Abstract
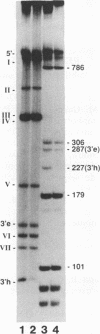
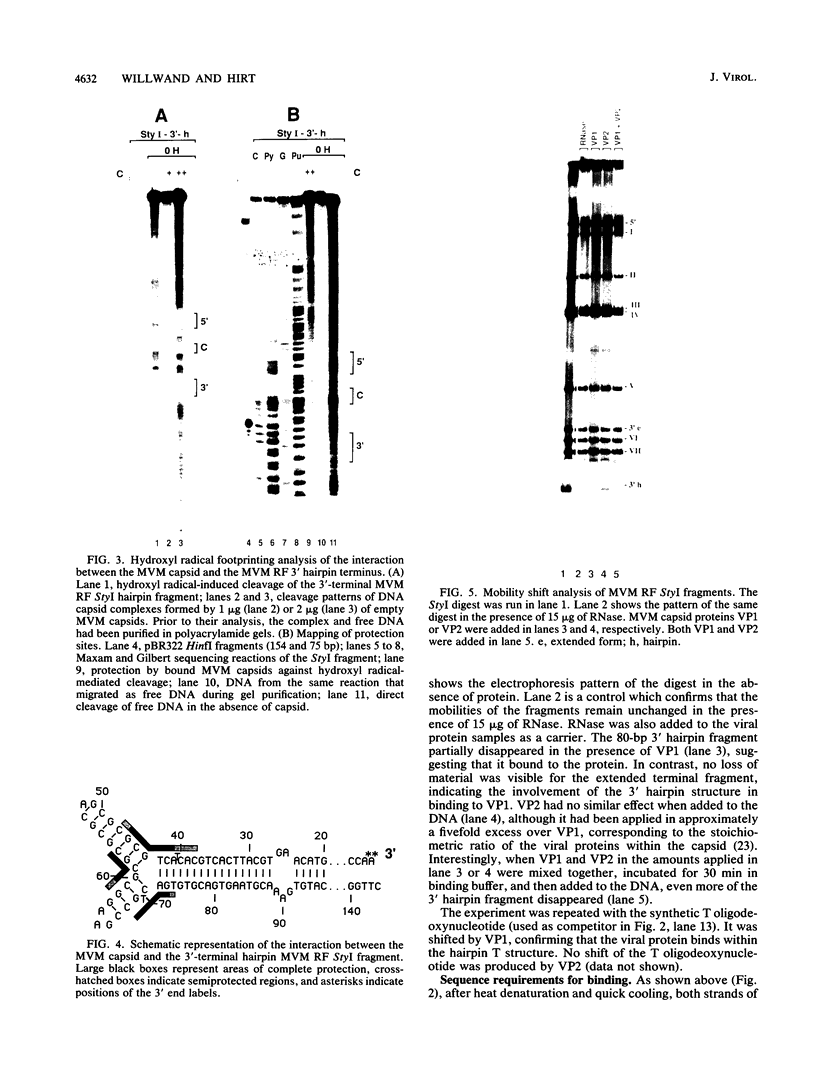
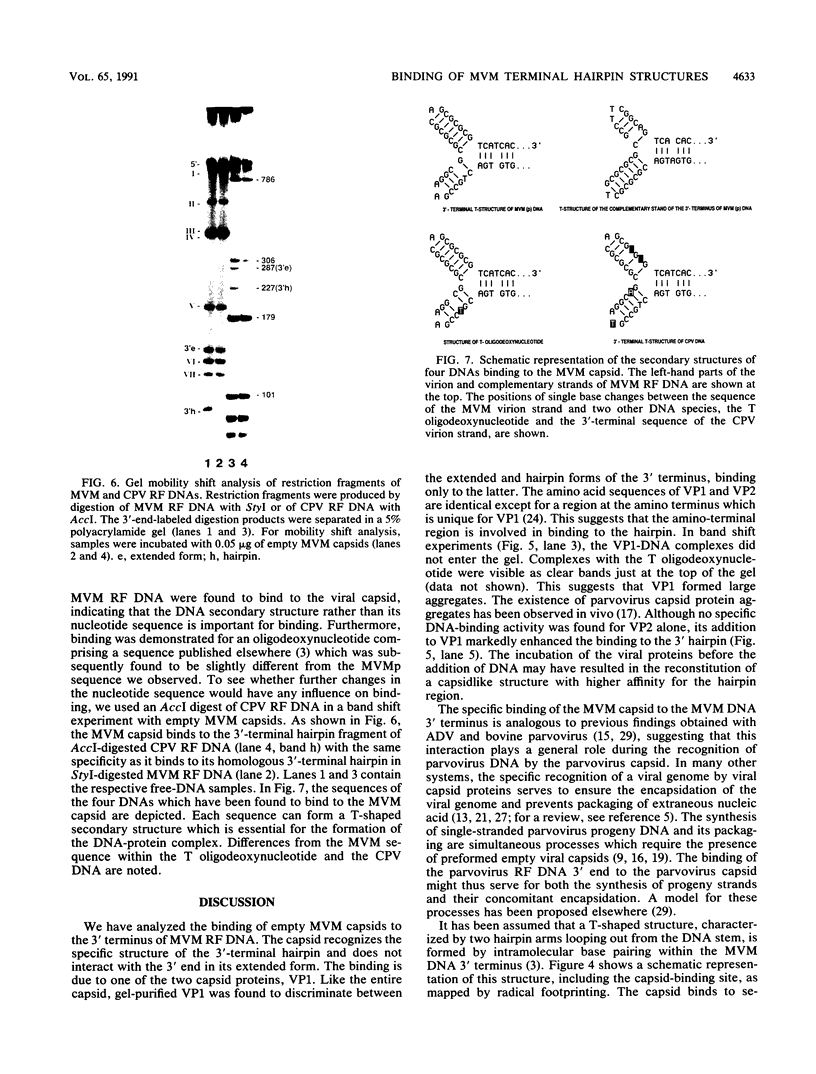
The terminal hairpin structures of the DNA of minute virus of mice (MVM) are essential for viral replication. Here we show that the hairpin 3' terminus of MVM replicative-form DNA binds specifically to empty MVM capsids. Binding of the same terminal DNA sequence in its linear double-stranded (extended) conformation was not observed. After heat denaturation and quick cooling of 3'-terminal extended-form fragments, not only the virion strand but also the complementary strand was found to bind to the capsid, presumably because each strand re-formed a similar hairpin structure. No binding affinity for the capsid was found to be associated with hairpin or extended 5' termini or with any other region of the viral DNA. Hydroxyl radical footprinting analyses revealed three protected nucleotide stretches forming a binding site at the branch point of the two 3'-terminal hairpin arms looping out from the DNA stem (T structure). Single base changes within this site did not affect the binding. In band shift experiments, specific binding to the T structure was demonstrated for VPI but not for VP2.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antonietti J. P., Sahli R., Beard P., Hirt B. Characterization of the cell type-specific determinant in the genome of minute virus of mice. J Virol. 1988 Feb;62(2):552–557. doi: 10.1128/jvi.62.2.552-557.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astell C. R., Gardiner E. M., Tattersall P. DNA sequence of the lymphotropic variant of minute virus of mice, MVM(i), and comparison with the DNA sequence of the fibrotropic prototype strain. J Virol. 1986 Feb;57(2):656–669. doi: 10.1128/jvi.57.2.656-669.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astell C. R., Smith M., Chow M. B., Ward D. C. Structure of the 3' hairpin termini of four rodent parvovirus genomes: nucleotide sequence homology at origins of DNA replication. Cell. 1979 Jul;17(3):691–703. doi: 10.1016/0092-8674(79)90276-9. [DOI] [PubMed] [Google Scholar]
- Bourguignon G. J., Tattersall P. J., Ward D. C. DNA of minute virus of mice: self-priming, nonpermuted, single-stranded genome with a 5'-terminal hairpin duplex. J Virol. 1976 Oct;20(1):290–306. doi: 10.1128/jvi.20.1.290-306.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotmore S. F., Tattersall P. The autonomously replicating parvoviruses of vertebrates. Adv Virus Res. 1987;33:91–174. doi: 10.1016/s0065-3527(08)60317-6. [DOI] [PubMed] [Google Scholar]
