Abstract
By using site-specific protein-DNA photocrosslinking, we define the positions of TATA-binding protein, transcription factor IIB, transcription factor IIF, and subunits of RNA polymerase II (RNAPII) relative to promoter DNA within the human transcription preinitiation complex. The results indicate that the interface between the largest and second-largest subunits of RNAPII forms an extended, ≈240 Å channel that interacts with promoter DNA both upstream and downstream of the transcription start. By using electron microscopy, we show that RNAPII compacts promoter DNA by the equivalent of ≈50 bp. Together with the published structure of RNAPII, the results indicate that RNAPII wraps DNA around its surface and suggest a specific model for the trajectory of the wrapped DNA.
Keywords: DNA compaction, DNA wrapping, DNA bending, protein-DNA photocrosslinking, electron microscopy
Transcription initiation at a eukaryotic protein-encoding gene involves assembly on promoter DNA of a complex consisting of RNA polymerase II (RNAPII) and six general transcription factors: IIA, IIB, IID (or TATA-element binding protein, TBP), IIE, IIF, and IIH (1–3). A subcomplex containing TBP, IIB, IIF, RNAPII, and promoter DNA is stable (4, 5), and, under certain conditions (e.g., conditions that promote DNA melting), is fully competent for transcription initiation (6–11). Results of DNA footprinting experiments indicate that the TBP-IIB-IIF-RNAPII-promoter complex with linear DNA at 30°C involves interactions both upstream and downstream of the transcription start (positions −42 to +17; H. Lu and D.R., unpublished data) and involves unmelted DNA (11). Thus, the TBP-IIB-IIF-RNAPII-promoter complex with linear DNA at 30°C appears to correspond to the RNA polymerase-promoter “intermediate complex” characterized in studies of Escherichia coli RNA polymerase (RPi or RPc2; refs. 12–15).
The TBP-IIB-IIF-RNAPII-promoter complex contains at least 14 distinct polypeptides (one in TBP, one in IIB, two in IIF, and at least 10 in RNAPII) and has a molecular mass in excess of 700 kDa (1–3). High-resolution structures have been determined for the TBP-DNA and TBP-IIB-DNA complexes (16–18). However, the TBP-IIB-IIF-RNAPII-promoter complex is too large for high-resolution structure determination by current methods. Therefore, information about the structure of the TBP-IIB-IIF-RNAPII-promoter complex must rely on low-resolution structure determination (19, 20) supplemented by biochemical and imaging data.
In the work in this report, we have used site-specific protein-DNA photocrosslinking and electron microscopy to define protein-DNA interactions within the human TBP-IIB-IIF-RNAPII-promoter complex.
MATERIALS AND METHODS
Derivatized Promoter DNA Fragments.
Derivatized promoter DNA fragments were prepared essentially as in ref. 21. Oligodeoxyribonucleotides containing phosphorothioate 5′ to the third nucleotide were synthesized by using solid-phase β-cyanoethylphosphoramidite chemistry and tetraethylthiuram disulfide (Applied Biosystems), purified on OPC (Applied Biosystems), and derivatized with azidophenacyl bromide (Sigma; ref. 22). Derivatized oligodeoxyribonucleotides (10 pmol) were radiophosphorylated by reaction with T4 polynucleotide kinase (10 units; Ambion) and [γ32P]-ATP (75 pmol; 200 Bq/fmol; New England Nuclear) in 20 μl of 50 mM Tris⋅HCl, pH 7.5/2.5 mM KCl/10 mM MgCl2/0.5 mM β-mercaptoethanol/2.5% glycerol for 20 min at 37°C, followed by 5 min at 65°C, and were desalted on CHROMA SPIN-10 (CLONTECH). Radiophosphorylated derivatized oligodeoxyribonucleotides (10 pmol), M13 universal sequencing primer (10 pmol; 5′-TGACCGGCAGCAAAATG-3′), and M13mp18-AdMLP or M13mp19-AdMLP ssDNA (0.4 pmol; ref. 21), were annealed in 40 μl of 20 mM Tris⋅HCl, pH 8.0/25 mM KCl/4 mM MgCl2/1 mM EDTA, by using a temperature gradient of 65°C to 25°C in 90 min. They were extended and ligated by the addition of T4 DNA polymerase [3 units; New England Biolabs; desalted into 10 mM potassium phosphate (pH 6.5) and 50% glycerol], T4 DNA ligase (5 units; Boehringer; desalted into 20 mM Tris⋅HCl, pH 7.5/60 mM KCl/1 mM EDTA/5 mM β-mercaptoethanol/50% glycerol), 2′-deoxynucleoside 5′-triphosphates (40 nmol each; Pharmacia), and ATP (100 nmol; Pharmacia), and incubation for 15 min at 25°C followed by 30 min at 37°C. The resulting derivatized M13mp18-AdMLP or M13mp19-AdMLP double-stranded DNA was desalted on CHROMA SPIN-100 (CLONTECH) and digested with 10 units of EcoRI (GIBCO-BRL) and 10 units of SphI (Boehringer) in 60 mM Tris⋅HCl, pH 8.0/100 mM NaCl/10 mM KCl/10 mM MgCl2/1 mM EDTA/5% glycerol for 60 min at 37°C. Derivatized promoter DNA fragments were isolated by 8% PAGE, followed by elution of excised gel slices in 500 μl of 10 mM Tris⋅HCl, pH 8.0/200 mM NaCl/1 mM EDTA for 15 hr at 30°C, followed by ethanol precipitation.
