Abstract
The importance of the amyloid precursor protein (APP) in the pathogenesis of Alzheimer’s disease (AD) became apparent through the identification of distinct mutations in the APP gene, causing early onset familial AD with the accumulation of a 4-kDa peptide fragment (βA4) in amyloid plaques and vascular deposits. However, the physiological role of APP is still unclear. In this work, Drosophila melanogaster is used as a model system to analyze the function of APP by expressing wild-type and various mutant forms of human APP in fly tissue culture cells as well as in transgenic fly lines. After expression of full-length APP forms, secretion of APP but not of βA4 was observed in both systems. By using SPA4CT, a short APP form in which the signal peptide was fused directly to the βA4 region, transmembrane domain, and cytoplasmic tail, we observed βA4 release in flies and fly-tissue culture cells. Consequently, we showed a γ-secretase activity in flies. Interestingly, transgenic flies expressing full-length forms of APP have a blistered-wing phenotype. As the wing is composed of interacting dorsal and ventral epithelial cell layers, this phenotype suggests that human APP expression interferes with cell adhesion/signaling pathways in Drosophila, independently of βA4 generation.
The amyloid precursor protein (APP) is a ubiquitously expressed, integral membrane protein (1, 2). APP belongs to a protein family with two other members in mammals: the amyloid precursor-like protein 1 and 2 (APLP1 and APLP2, refs. 3–5). The central role of APP in the pathogenesis of Alzheimer’s disease (AD, ref. 6) and its evolutionary conservation suggest important biological functions for the protein. Two homologues have been found in invertebrates: the amyloid protein-like protein 1 (APL-1) in Caenorhabditis elegans (7) and the amyloid precursor protein-like protein (APPL) in Drosophila melanogaster (8). Flies deficient for expression of APPL show phototaxis impairment and can be rescued by the expression of human APP (9). None of the homologous members of the APP protein family exhibit sequence similarities within the β-amyloid region of APP that encodes the characteristic 4-kDa proteinaceous component in vascular deposits and amyloid plaques of AD, βA4 (6). The βA4 peptide is cleaved from APP by unknown proteases termed β-secretase, which generates the N terminus and γ-secretase, which releases the C terminus by cleaving within the transmembrane domain of APP. Differential cleavage by γ-secretase produces βA4 of 40 or 42 amino acid residues in length with the 42-aa peptide being increased by distinct mutations in the APP and presenilin genes causing early onset familial AD (reviewed in ref. 6). A third cleavage of APP by α-secretase occurs within the βA4 domain and precludes βA4 formation. The 110- to 130-kDa ectodomain of APP generated by α- or β-secretase is secreted into the extracellular space.
Despite the availability of APPL-deficient flies (9), APP-null mutants, or transgenic mice expressing human APP (10–12), the physiological role of APP remains obscure. The structure of APP indicated a possible function as receptor (1) involved in cell–cell and cell–matrix adhesion. The discovery and characterization of cytoplasmic APP-binding proteins such as Fe65 and X11 support a possible signaling-receptor function for APP (13). It has also been suggested that secreted APP forms may function as growth factors in fibroblasts (14) or as mediators of neurite outgrowth in PC12 cells (15).
In this study, we investigated the processing of APP in insect cells and the role of βA4 in APP function in transgenic D. melanogaster. By analyzing the processing of APP and its derivatives, we were able to establish a γ-secretase activity in insects. Depending on the gene expression of full-length APP and the derivatives thereof, we obtained a distinct blistered-wing phenotype in transgenic flies, independent of βA4 production, suggesting a physiological function of APP in the regulation of cell–cell adhesion.
MATERIALS AND METHODS
DNA Constructs.
The following APP695 constructs were introduced into the pUAST vector (16): (i) APP695, (ii) APP695-Swedish with the familial mutation in codons 670 K → N and 671 M → L (ref. 17; for the numbering of APP695 see ref. 1), (iii) SPA4CT-C-myc (ref. 18; also termed SPC99-C-myc), (iv) APP695 ΔCT-N-myc (19), and (v) chimeric APP695/APLP2-N-myc. For the latter construct, the BglII–ClaI fragment covering the βA4 region and the entire C terminus of APP695 was replaced by the homologous region of APLP2. To introduce the BglII and ClaI restriction sites in APLP2, a corresponding fragment was amplified by PCR with the following primers: APLP2-s-BglII (GGAAGATCTCCGATGTTAAGGAAATGATTT) and APLP2-as-ClaI (CCCATCGATGGGATCTTCCGGCCCACCTGC). The products were shown to be correct by sequencing. The c-myc encoding epitope (EQKLISEEDL) was inserted into the constructs as described (ref. 19; Fig. 1). The pUAST vector and the APP695-constructs were linearized with NotI and KpnI by partial digestion and ligated.
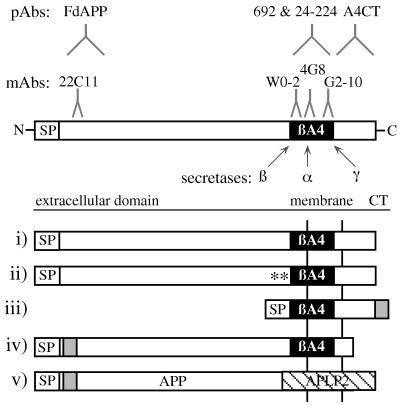
Figure 1.