- Gough G. W., Lilley D. M. DNA bending induced by cruciform formation. Nature. 1985 Jan 10;313(5998):154–156. doi: 10.1038/313154a0. [DOI] [PubMed] [Google Scholar]
- Hermonat P. L., Labow M. A., Wright R., Berns K. I., Muzyczka N. Genetics of adeno-associated virus: isolation and preliminary characterization of adeno-associated virus type 2 mutants. J Virol. 1984 Aug;51(2):329–339. doi: 10.1128/jvi.51.2.329-339.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzberg R. P., Dervan P. B. Cleavage of DNA with methidiumpropyl-EDTA-iron(II): reaction conditions and product analyses. Biochemistry. 1984 Aug 14;23(17):3934–3945. doi: 10.1021/bi00312a022. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Lefebvre R. B., Riva S., Berns K. I. Conformation takes precedence over sequence in adeno-associated virus DNA replication. Mol Cell Biol. 1984 Jul;4(7):1416–1419. doi: 10.1128/mcb.4.7.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever A., Gottlinger H., Haseltine W., Sodroski J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J Virol. 1989 Sep;63(9):4085–4087. doi: 10.1128/jvi.63.9.4085-4087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Metcalf J. B., Bates R. C., Lederman M. Interaction of virally coded protein and a cell cycle-regulated cellular protein with the bovine parvovirus left terminus ori. J Virol. 1990 Nov;64(11):5485–5490. doi: 10.1128/jvi.64.11.5485-5490.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller D. E., Siegl G. Maturation of parvovirus LuIII in a subcellular system. I. Optimal conditions for in vitro synthesis and encapsidation of viral DNA. J Gen Virol. 1983 May;64(Pt 5):1043–1054. doi: 10.1099/0022-1317-64-5-1043. [DOI] [PubMed] [Google Scholar]
- Paradiso P. R. Analysis of the protein-protein interactions in the parvovirus H-1 capsid. J Virol. 1983 Apr;46(1):94–102. doi: 10.1128/jvi.46.1.94-102.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senapathy P., Tratschin J. D., Carter B. J. Replication of adeno-associated virus DNA. Complementation of naturally occurring rep- mutants by a wild-type genome or an ori- mutant and correction of terminal palindrome deletions. J Mol Biol. 1984 Oct 15;179(1):1–20. doi: 10.1016/0022-2836(84)90303-6. [DOI] [PubMed] [Google Scholar]
- Siegl G., Bates R. C., Berns K. I., Carter B. J., Kelly D. C., Kurstak E., Tattersall P. Characteristics and taxonomy of Parvoviridae. Intervirology. 1985;23(2):61–73. doi: 10.1159/000149587. [DOI] [PubMed] [Google Scholar]