Electrophoretic mobility shift experiments (23) established that the derivatized promoter DNA fragments retain the ability to form the TBP-IIB-IIF-RNAPII-promoter complex (data not shown).
Site-Specific Protein-DNA Photocrosslinking.
Reaction mixtures for complex formation contained 20 μl of 2 nM derivatized promoter DNA fragment (200 Bq/fmol), 5 nM human TBP (24), 5 nM human IIB (24), 5 nM human IIF (24), 10 nM human RNAPII (form “IIA”; ref. 24), 10 mM Hepes⋅NaOH (pH 7.9), 60 mM KCl, 4 mM MgCl2, 0.2 mM EDTA, 25 mg/ml poly(dG-dC) (MWav = 700 kDa), 2.6% polyethylene glycol (MWav = 8 kDa), and 5% glycerol. After 40 min at 30°C, reaction mixtures were UV-irradiated for 2 min at 25°C (900 erg/mm2 per sec; samples in polystyrene microcentrifuge tubes held inside 13 × 100-mm borosilicate glass culture tubes) by using a Rayonet RPR-100 photochemical reactor equipped with 16 RPR-3500 Å tubes and a RMA400 sample holder (Southern New England Ultraviolet, Hamden, CT). After UV irradiation, nuclease digestion was performed as in ref. 25, by using 12 units of DNase I, 20 units of nuclease S1, and a mixture of protease inhibitors (350 μg/ml phenylmethylsulfonyl fluoride, 0.5 μg/ml leupeptin, 0.7 μg/ml pepstatin, 1 μg/ml aprotinin, and 100 μg/ml chymostatin). Reactions were terminated by the addition of 0.2 vol of 10 M urea/250 mM Tris⋅HCl, pH 6.8/10% SDS/25% β-mercaptoethanol/50% glycerol/1% bromophenol blue, followed by heating 3 min at 65°C. Products were analyzed by 7–15% gradient SDS/PAGE. Identities of crosslinked polypeptides were confirmed by using derivatives with different molecular masses: TBPc (21), IIBc (21), IIF with hexahistidine-tagged RAP30 (IIF-RAP30*; ref. 26), and IIF with hexahistidine-tagged RAP74(1–409) (IIF-RAP74*; ref. 27) (Fig. 1C and data not shown). Standard photocrosslinking experiments were performed for each of the 80 derivatized promoter DNA fragments (2–5 independent experiments per DNA fragment).
Figure 1.
Results of protein-DNA photocrosslinking within the TBP-IIB-IIF-RNAPII-promoter complex. (A) Representative data from standard photocrosslinking experiments (positions −24 to −8 of DNA template strand). (B) Representative data from control “in-gel” photocrosslinking experiments (positions −24 to −8 of DNA template strand). (C) Representative data from control experiments by using IIF-RAP30* and IIF-RAP74* (positions −18 and −14 of DNA template strand; see Materials and Methods). (D) Summary of results (results for nontemplate strand above sequence; results for template strand beneath sequence). Phosphates analyzed are indicated by ∗. Sites exhibiting reproducible crosslinking are indicated by solid bars; sites exhibiting less reproducible crosslinking are indicated by shaded bars (scoring based on data from 2–5 independent standard photocrosslinking experiments). The TATA element and transcription start are indicated by shading.
