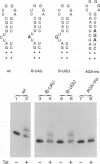
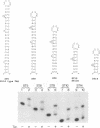
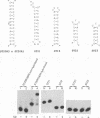
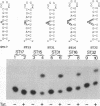
Abstract
The human immunodeficiency virus type 1 Tat protein binds to an RNA stem-loop structure called TAR which is present at the 5' end of all human immunodeficiency virus type 1 transcripts. This binding is centered on a bulge within the stem of TAR and is an essential step in the trans-activation process which results in a dramatic increase in viral gene expression. By analysis of a series of TAR derivatives produced by transcription or direct chemical synthesis, we determined the structural and chemical requirements for Tat binding. Tat binds well to structures which have a bulge of two to at least five unpaired bases bounded on both sides by a double-stranded RNA stem. This apparent flexibility in bulge size is in contrast to an absolute requirement for an unpaired uridine (U) in the 5'-most position of the bulge (+23). Substitution of the U with either natural bases or chemical analogs demonstrated that the imido group at the N-3 position and, possibly, the carbonyl group at the C-4 position of U are critical for Tat binding. Cytosine (C), which differs from U at only these positions, is not an acceptable substitute. Furthermore, methylation at N-3 abolishes binding. While methylation of U at the C-5 position has little effect on binding, fluorination reduces it, possibly because of its effects on relative tautomer stability at the N-3 and C-4 positions. Thus, we have identified key moieties in the U residue that are of importance for the binding of Tat to TAR RNA. We hypothesize that the invariant U is involved in hydrogen bond interactions with either another part of TAR or the TAR-binding domain in Tat.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arya S. K., Guo C., Josephs S. F., Wong-Staal F. Trans-activator gene of human T-lymphotropic virus type III (HTLV-III). Science. 1985 Jul 5;229(4708):69–73. doi: 10.1126/science.2990040. [DOI] [PubMed] [Google Scholar]
- Berkhout B., Jeang K. T. trans activation of human immunodeficiency virus type 1 is sequence specific for both the single-stranded bulge and loop of the trans-acting-responsive hairpin: a quantitative analysis. J Virol. 1989 Dec;63(12):5501–5504. doi: 10.1128/jvi.63.12.5501-5504.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calnan B. J., Biancalana S., Hudson D., Frankel A. D. Analysis of arginine-rich peptides from the HIV Tat protein reveals unusual features of RNA-protein recognition. Genes Dev. 1991 Feb;5(2):201–210. doi: 10.1101/gad.5.2.201. [DOI] [PubMed] [Google Scholar]
- Calnan B. J., Tidor B., Biancalana S., Hudson D., Frankel A. D. Arginine-mediated RNA recognition: the arginine fork. Science. 1991 May 24;252(5009):1167–1171. doi: 10.1126/science.252.5009.1167. [DOI] [PubMed] [Google Scholar]
- Caruthers M. H., Barone A. D., Beaucage S. L., Dodds D. R., Fisher E. F., McBride L. J., Matteucci M., Stabinsky Z., Tang J. Y. Chemical synthesis of deoxyoligonucleotides by the phosphoramidite method. Methods Enzymol. 1987;154:287–313. doi: 10.1016/0076-6879(87)54081-2. [DOI] [PubMed] [Google Scholar]
- Cordingley M. G., LaFemina R. L., Callahan P. L., Condra J. H., Sardana V. V., Graham D. J., Nguyen T. M., LeGrow K., Gotlib L., Schlabach A. J. Sequence-specific interaction of Tat protein and Tat peptides with the transactivation-responsive sequence element of human immunodeficiency virus type 1 in vitro. Proc Natl Acad Sci U S A. 1990 Nov;87(22):8985–8989. doi: 10.1073/pnas.87.22.8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen B. R. The HIV-1 Tat protein: an RNA sequence-specific processivity factor? Cell. 1990 Nov 16;63(4):655–657. doi: 10.1016/0092-8674(90)90129-3. [DOI] [PubMed] [Google Scholar]