Schematic representation of APP695 as well as the different APP695 derivatives used in the study. The α-, β-, and γ-secretase cleavage sites are indicated by arrows and epitope sites of antibodies used by a stylized antibody “Y”. Constructs: i, APP695; ii, APP695-Swedish; iii, SPA4CT-C-myc; iv, APP695 ΔCT-N-myc; v, APP695/APLP2-N-myc; N, N terminus; C, C terminus; SP, signal peptide; CT, cytoplasmic tail; ∗∗, Swedish double mutation; black box, βA4 region; and gray bar, myc-tag.
Transient Transfection of Schneider Cells SL-2.
A D. melanogaster embryonic cell line, Schneider cells SL-2 (20) grown in Schneider’s medium at 25°C, was transfected transiently with Lipofectin, according to the manufacturer’s protocol (GIBCO/BRL). The expression of the different APP forms in the pUAST vector was induced on cotransfection with a vector expressing the yeast transactivator GAL4 under control of the actin promoter (21). Transiently transfected SL-2 cells were cultured on 10-cm dishes and usually harvested after 48 h. Cells were lysed in 500 μl of lysis buffer (50 mM Tris⋅HCl, pH 7.5/150 mM NaCl/1% Nonidet P-40, vol/vol). Complete protease-inhibitor mixture (Boehringer Mannheim) was added according to the manufacturer’s protocol. Cell lysates and conditioned medium (CM) were investigated in immunoprecipitations and Western blot analyses (19, 22).
Transgenic Fly Lines.
Strains Oregon R as wild-type control and the transformation host white (w1118) were raised on standard fly food at 18°C or 25°C with 60–70% relative humidity. Each of the APP695-constructs described above was coinjected with the helper vector pUChsπΔ2–3 into w1118 embryos. Transformants were selected, and lines with single-copy inserts were established. Transgenic lines were mated with different GAL4 driver strains. For extract preparations, 5–10 flies or 10 fly bodies and 100 fly heads were homogenized separately in 1.5-ml reaction tubes with a micropestle in 500 μl of RIPA buffer [50 mM Tris⋅HCl, pH 8.0/1% SDS (wt/vol)/0.5% sodium deoxycholate (wt/vol)/1% Triton X-100 (vol/vol)/150 mM NaCl]. Complete protease-inhibitor mixture (Boehringer Mannheim) was added according to the manufacturer’s protocol. After homogenization, the fly debris was centrifuged at 18,000 × g, and the supernatant was loaded directly on a gel or used for immunoprecipitation.
Immunoprecipitation and Western Blotting.
For detection of APP695, APP695-Swedish, APP695 ΔCT-N-myc, and chimeric APP695/APLP2-N-myc (the full-length APP695 derivatives) cell lysates, CM, and fly homogenates were immunoprecipitated with polyclonal antiserum FdAPP (22). For detection of SPA4CT-C-myc cell lysates, CM and fly homogenates were immunoprecipitated with polyclonal antiserum A4CT (18). α-Secretory APP695 was immunoprecipitated with mAb W0-2 (23) from CM of transfected SL-2 cells. βA4 was immunoprecipitated with mAbs W0-2, G2–10 (23), and 4G8 (24) and with polyclonal rabbit antisera 692 and 24–224 (obtained from G. Multhaup, University of Heidelberg). Expressed constructs carrying a myc tag were immunoprecipitated in cell-culture CM or in cell lysates (C-terminal myc tag) with polyclonal anti-myc antiserum (Eurogentec, Brussels). Immunoprecipitations with mAbs were performed in the presence of protein G-agarose. With polyclonal sera (Boehringer Mannheim), protein A-Sepharose (Pharmacia) was used according to the manufacturer’s protocol. Precipitates of full-length APP695 derivatives were analyzed on SDS/7.5% PAGE, and precipitates of SPA4CT-C-myc and of βA4 were analyzed on SDS/16.5% PAGE. For Western blotting, full-length APP695 derivatives were transferred to 0.45-μm nitrocellulose membranes at 190 mA for 4 h, and SPA4CT-C-myc and βA4 were transferred to 0.45-μm nitrocellulose membranes at 380 mA for 45 min. To enhance the βA4 signal, the nitrocellulose membrane was boiled in PBS for 5 min as described (23). Full-length APP695 derivatives were visualized with mAb 22C11 (22), and SPA4CT-C-myc and βA4 were visualized with mAb W0-2 (23), all with the enhanced-chemiluminescence detection system (Amersham).
RESULTS
APP695 and Derivatives Expressed in Drosophila Schneider Cell Culture.