- Stow N. D., McMonagle E. C., Davison A. J. Fragments from both termini of the herpes simplex virus type 1 genome contain signals required for the encapsidation of viral DNA. Nucleic Acids Res. 1983 Dec 10;11(23):8205–8220. doi: 10.1093/nar/11.23.8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straus S. E., Sebring E. D., Rose J. A. Concatemers of alternating plus and minus strands are intermediates in adenovirus-associated virus DNA synthesis. Proc Natl Acad Sci U S A. 1976 Mar;73(3):742–746. doi: 10.1073/pnas.73.3.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattersall P., Cawte P. J., Shatkin A. J., Ward D. C. Three structural polypeptides coded for by minite virus of mice, a parvovirus. J Virol. 1976 Oct;20(1):273–289. doi: 10.1128/jvi.20.1.273-289.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattersall P., Shatkin A. J., Ward D. C. Sequence homology between the structural polypeptides of minute virus of mice. J Mol Biol. 1977 Apr 25;111(4):375–394. doi: 10.1016/s0022-2836(77)80060-0. [DOI] [PubMed] [Google Scholar]
- Tattersall P., Ward D. C. Rolling hairpin model for replication of parvovirus and linear chromosomal DNA. Nature. 1976 Sep 9;263(5573):106–109. doi: 10.1038/263106a0. [DOI] [PubMed] [Google Scholar]
- Travers A. A. DNA conformation and protein binding. Annu Rev Biochem. 1989;58:427–452. doi: 10.1146/annurev.bi.58.070189.002235. [DOI] [PubMed] [Google Scholar]
- Vlazny D. A., Kwong A., Frenkel N. Site-specific cleavage/packaging of herpes simplex virus DNA and the selective maturation of nucleocapsids containing full-length viral DNA. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1423–1427. doi: 10.1073/pnas.79.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willwand K., Kaaden O. R. Capsid protein VP1 (p85) of Aleutian disease virus is a major DNA-binding protein. Virology. 1988 Sep;166(1):52–57. doi: 10.1016/0042-6822(88)90145-6. [DOI] [PubMed] [Google Scholar]
- Willwand K., Kaaden O. R. Proteins of viral and cellular origin bind to the Aleutian disease virus (ADV) DNA 3'-terminal hairpin: presentation of a scheme for encapsidation of ADV DNA. J Virol. 1990 Apr;64(4):1598–1605. doi: 10.1128/jvi.64.4.1598-1605.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorbas H., Rogge L., Meisterernst M., Winnacker E. L. Hydroxyl radical footprints reveal novel structural features around the NF I binding site in adenovirus DNA. Nucleic Acids Res. 1989 Oct 11;17(19):7735–7748. doi: 10.1093/nar/17.19.7735. [DOI] [PMC free article] [PubMed] [Google Scholar]