“In-Gel” Site-Specific Protein-DNA Photocrosslinking.
Reaction mixtures identical to those in standard photocrosslinking experiments were electrophoresed in 45 mM Tris-borate (pH 8.0), 0.1 mM EDTA, through 5% polyacrylamide (19:1 acrylamide/N,N′-bisacrylylcystamine, ref. 28) slab gels (27 × 15 × 0.15 cm; 7 V/cm; 5 hr at 25°C). After electrophoresis, gels with both glass plates in place were mounted vertically in a Rayonet RPR-100 photochemical reactor and UV-irradiated at 350 nm for 4 min at 25°C (900 erg/mm2 per sec). Gels were autoradiographed 2 hr at 25°C, and excised gel slices containing complexes were solubilized by the addition of 8 μl of 1 M DTT (5 min at 37°C). Nuclease digestions were performed as in standard photocrosslinking experiments, by using 24 units of DNase I (20 min at 37°C) and 40 units of nuclease S1 (15 min at 37°C), omitting urea and β-mercaptoethanol in the termination step, and heating 10 min at 65°C in the termination step. Crosslinked polypeptides were identified by 7–15% gradient SDS/PAGE and autoradiography. “In-gel” photocrosslinking experiments were performed for 55 of the 80 derivatized promoter DNA fragments.
Electron Microscopy.
Samples were prepared and imaged as in ref. 29 (direct-mounting method) by using reaction mixtures containing 10 nM DNA fragment (prepared by PCR of pΔ50; ref. 30), 7 nM human TBP (24), 10 nM human IIB (24), 4 nM human IIF (24), 10 nM human RNAPII (form “IIA”; ref. 24), 0.6 mM ATP, 10 mM Tris⋅HCl (pH 7.9), 10 mM Hepes-OH, 50 mM KCl, 10 mM MgCl2, 8 mM ammonium sulfate, 0.1 mM EDTA, 10% (wt/vol) glycerol, and 1.7% (wt/vol) polyethylene glycol. Contour lengths were measured from micrographs by using a digitizing tablet (Summagraphics, Fairfield, CT) and software written by J.D.G. Bend angles were measured from micrographs by using a charged-coupled device camera (Cohu, San Diego) and an angle-calculation routine in image (National Institutes of Health).
Molecular Modeling.
Atomic coordinates for TBP-IIB-TATA at 2.7 Å resolution (18) were obtained from S. Burley (The Rockefeller University, New York). Modeled atomic coordinates for the DNA segments upstream and downstream of the TATA element were generated in insightii (Molecular Simulations, San Diego). Electron density data for yeast RNAPII Δ4/7 at 16 Å resolution (19) were obtained from S. Darst (The Rockefeller University), edited to eliminate lattice-neighbor density, converted to insightii contour-file format, and rendered as an insightii contour object (1.6 σ electron-density isocontour). Atomic structures and contour objects were viewed and manipulated in insightii.
RESULTS
Site-Specific Protein-DNA Photocrosslinking.
To define positions of polypeptides relative to promoter DNA within the human TBP-IIB-IIF-RNAPII-promoter complex, we have performed site-specific protein-DNA photocrosslinking (ref. 21; Fig. 1). We constructed 80 site-specifically derivatized DNA fragments, each containing a phenyl-azide photoactivatible crosslinking agent incorporated at a single, defined phosphate of the adenovirus major late promoter (positions −55 to +25; Fig. 1D). For each of the 80 DNA fragments, we then formed the TBP-IIB-IIF-RNAPII-promoter complex, UV-irradiated the TBP-IIB-IIF-RNAPII-promoter complex, and determined the polypeptide or polypeptides at which crosslinking occurred.
Representative data are presented in Fig. 1 A-C, and data are summarized in Fig. 1D. Three lines of evidence indicate that the crosslinks summarized in Fig. 1D are specific, i.e., that they require formation of the specific TBP-IIB-IIF-RNAPII-promoter complex. First, the crosslinks are position-dependent. Second, for each polypeptide, except RPB2, the crosslinks are TBP-dependent (data not shown). Third, standard photocrosslinking experiments and “in-gel” photocrosslinking experiments—with preisolation of specific TBP-IIB-IIF-RNAPII-promoter complexes by nondenaturing PAGE and UV irradiation in situ—yield similar patterns of crosslinking (compare Fig. 1 A and B).
TBP and IIB.