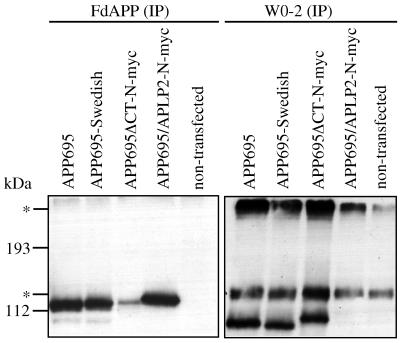
To analyze APP expression and metabolism in the Drosophila system, we transiently expressed wild-type human APP and mutant derivatives in SL-2 tissue-culture cells. The different forms used in this study are depicted in Fig. 1. To express the various APP forms, SL-2 cells were cotransfected with pPAC/GAL4 expressing the yeast transcriptional activator GAL4 under the control of a strong actin promoter. Immunoprecipitations of cell lysates with the APP-specific polyclonal antiserum FdAPP (22) showed expression of APP695, APP695-Swedish, APP695ΔCT-N-myc, and the chimeric APP695/APLP2-N-myc construct in SL-2 cells, whereas nontransfected cells were devoid of signals (Fig. 2).
Figure 2.
Transiently transfected SL-2 cells show expression and secretion of APP695 and derivatives. Cell lysate was immunoprecipitated with polyclonal serum FdAPP and CM with mAb W0-2. Nontransfected cells did not show an APP protein band. The construct APP695/APLP2-N-myc lacks the epitope of mAb W0-2. Immunoprecipitates were electrophoresed on SDS/7.5% PAGE, immunoblotted, and detected with mAb 22C11. The asterisks indicate background protein bands. The upper band might represent glycosylated forms of APP (22). The lower protein band shows a higher molecular mass than APP and derivatives, and it might represent bovine APP from the fetal calf serum that was supplied to the media. At least mAb W0-2 is able to detect bovine βA4 from fetal calf serum (data not shown).
After immunoprecipitations with mAb W0-2 of CM, secreted APP695, APP695-Swedish, and APP695ΔCT-N-myc became readily detectable (Fig. 2). The APP695ΔCT-N-myc band was shifted slightly to a higher molecular mass because of the myc tag. Because antibody W0-2 recognizes amino acids 5–9 from the βA4, secretory forms of APP produced by β- or the recently postulated δ-secretase (25) could not be detected. α-Secreted APP695, produced mainly by cleavage C-terminal to lysine residue 16 of βA4, is detectable with mAb W0-2. As expected, cells transfected with the chimeric APP695/APLP2-N-myc and nontransfected SL-2 cells did not give rise to bands with W0-2 (Fig. 2). APP695/APLP2-N-myc thus served as an internal antibody control, because it lacks the βA4 domain recognized by W0-2. By analyzing CM of cells expressing this construct with polyclonal serum FdAPP, we also detected secretion (data not shown).
SPA4CT-C-myc Expressing SL-2 Cells Release a 4-kDa Peptide Detectable with βA4-Specific Antibodies.
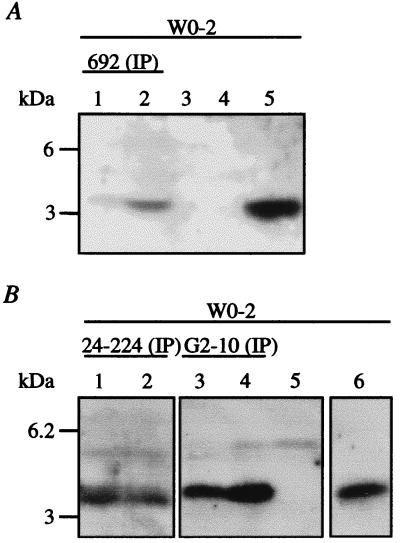
Because the production of βA4 from full-length APP indicates the presence of β- and γ-secretase, and production of βA4 from SPA4CT indicates γ-secretase activity (18), we analyzed the βA4 content in CM of SL-2 cells expressing the full-length APP derivatives and SPA4CT-C-myc by using βA4-specific antibodies for immunoprecipitations. No βA4 was detectable in CM of SL-2 cells expressing APP695, APP695-Swedish, APP695ΔCT-N-myc, or chimeric APP695/APLP2-N-myc. However, βA4 was readily detectable as a 4-kDa band in immunoprecipitates of CM from SL-2 cells expressing SPA4CT-C-myc (Fig. 3). The same 4-kDa band was obtained with different βA4-specific mAbs (Fig. 1), polyclonal serum 692 (Fig. 3A), polyclonal serum 24–224 (Fig. 3B), and mAb G2–10 (Fig. 3B), suggesting that this band represents βA4 secreted by the insect cells. This band was not detected in CM of nontransfected cells (Fig. 3) or of cells transfected only with the vector pPAC/Gal4.
Figure 3.
Transiently transfected SL-2 cells expressing SPA4CT-C-myc show secretion of βA4. Immunoprecipitations of CM with βA4-specific polyclonal serum 692 (A), polyclonal serum 24–224 (B), and mAb G2–10 (B) precipitated the same 4-kDa band of identical electrophoretic mobility as 200 pg of synthetic βA4 (A, lane 5; B, lane 6). For every immunoprecipitation, two identically transfected cell-culture dishes were analyzed. As controls, CM of SL-2 cells transfected with only pPAC/GAL4 (A, lane 3) and nontransfected cells (A, lane 4) were analyzed. Nontransfected cells do not show protein bands at lower ranges (B, lane 5). Samples were electrophoresed on SDS/16.5% PAGE and immunoblotted with mAb W0-2.
