7.2608-2616.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraboulos A., Scott M., Semenov A., Avrahami D., Laszlo L., Prusiner S. B. Cholesterol depletion and modification of COOH-terminal targeting sequence of the prion protein inhibit formation of the scrapie isoform. J. Cell Biol. 1995;129:121–132. doi: 10.1083/jcb.129.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telling G. C., Haga T., Torchia M., Tremblay P., DeArmond S. J., Prusiner S. B. Interactions between wild-type and mutant prion proteins modulate neurodegeneration in transgenic mice. Genes Dev. 1996;10:1736–1750. doi: 10.1101/gad.10.14.1736. [DOI] [PubMed] [Google Scholar]
- Tsai B., Rodighiero C., Lencer W. I., Rapoport T. A. Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell. 2001;104:937–948. doi: 10.1016/s0092-8674(01)00289-6. [DOI] [PubMed] [Google Scholar]
- Vidugiriene J., Vainauskas S., Johnson A. E., Menon A. K. Endoplasmic reticulum proteins involved in glycosylphosphatidylinositol-anchor attachment: photocrosslinking studies in a cell-free system. Eur. J. Biochem. 2001;268:2290–2300. doi: 10.1046/j.1432-1327.2001.02106.x. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Signal sequences. The limits of variation. J. Mol. Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- Wadsworth J. D., Collinge J. Update on human prion disease. Biochim. Biophys. Acta. 2007;1772:598–609. doi: 10.1016/j.bbadis.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Wainwright L. J., Field M. C. Quality control of glycosylphosphatidylinositol anchor attachment in mammalian cells: a biochemical study. Biochem. J. 1997;321(Pt 3):655–664. doi: 10.1042/bj3210655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waneck G. L., Stein M. E., Flavell R. A. Conversion of a PI-anchored protein to an integral membrane protein by a single amino acid mutation. Science. 1988;241:697–699. doi: 10.1126/science.3399901. [DOI] [PubMed] [Google Scholar]
- Wang X., Wang F., Sy M. S., Ma J. Calpain and other cytosolic proteases can contribute to the degradation of retro-translocated prion protein in the cytosol. J. Biol. Chem. 2005;280:317–325. doi: 10.1074/jbc.M410649200. [DOI] [PubMed] [Google Scholar]
- Winklhofer K. F., Heske J., Heller U., Reintjes A., Muranyi W., Moarefi I., Tatzelt J. Determinants of the in vivo folding of the prion protein. A bipartite function of helix 1 in folding and aggregation. J. Biol. Chem. 2003;278:14961–14970. doi: 10.1074/jbc.M209942200. [DOI] [PubMed] [Google Scholar]
- Yanai A., Meiner Z., Gahali I., Gabizon R., Taraboulos A. Subcellular trafficking abnormalities of a prion protein with a disrupted disulfide loop. FEBS Lett. 1999;460:11–16. doi: 10.1016/s0014-5793(99)01316-2. [DOI] [PubMed] [Google Scholar]
- Yedidia Y., Horonchik L., Tzaban S., Yanai A., Taraboulos A. Proteasomes and ubiquitin are involved in the turnover of the wild-type prion protein. EMBO J. 2001;20:5383–5391. doi: 10.1093/emboj/20.19.5383. [DOI] [PMC free article] [PubMed] [Google Scholar]