- Dayton A. I., Sodroski J. G., Rosen C. A., Goh W. C., Haseltine W. A. The trans-activator gene of the human T cell lymphotropic virus type III is required for replication. Cell. 1986 Mar 28;44(6):941–947. doi: 10.1016/0092-8674(86)90017-6. [DOI] [PubMed] [Google Scholar]
- Delling U., Roy S., Sumner-Smith M., Barnett R., Reid L., Rosen C. A., Sonenberg N. The number of positively charged amino acids in the basic domain of Tat is critical for trans-activation and complex formation with TAR RNA. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6234–6238. doi: 10.1073/pnas.88.14.6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C., Ernberg I., Gait M. J., Green S. M., Heaphy S., Karn J., Lowe A. D., Singh M., Skinner M. A. HIV-1 tat protein stimulates transcription by binding to a U-rich bulge in the stem of the TAR RNA structure. EMBO J. 1990 Dec;9(12):4145–4153. doi: 10.1002/j.1460-2075.1990.tb07637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C., Ernberg I., Gait M. J., Green S. M., Heaphy S., Karn J., Lowe A. D., Singh M., Skinner M. A., Valerio R. Human immunodeficiency virus 1 tat protein binds trans-activation-responsive region (TAR) RNA in vitro. Proc Natl Acad Sci U S A. 1989 Sep;86(18):6925–6929. doi: 10.1073/pnas.86.18.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H. Phy M: an RNase activity specific for U and A residues useful in RNA sequence analysis. Nucleic Acids Res. 1980 Jul 25;8(14):3133–3142. doi: 10.1093/nar/8.14.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eadie J. S., McBride L. J., Efcavitch J. W., Hoff L. B., Cathcart R. High-performance liquid chromatographic analysis of oligodeoxyribonucleotide base composition. Anal Biochem. 1987 Sep;165(2):442–447. doi: 10.1016/0003-2697(87)90294-6. [DOI] [PubMed] [Google Scholar]
- Feng S., Holland E. C. HIV-1 tat trans-activation requires the loop sequence within tar. Nature. 1988 Jul 14;334(6178):165–167. doi: 10.1038/334165a0. [DOI] [PubMed] [Google Scholar]
- Fisher A. G., Feinberg M. B., Josephs S. F., Harper M. E., Marselle L. M., Reyes G., Gonda M. A., Aldovini A., Debouk C., Gallo R. C. The trans-activator gene of HTLV-III is essential for virus replication. 1986 Mar 27-Apr 2Nature. 320(6060):367–371. doi: 10.1038/320367a0. [DOI] [PubMed] [Google Scholar]
- Garcia J. A., Harrich D., Soultanakis E., Wu F., Mitsuyasu R., Gaynor R. B. Human immunodeficiency virus type 1 LTR TATA and TAR region sequences required for transcriptional regulation. EMBO J. 1989 Mar;8(3):765–778. doi: 10.1002/j.1460-2075.1989.tb03437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatignol A., Kumar A., Rabson A., Jeang K. T. Identification of cellular proteins that bind to the human immunodeficiency virus type 1 trans-activation-responsive TAR element RNA. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7828–7832. doi: 10.1073/pnas.86.20.7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor R., Soultanakis E., Kuwabara M., Garcia J., Sigman D. S. Specific binding of a HeLa cell nuclear protein to RNA sequences in the human immunodeficiency virus transactivating region. Proc Natl Acad Sci U S A. 1989 Jul;86(13):4858–4862. doi: 10.1073/pnas.86.13.