The major βA4 species produced by the insect cells was the 40-aa peptide as shown by immunoprecipitation with our 40-specific mAb G2–10 (Fig. 3B). βA4 peptides ending at residue 42, however, could not be detected with our 42-specific mAb G2–11 (data not shown). Therefore, the major βA4 form produced by insect cells seems to end after 40 aa. If the 42-aa species was also produced, the amounts were below our detection limit (23).
Transgenic Drosophila Expressing APP695 and Derivatives.
A well established method to modulate the expression of transgenes subcloned into the pUAST vector in Drosophila is to cross these flies with fly lines transgenic for a particular GAL4 driver (16). In this study, we used mainly the GAL4-strain Mz-1087.hx, in which the expression of transgenes occurs preferentially during larval stages in salivary glands, midgut, and imaginal discs including the wing discs (kindly obtained from J. Urban and G. Technau, University of Mainz, Germany). The wing discs show a rather homogenous expression of GAL4. In adult flies, expression of GAL4 transgenes was strongest in fly heads.
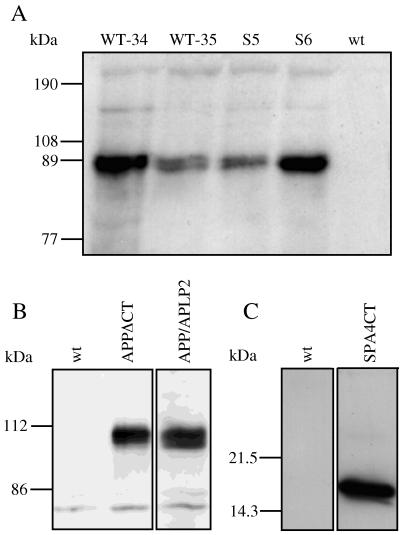
Transgenic Drosophila were investigated for the expression of APP695 and derivatives after crossing with the GAL4-strain Mz-1087.hx. Homogenates (soluble fraction) of 5–10 flies were analyzed directly on SDS/PAGE. In Fig. 4A, we show high protein-expression levels of APP695 (WT-34, WT-35) and APP695-Swedish (S5, S6) for two different transgenic fly lines. In contrast, controls (wild-type flies) showed no APP band. For each construct, we always assessed at least two different lines for transgene expression. Strong expression of APP695/APLP2-N-myc and APP695ΔCT-N-myc could also be detected in transgenic flies (Fig. 4B). The corresponding bands were absent in extracts of wild-type flies (Fig. 4B). The APP bands shown in Fig. 4 A and B appear to be double bands and may represent differences in glycosylation, which occurs in insect cells but to a lesser extent than in mammalian cells (26). SPA4CT-C-myc was also expressed in transgenic flies (Fig. 4C).
Figure 4.
Transgenic D. melanogaster expressing APP695 and derivatives. (A) Five to 10 flies were homogenized in 500 μl of RIPA buffer, and 40–50 μl of homogenate was loaded on SDS/7.5% PAGE and immunoblotted with mAb 22C11. Two different fly lines were investigated for APP695 expression, WT-34 and WT-35, and for APP695-Swedish expression, S5 and S6. In each case, a distinct protein band of appropriate size is seen. This band is absent in wild-type flies (wt). (B) The constructs APP695/APLP2-N-myc (APP/APLP2) and APPΔCT-N-myc (APPΔCT) were expressed; wt was used as a negative control. (C) Five to 10 flies were homogenized in 500 μl of RIPA buffer, and 40–50 μl of homogenate was loaded directly on SDS/16.5% PAGE and immunoblotted with mAb W0-2. SPA4CT-C-myc is expressed in transgenic flies (SPA4CT) and not detected in wt.
Transgenic Drosophila Expressing SPA4CT-C-myc Release a 4-kDa Peptide Detectable with Anti-βA4 Antibodies.
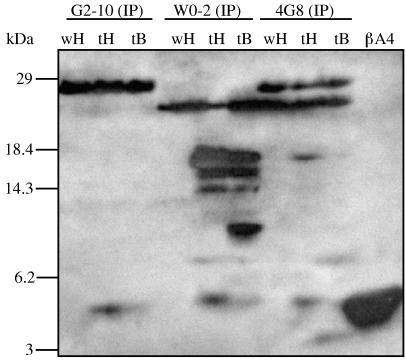
Because one aim of this study was to investigate whether insects are able to produce βA4 from human APP, we tested all transgenic lines for βA4 release by using homogenized flies, fly heads, larvae, or embryos. Flies transgenic for APP695/APLP2-N-myc and wild-type flies served as controls. βA4 could not be detected in lines expressing APP695, APP695-Swedish, or APP695ΔCT-N-myc by direct SDS/PAGE or by SDS/PAGE of immunoprecipitates (data not shown), confirming our observations made with SL-2 cells. However, we found, as we did in transfected SL-2 cells, that flies expressing SPA4CT-C-myc produce βA4 (Fig. 5). We separately investigated homogenates from fly heads and bodies by immunoprecipitation with mAbs G2–10, W0-2, and 4G8, which are specific for three different regions of βA4 (Fig. 1). In immunoprecipitations of fly-head homogenates, all three antibodies yielded the same 4-kDa peptide band (Fig. 5, tH), which showed the same electrophoretic mobility as synthetic βA4. The same band was also present, although weakly, in fly-body homogenates (tB). In the wild-type controls (wH), bands of comparable size were not detectable. SPA4CT-C-myc protein could be visualized with mAbs 4G8 (tH, tB) and W0-2 (tH) as a band migrating below the 18.4-kDa protein marker. Immunoprecipitation with mAb W0-2 showed one additional band in the range between 14 and 18 kDa in fly heads and bodies, which seems to be a C-terminal shortened breakdown product of SPA4CT (tH, tB). In addition, W0-2 precipitated from fly bodies (tB) a distinct 8-kDa protein band, which may represent a caspase cleavage product of SPA4CT-C-myc.