TBP and IIB exhibit patterns of crosslinking in the TBP-IIB-IIF-RNAPII-promoter complex similar to those previously reported for the TBP-DNA and TBP-IIB-DNA complexes (21) and in agreement with the crystallographic structures of the TBP-DNA and TBP-IIB-DNA complexes (16–18) (Fig. 1D). Thus, TBP crosslinked between positions −33 and −22, and IIB crosslinked between positions −39 and −12. The cross-groove patterns of crosslinking indicate that TBP interacts with the DNA minor groove of the TATA element and that IIB interacts with the DNA major groove immediately upstream of the TATA element and the DNA minor groove immediately downstream of the TATA element.
IIF.
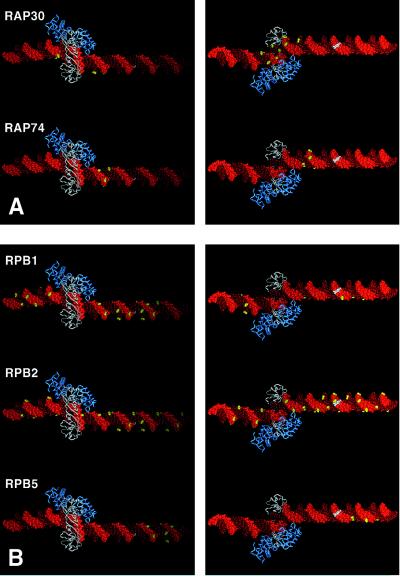
Human IIF consists of two polypeptides: RAP30 and RAP74 (1–2). Both RAP30 and RAP74 crosslink to DNA, with RAP30 crosslinking between positions −26 and −12 (and, to a lesser extent, positions −37 and −31), and with RAP74 crosslinking between positions −16 and −10 (Figs. 1D and 2A). The cross-groove patterns of crosslinking indicate that RAP30 interacts with the DNA major groove immediately downstream of the TATA element (and possibly with the TATA element itself), and that RAP74 interacts with the major groove ≈10 bp downstream of the TATA element (Fig. 2A). These results are in general agreement with results indicating crosslinking of RAP30 at −19 and RAP74 at −15 and −5 (31, 32).
Figure 2.
Structural interpretation of protein-DNA photocrosslinking within the TBP-IIB-IIF-RNAPII-promoter complex. (A) Sites at which polypeptides of IIF crosslink to DNA (yellow). (B) Sites at which polypeptides of RNAPII crosslink to DNA (yellow). For each polypeptide, two views are shown of the TBP-IIB-promoter complex along the vector of the TBP-induced DNA bend: a “top” view, with DNA ends receding from viewer (Left) and a “bottom” view, with DNA ends approaching viewer (Right). TBP, white; IIB, blue; DNA nontemplate strand, light red; DNA template strand, dark red; the transcription start, white.
In experiments with one preparation of IIF, RAP74 also crosslinked to DNA at positions +1, +3, +9, +11, and +19 of the nontemplate strand and positions +7, +11, +13, +15, and +17 of the template strand, possibly indicating binding of a second molecule of RAP74 to the complex (data not shown). This preparation of IIF may have contained an effective stoichiometric excess of RAP74 over RAP30.
Forget et al. (32) have reported that RAP74 can crosslink to a DNA fragment triply derivatized at positions −48, −46, and −45 of the nontemplate strand and to a DNA fragment doubly derivatized at positions −40 and −39 of the nontemplate strand. They have proposed that RAP74 interacts with DNA both upstream and downstream of the TATA element. In contrast to Forget et al., we have not observed RAP74-DNA crosslinking upstream of the TATA element.
RNAPII.
Human RNAPII consists of at least 10 distinct polypeptides (1, 2). Only three of these polypeptides crosslink to DNA: RPB1 (the largest polypeptide; homologous to E. coli β′), RPB2 (the second-largest polypeptide; homologous to E. coli β), and RPB5 (the fifth-largest polypeptide; shared by RNAPI, RNAPII, and RNAPIII) (Figs. 1D and 2B). RPB1 crosslinks over an extensive region, from positions −53 to +9, and crosslinks to a single face of the DNA helix (Fig. 2B, Top Left). RPB2 also crosslinks over an extensive region, from positions −49 to +19, and, except between positions −4 and +3, crosslinks to a single face of the DNA helix—the face opposite that to which RPB1 crosslinks (Fig. 2B, Middle Right). RPB5 crosslinks between positions +5 and +15 on a single face of the DNA helix—the face to which RPB1 crosslinks (Fig. 2B, Bottom Left).