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentz R., Chen C. H., Rosen C. A. Bioassay for trans-activation using purified human immunodeficiency virus tat-encoded protein: trans-activation requires mRNA synthesis. Proc Natl Acad Sci U S A. 1989 Feb;86(3):821–824. doi: 10.1073/pnas.86.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauber J., Cullen B. R. Mutational analysis of the trans-activation-responsive region of the human immunodeficiency virus type I long terminal repeat. J Virol. 1988 Mar;62(3):673–679. doi: 10.1128/jvi.62.3.673-679.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberger C. Fluorinated pyrimidines. Prog Nucleic Acid Res Mol Biol. 1965;4:1–50. doi: 10.1016/s0079-6603(08)60783-7. [DOI] [PubMed] [Google Scholar]
- Jaeger J. A., Turner D. H., Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger J. A., Turner D. H., Zuker M. Predicting optimal and suboptimal secondary structure for RNA. Methods Enzymol. 1990;183:281–306. doi: 10.1016/0076-6879(90)83019-6. [DOI] [PubMed] [Google Scholar]
- Jakobovits A., Smith D. H., Jakobovits E. B., Capon D. J. A discrete element 3' of human immunodeficiency virus 1 (HIV-1) and HIV-2 mRNA initiation sites mediates transcriptional activation by an HIV trans activator. Mol Cell Biol. 1988 Jun;8(6):2555–2561. doi: 10.1128/mcb.8.6.2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koontz S. W., Schimmel P. R. Aminoacyl-tRNA synthetase-catalyzed cleavage of the glycosidic bond of 5-halogenated uridines. J Biol Chem. 1979 Dec 25;254(24):12277–12280. [PubMed] [Google Scholar]
- Marciniak R. A., Calnan B. J., Frankel A. D., Sharp P. A. HIV-1 Tat protein trans-activates transcription in vitro. Cell. 1990 Nov 16;63(4):791–802. doi: 10.1016/0092-8674(90)90145-5. [DOI] [PubMed] [Google Scholar]
- Marciniak R. A., Garcia-Blanco M. A., Sharp P. A. Identification and characterization of a HeLa nuclear protein that specifically binds to the trans-activation-response (TAR) element of human immunodeficiency virus. Proc Natl Acad Sci U S A. 1990 May;87(9):3624–3628. doi: 10.1073/pnas.87.9.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerovitch K., Pelletier J., Sonenberg N. A cellular protein that binds to the 5'-noncoding region of poliovirus RNA: implications for internal translation initiation. Genes Dev. 1989 Jul;3(7):1026–1034. doi: 10.1101/gad.3.7.1026. [DOI] [PubMed] [Google Scholar]
- Muesing M. A., Smith D. H., Capon D. J. Regulation of mRNA accumulation by a human immunodeficiency virus trans-activator protein. Cell. 1987 Feb 27;48(4):691–701. doi: 10.1016/0092-8674(87)90247-9. [DOI] [PubMed] [Google Scholar]
- Müller W. E., Okamoto T., Reuter P., Ugarkovic D., Schröder H. C. Functional characterization of Tat protein from human immunodeficiency virus. Evidence that Tat links viral RNAs to nuclear matrix. J Biol Chem. 1990 Mar 5;265(7):3803–3808. [PubMed] [Google Scholar]
- Ogilvie K. K., Usman N., Nicoghosian K., Cedergren R. J. Total chemical synthesis of a 77-nucleotide-long RNA sequence having methionine-acceptance activity. Proc Natl Acad Sci U S A. 1988 Aug;85(16):5764–5768. doi: 10.1073/pnas.85.16.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin N. T., Cohen E. A., Darveau A., Rosen C., Haseltine W., Sonenberg N. Mutational analysis of the 5' non-coding region of human immunodeficiency virus type 1: effects of secondary structure on translation. EMBO J. 1988 Sep;7(9):2831–2837. doi: 10.1002/j.1460-2075.1988.tb03139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi J. D., Wyatt J. R., Tinoco I., Jr Solution conformation of an RNA hairpin loop. Biochemistry. 1990 May 1;29(17):4215–4226. doi: 10.1021/bi00469a026. [DOI] [PubMed] [Google Scholar]