Figure 5.
Transgenic D. melanogaster expressing SPA4CT-C-myc secrete a 4-kDa peptide detectable with different βA4-specific mAbs. Immunoprecipitates of soluble homogenates from 100 wild-type fly heads (wH), 100 transgenic fly heads (tH), and 10 transgenic fly bodies (tB) with mAb G2–10, W0-2, and 4G8 are shown after SDS/16.5% PAGE in an immunoblot detected with mAb W0-2. A 4-kDa βA4 was precipitated with mAb G2–10 (tH), mAb W0-2 (tH), and mAb 4G8 (tH). The 4-kDa band is in the same range as synthetic βA4 (500 pg). Expression of SPA4CT-C-myc was detectable as an 18-kDa band with antibodies mAb W0-2 (tH, tB) and mAb 4G8 (tH).
Transgenic Drosophila Expressing Full-Length APP Derivatives Develop a Blistered-Wing Phenotype.
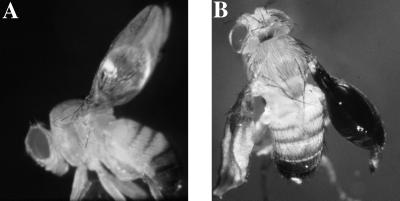
During the establishment of transgenic flies expressing APP, we noted that, in certain lines, wing abnormalities were independent of the site of integration but dependent on the transgene. The most striking result was a dominant blistered-wing phenotype that occurred only in flies transgenic for APP695, APP695-Swedish, and APP695/APLP2-N-myc. These flies develop a wing deformation in one or both wings. Large blisters often occupied >50% of the wing surface (Fig. 6). In extreme cases, wings were formed like balloons (Fig. 6 B). The blisters appear transparent or black, were filled with hemolymph, and occurred in 5–20% of transgenic animals that were raised at a temperature of 25°C. The pattern of the wing veins remained normal. To assess whether this phenotype was specific for the Mz-1087.hx line, we crossed the APP695/APLP2-N-myc with different GAL4-expressing driver lines that have been described (16). Lines expressing GAL4 in the wing disc always had wing blisters (Table 1). This phenotype points to a dysfunction in cell–cell adhesion between the two epithelial cell layers forming the fly wing (27). We observed no other obvious phenotypic consequences in these lines, although some combinations show a decreased viability.
Figure 6.
Transgenic Drosophila expressing full-length APP derivatives develop a blistered-wing phenotype. Flies expressing APP695, APP695-Swedish, or APP695/APLP2-N-myc develop abnormal wings with distinct blisters. Two examples are shown with different extents of the wing phenotype. (A) A round blister in the proximal part of the wing is shown. (B) In the most severe phenotype, the two epithelial wing-cell layers are not connected anymore. The whole wing looks like a black balloon filled with hemolymph.
Table 1.
Wing blisters induced by different GAL4 driver lines
| GAL4 line* | Wing disc pattern | Phenotype |
|---|---|---|
| 1087 | Strong expression in entire disc | Wing blisters |
| 69B | Expression in entire disc | Wing blisters |
| 30A | Expression in prospective wing base | No effect |
| T80 | Expression in entire disc | Wing blisters |
GAL4 driver lines, except Mz-1087.hx, are described in ref. 16. The stock number in the Bloomington Drosophila Stock Center is B-1795 for 30A, B-1774 for 69B, and B-1878 for T80. Mz-1087.hx was kindly provided by J. Urban and G. Technau, University of Mainz.
Lines with a C-terminally truncated protein (APP695ΔCT-N-myc) showed no blistered-wing phenotype. This result cannot be attributed to a lower level of protein expression in these lines, as corresponding immunostainings of imaginal discs showed a protein level comparable to that of lines expressing full-length forms of APP (data not shown). Similarly, flies transgenic for SPA4CT-C-myc did not develop a blistered-wing phenotype. Because these are the only flies with detectable production of βA4, we assume that the wing blisters are independent of βA4 production. Additionally, flies expressing APP695/APLP2-N-myc do not possess a βA4 region, but these flies still show distinct wing blisters, establishing that the βA4 region is not essential for this phenotype.
DISCUSSION
In the present study, transgenic D. melanogaster was established as a model to analyze basic functions of human APP695. By using this fly system, we were able to show the processing of APP by α- and γ-secretase. Additionally, we observed a rather intriguing wing phenotype in certain transgenic fly lines, pointing to interesting functions of APP in vivo. Secreted APP695 forms had already been produced in Spodoptera frugiperda (Sf9) insect cells infected with APP-recombinant baculoviruses (28). In this system, secretion of the N-terminal APP ectodomain fragment was caused by α-cleavage within the βA4 sequence C-terminal to glutamine-15 or lysine-16. The C-terminal counterpart of this cleavage was isolated from mammalian cells and started with βA4-residue leucine-17 (29). These data confirmed processing of APP in Sf9 cells identical to that in mammalian cells. Similarly, for APPL, the endogenous fly homologue of APP, a secretion was observed in primary cell cultures of embryos and in SL-2 cells (9).
We analyzed the cleavage site in the APP ectodomain with antibody W0-2. This antibody recognizes residues 5–9 of the βA4 region. Because we were able to detect secreted APP in CM of transfected SL-2 cells with mAb W0-2, the secretory cleavage occurred C-terminal of residue 9 of the βA4 region that would be compatible with cleavage by α-secretase. Cleavages by β- or δ-secretase occur N-terminal of the W0-2 epitope and were therefore excluded (25). Additionally, we specifically addressed the question of whether amyloidogenic processing occurs in flies transgenic for human APP and whether βA4 release is detectable. Although this has not been observed yet (30), we clearly show that in transfected SL-2 cells and in flies transgenic for SPA4CT-C-myc, a 4-kDa βA4 band, recognized by different βA4-specific antibodies after immunoprecipitation, is produced. As we were not able to detect this 4-kDa peptide as a direct cleavage product from full-length APP695 and its derivatives, we assume that the postulated β-secretase might somehow be altered in insects or might cleave APP to a nondetectable extent. Because, in mammalian cells, SPA4CT-C-myc is processed by signal peptidase to A4CT-C-myc, which begins with the N terminus of βA4, a single γ-secretase cleavage was sufficient to release βA4 from the A4CT-C-myc stub (18). As the signal peptide of human APP695 was recognized and properly cleaved in Sf9 cells (28), detection of βA4 in CM of SL-2 cells and homogenates of transgenic flies expressing SPA4CT strongly suggests the presence of γ-secretase activity in Drosophila. By using antibodies that specifically react with the C-terminal eight amino acid residues of βA4 1–40 (G2–10) or βA4 1–42 (G2–11), respectively, we identified βA4 1–40 as the major form produced in this system. βA4 1–42 was either not produced or produced at levels below the detection limits of G2–11 (23).
Flies transgenic for the full-length APP forms of APP695, APP695-Swedish, and chimeric APP/APLP2-N-myc developed a striking blistered-wing phenotype. This phenotype is not caused by position effects; different integrants for a given construct showed the same effects. In addition, the phenotype cannot be caused by mutations in wing-specific genes induced by the integration of the transgenes. Homozygous lines lacking the GAL4 driver never exhibited the wing phenotype, and only after GAL4-induction in appropriate lines and crosses was a wing phenotype observed. We also do not consider it very likely that the wing phenotype is an unspecific effect caused by the overproduction of a transmembrane protein. Lines, like APPΔCT-N-myc, expressed comparable amounts of membrane APP but did not produce the phenotype. This construct showed also that in addition to the ectodomain and transmembrane domain, the cytoplasmic domain of APP is necessary for blistered-wing formation. The latter may indicate that a transmembrane signaling function of APP is imposed on the blistered-wing phenotype in a dominant function presumably by interacting and inhibiting an appropriate partner on the cell membrane.
We suggest that the blistered-wing phenotype resulting from ectopic APP expression in Drosophila reflects a physiological function of APP in the regulation of cell adhesion and signaling. Wing morphogenesis in Drosophila is characterized by two periods of apposition and separation of the dorsal and a ventral cell layer of the wing before the epithelia become fused permanently (31). We presume that APP negatively interacts with factors involved in the adhesion of the two epithelia during wing development. In Drosophila, several mutations affecting wing morphology, and particularly the adhesion of the two epithelial cell layers, have been isolated. Mutant flies deficient for the expression of most integrin subunits develop distinct wing blisters. Integrins are heterodimeric receptors that may be ligand-bound to extracellular matrix components and may have a signaling function (32). Fly integrins are especially required for the function of the cell–matrix–cell junction, where one surface of the developing wing adheres to the other (33). This function of integrins supports our suggestion of APP’s involvement in cell adhesion. In this context, APP might be an antagonist for integrins, such that APP would bind, either directly or via other components, the same extracellular matrix components as integrins. Indeed, recent investigations in primary rat neurons confirmed a colocalization of APP and integrins (34, 35). The phenotype we observed in Drosophila might be caused by a specific interaction of human APP with evolutionarily conserved protein partners, resulting in a deterioration of the adhesion/signaling pathway supported by the integrins. A particularly intriguing result in this context is the recent finding that one of the integrin subunits is involved in short-term memory in Drosophila (36), thus, potentially uncovering an interesting link between basic APP functions, integrin-mediated adhesion processes, and memory mechanisms. However, other pathways have to be considered. For example, expression of a mutant form of a G protein involved in cAMP signaling results in a comparable phenotype (37). Similarly, the blistered gene, encoding a protein related to human serum-response factor (38), represents an additional candidate for a pathway in which APP might be involved.