- Rosen C. A., Pavlakis G. N. Tat and Rev: positive regulators of HIV gene expression. AIDS. 1990 Jun;4(6):499–509. [PubMed] [Google Scholar]
- Rosen C. A., Sodroski J. G., Haseltine W. A. The location of cis-acting regulatory sequences in the human T cell lymphotropic virus type III (HTLV-III/LAV) long terminal repeat. Cell. 1985 Jul;41(3):813–823. doi: 10.1016/s0092-8674(85)80062-3. [DOI] [PubMed] [Google Scholar]
- Roy S., Delling U., Chen C. H., Rosen C. A., Sonenberg N. A bulge structure in HIV-1 TAR RNA is required for Tat binding and Tat-mediated trans-activation. Genes Dev. 1990 Aug;4(8):1365–1373. doi: 10.1101/gad.4.8.1365. [DOI] [PubMed] [Google Scholar]
- Roy S., Parkin N. T., Rosen C., Itovitch J., Sonenberg N. Structural requirements for trans activation of human immunodeficiency virus type 1 long terminal repeat-directed gene expression by tat: importance of base pairing, loop sequence, and bulges in the tat-responsive sequence. J Virol. 1990 Mar;64(3):1402–1406. doi: 10.1128/jvi.64.3.1402-1406.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaie M. R., Benter T., Wong-Staal F. Site-directed mutagenesis of two trans-regulatory genes (tat-III,trs) of HIV-1. Science. 1988 Feb 19;239(4842):910–913. doi: 10.1126/science.3277284. [DOI] [PubMed] [Google Scholar]
- Scaringe S. A., Francklyn C., Usman N. Chemical synthesis of biologically active oligoribonucleotides using beta-cyanoethyl protected ribonucleoside phosphoramidites. Nucleic Acids Res. 1990 Sep 25;18(18):5433–5441. doi: 10.1093/nar/18.18.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby M. J., Bain E. S., Luciw P. A., Peterlin B. M. Structure, sequence, and position of the stem-loop in tar determine transcriptional elongation by tat through the HIV-1 long terminal repeat. Genes Dev. 1989 Apr;3(4):547–558. doi: 10.1101/gad.3.4.547. [DOI] [PubMed] [Google Scholar]
- Sinha N. D., Biernat J., McManus J., Köster H. Polymer support oligonucleotide synthesis XVIII: use of beta-cyanoethyl-N,N-dialkylamino-/N-morpholino phosphoramidite of deoxynucleosides for the synthesis of DNA fragments simplifying deprotection and isolation of the final product. Nucleic Acids Res. 1984 Jun 11;12(11):4539–4557. doi: 10.1093/nar/12.11.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodroski J., Rosen C., Wong-Staal F., Salahuddin S. Z., Popovic M., Arya S., Gallo R. C., Haseltine W. A. Trans-acting transcriptional regulation of human T-cell leukemia virus type III long terminal repeat. Science. 1985 Jan 11;227(4683):171–173. doi: 10.1126/science.2981427. [DOI] [PubMed] [Google Scholar]
- Starzyk R. M., Koontz S. W., Schimmel P. A covalent adduct between the uracil ring and the active site of an aminoacyl tRNA synthetase. Nature. 1982 Jul 8;298(5870):136–140. doi: 10.1038/298136a0. [DOI] [PubMed] [Google Scholar]
- Tang R. S., Draper D. E. Bulge loops used to measure the helical twist of RNA in solution. Biochemistry. 1990 Jun 5;29(22):5232–5237. doi: 10.1021/bi00474a003. [DOI] [PubMed] [Google Scholar]
- Weeks K. M., Ampe C., Schultz S. C., Steitz T. A., Crothers D. M. Fragments of the HIV-1 Tat protein specifically bind TAR RNA. Science. 1990 Sep 14;249(4974):1281–1285. doi: 10.1126/science.2205002. [DOI] [PubMed] [Google Scholar]
- Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989 Apr 7;244(4900):48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]