Thus, Drosophila seems to be a formidable animal-model system for studying basic functions of human APP and may be suited for studying APP transmembrane signaling mechanisms potentially related to AD. The α- and γ-secretase activities described here establish the usefulness of Drosophila as a model system for isolating the unknown secretases. Additionally, possible inhibitors of γ-secretase might be tested more efficiently in the fly system; these inhibitors may become therapeutic tools in diminishing the deleterious effects of βA4 generation in AD. To analyze physiological functions of APP, the fly model seems especially suitable, because we can find a specific phenotype depending on the ectopic expression of full-length APP. Thus, the possibility of testing genetic interactions between APP-expressing lines and the comparable mutants engaged in wing morphology should allow us to uncover the biological pathways affected by human APP.
Acknowledgments
We thank J. Urban and G. Technau (University of Mainz) and Gunter Merdes (Deutsches Krebsforschungszentrum, Heidelberg) for fly stocks and G. Multhaup and D. Beher for providing antibodies. This work was supported in part by a fellowship from the Deutsche Akademische Austauschdienst (to A.F.). C.L.M., K.B., and R.P. were supported by the Deutsche Forschungsgemeinschaft, Fonds der Chemischen Industrie, and the Erna Struckmann Foundation.
ABBREVIATIONS
- AD
Alzheimer’s disease
- APLP
amyloid precursor-like protein
- APP
amyloid precursor protein
- APPL
amyloid precursor protein-like protein
- βA4
a 4-kDa peptide, a constituent of Alzheimer’s disease amyloid
- CM
conditioned medium
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Kang J, Lemaire H G, Unterbeck A, Salbaum J M, Masters C L, Grzeschik K H, Multhaup G, Beyreuther K, Mueller-Hill B. Nature (London) 1987;325:733–736. doi: 10.1038/325733a0. [DOI] [PubMed] [Google Scholar]
- 2.Sandbrink R, Masters C L, Beyreuther K. J Biol Chem. 1994;269:14227–14234. [PubMed] [Google Scholar]
- 3.Wasco W, Bupp K, Magendanzt M, Gusella J F, Tanzi R E, Solomon F. Proc Natl Acad Sci USA. 1992;89:10758–10762. doi: 10.1073/pnas.89.22.10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wasco W, Gurubhagavatula S, Paradis M D, Romano D M, Sisodia S S, Hyman B T, Neve R L, Tanzi R E. Nat Genet. 1993;5:95–100. doi: 10.1038/ng0993-95. [DOI] [PubMed] [Google Scholar]
- 5.Paliga K, Weidemann A, Peraus G, Kreger S, Dürrwang U, Hesse L, Multhaup G, Masters C L, Beyreuther K. Eur J Biochem. 1997;250:354–363. doi: 10.1111/j.1432-1033.1997.0354a.x. [DOI] [PubMed] [Google Scholar]
- 6.Selkoe D J. J Biol Chem. 1996;271:18295–18298. doi: 10.1074/jbc.271.31.18295. [DOI] [PubMed] [Google Scholar]
- 7.Daigle I, Li C. Proc Natl Acad Sci USA. 1993;90:12045–12049. doi: 10.1073/pnas.90.24.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosen D R, Martin-Morris L, Luo L Q, White K. Proc Natl Acad Sci USA. 1989;86:2478–2482. doi: 10.1073/pnas.86.7.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo L, Tully T, White K. Neuron. 1992;9:595–605. doi: 10.1016/0896-6273(92)90024-8. [DOI] [PubMed] [Google Scholar]
- 10.Zheng H, Jiang M H, Trumbauer M E, Sirinathsinghji D J S, Hopkins R, Smith D W, Heavens R P, Dawson G R, Boyce S, Conner M W, et al. Cell. 1995;81:525–531. doi: 10.1016/0092-8674(95)90073-x. [DOI] [PubMed] [Google Scholar]
- 11.Games D, Adams D, Alessandrini R, Barbour R, Berthelette P, Blackwell C, Carr T, Clemens J, Donaldson T, Gillespie F, et al. Nature (London) 1995;373:523–527. doi: 10.1038/373523a0. [DOI] [PubMed] [Google Scholar]
- 12.Hsiao K K, Chapman P, Nilsen S, Eckman C, Harigaya Y, Younkin S, Yang F, Cole G. Science. 1996;274:99–102. doi: 10.1126/science.274.5284.99. [DOI] [PubMed] [Google Scholar]
- 13.Fiore F, Zambrano N, Minopoli G, Donini V, Duilio A, Russo T. J Biol Chem. 1995;270:30853–30856. doi: 10.1074/jbc.270.52.30853. [DOI] [PubMed] [Google Scholar]
- 14.Saitoh T, Sundsmo M, Roch J-M, Kimura N, Cole G, Schubert D, Oltersdorf T, Schenk D B. Cell. 1989;58:615–622. doi: 10.1016/0092-8674(89)90096-2. [DOI] [PubMed] [Google Scholar]
- 15.Milward E A, Papadopoulos R, Fuller S J, Moir R D, Small D, Beyreuther K, Masters C L. Neuron. 1992;9:129–137. doi: 10.1016/0896-6273(92)90228-6. [DOI] [PubMed] [Google Scholar]
- 16.Brand A H, Perrimon N. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 17.Mullan M, Crawford F, Axelman K, Houlden H, Lilius L, Winblad B, Lannfelt L. Nat Genet. 1992;1:345–347. doi: 10.1038/ng0892-345. [DOI] [PubMed] [Google Scholar]
- 18.Dyrks T, Dyrks E, Masters C, Beyreuther K. FEBS Lett. 1992;309:20–24. doi: 10.1016/0014-5793(92)80730-5. [DOI] [PubMed] [Google Scholar]
- 19.Tienari P J, De Strooper B, Ikonen E, Simons M, Weidemann A, Czech C, Hartmann T, Ida N, Multhaup G, Masters C L, et al. EMBO J. 1996;15:5218–5229. [PMC free article] [PubMed] [Google Scholar]
- 20.Schneider I. J Embryol Exp Morph. 1972;27:353–365. [PubMed] [Google Scholar]
- 21.Krasnow M A, Saffman E E, Kornfeld K, Hogness D S. Cell. 1989;57:1031–1043. doi: 10.1016/0092-8674(89)90341-3. [DOI] [PubMed] [Google Scholar]
- 22.Weidemann A, König G, Bunke D, Fischer P, Salbaum J M, Masters C L, Beyreuther K. Cell. 1989;57:115–126. doi: 10.1016/0092-8674(89)90177-3. [DOI] [PubMed] [Google Scholar]
- 23.Ida N, Hartmann T, Pantel J, Schroder J, Zerfass R, Forstl H, Sandbrink R, Masters C L, Beyreuther K. J Biol Chem. 1996;271:22908–22914. doi: 10.1074/jbc.271.37.22908. [DOI] [PubMed] [Google Scholar]
- 24.Kim K, Miller D L, Chen M-C, Sapienza V J, Bai C, Grundke-Iqbal I, Currie J R, Wiesniewski H M. Neurosci Res Commun. 1988;2:121–130. [Google Scholar]
- 25.Simons M, De Strooper B, Multhaup G, Tienari P J, Dotti C G, Beyreuther K. J Neurosci. 1996;16:899–908. doi: 10.1523/JNEUROSCI.16-03-00899.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luckow V A, Summers M D. Bio/Technology. 1988;6:47–55. [Google Scholar]
- 27.Fristrom D, Fristrom J W. In: The Development of Drosophila melanogaster. Bate M, Arias A M, editors. Vol. 2. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 843–897. [Google Scholar]
- 28.Lowery D E, Pasternack J M, Gonzalez-DeWhitt P A, Zürcher-Neely H, Tomich C C, Altman R A, Fairbanks M B, Heinrikson R L, Younkin S G, Greenberg B D. J Biol Chem. 1991;266:19842–19850. [PubMed] [Google Scholar]
- 29.Ramabhadran T V, Gandy S E, Ghiso J, Czernik A J, Ferris D, Bhasin R, Goldgaber D, Frangione B, Greengard P. J Biol Chem. 1993;268:2009–2012. [PubMed] [Google Scholar]
- 30.Essalmani R, Guillaume J M, Mercken L, Octave J N. FEBS Lett. 1996;389:157–161. doi: 10.1016/0014-5793(96)00561-3. [DOI] [PubMed] [Google Scholar]
- 31.Brabant M C, Brower D L. Dev Biol. 1993;157:49–59. doi: 10.1006/dbio.1993.1111. [DOI] [PubMed] [Google Scholar]
- 32.Hynes R O. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 33.Brown N H. BioEssays. 1993;15:383–390. doi: 10.1002/bies.950150604. [DOI] [PubMed] [Google Scholar]
- 34.Storey E, Beyreuther K, Masters C L. Brain Res. 1996;735:217–231. doi: 10.1016/0006-8993(96)00608-7. [DOI] [PubMed] [Google Scholar]
- 35.Yamazaki T, Haass C, Saido T C, Omura S, Ihara Y. Biochemistry. 1997;36:8377–8383. doi: 10.1021/bi970209y. [DOI] [PubMed] [Google Scholar]
- 36.Grotewiel M S, Beck C D O, Wu K H, Zhu X-R, Davis L D. Nature (London) 1998;391:455–460. doi: 10.1038/35079. [DOI] [PubMed] [Google Scholar]
- 37.Wolfgang J W, Roberts I J H, Quan F, O’Kane C, Forte M. Proc Natl Acad Sci USA. 1996;93:14542–14547. doi: 10.1073/pnas.93.25.14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prout M, Damania Z, Soong J, Fristrom D, Fristrom J W. Genetics. 1997;146:275–285. doi: 10.1093/genetics/146.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]