Our results establish the existence of an extremely long RNAPII-DNA contact region (≈70 bp, ≈240 Å) and show that, throughout most of the RNAPII-DNA contact region, RPB1 and RPB2 interact with opposite faces of the DNA helix (Figs. 1D and 2B). The RNAPII-DNA contact is continuous except in the vicinity of the TATA element, where a break in the RNAPII-DNA contact permits access to promoter DNA by TBP, IIB, and IIF (Figs. 1D and 2). The results confirm DNA-footprinting results showing protection of an extended region (refs. 33 and 34; H. Lu and D.R., unpublished data) and establish that protection is due to direct RNAPII-DNA contact, rather than to RNAPII-induced changes in DNA conformation.
We infer that RPB1 and RPB2 form opposite edges of a ≈240 Å channel that interacts with promoter DNA. In view of the fact that 240 Å is 1.7 times the longest dimension of RNAPII (140 × 136 × 100 Å; ref. 19), we further infer that RNAPII must wrap DNA around its surface.
Electron Microscopy.
To provide independent information regarding the trajectory of DNA in the TBP-IIB-IIF-RNAPII-promoter complex, we have used electron microscopy to characterize the complex on a 487-bp DNA fragment containing the adenovirus major late promoter (TATA element 267 bp from upstream end of DNA fragment).
Electron micrographs of complexes show a roughly spherical protein mass with a diameter of 110 ± 20 Å (a diameter consistent with that expected based on the molecular masses of TBP, IIB, IIF, and RNAPII) flanked by an upstream DNA segment and a shorter downstream DNA segment (protein-free contour lengths 50 ± 9% and 31 ± 7% the contour length of the DNA fragment alone) (Fig. 3A). [The identities of the upstream and downstream DNA segments were determined by imaging complexes after digestion of the DNA fragment with HindIII, which cleaves 212 bp from the upstream end (data not shown).] The upstream edge of the protein mass is located ≈20 bp upstream of the TATA element. The downstream edge of the protein mass is located downstream of the TATA element and transcription start (Fig. 3B).
Figure 3.
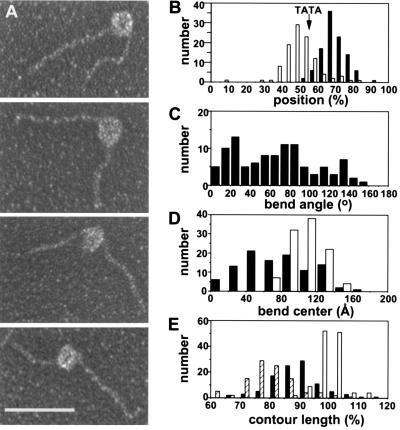
Electron microscopy of the TBP-IIB-IIF-RNAPII-promoter complex. (A) Representative images (upstream DNA segment at left in top two panels; downstream DNA segment at left in bottom two panels; see Materials and Methods). The bar represents 500 Å. (B) Observed positions of upstream and downstream edges of protein mass on DNA fragment. Open bars, positions of upstream edge (determined from protein-free contour lengths of upstream DNA segments). Solid bars, positions of downstream edge (determined from protein-free contour lengths of downstream DNA segments). Arrow, expected position of the TATA element. (C) Observed DNA bend angles (bend angle defined as angle of deviation from linear DNA trajectory). (D) Observed DNA bend centers (bend center defined as intersection of projected upstream and downstream DNA segments). Open bars, diameters of protein mass. Solid bars, distances between bend center and upstream edge of protein mass. (E) Observed contour lengths. Open bars, DNA alone. Hatched bars, protein-free DNA in complexes. Solid bars, protein-free DNA plus diameter of protein mass in complexes.
Analysis of the micrographs reveals three important features:
First, DNA in the complex is bent (Fig. 3 A, C, and D). The mean observed DNA bend angle is 70°, but the distribution is broad and multimodal [0°–160°; presumably due to different orientations of complexes on the sample grid, and/or to the influence of hydrodynamic forces upon adsorption to the sample grid (see ref. 27)] (Fig. 3C). The observed DNA bend center is located at or near the center of the protein mass (Fig. 3D).
Second, the protein-free DNA contour length in the complex—i.e., the sum of the contour lengths of the upstream and downstream DNA segments—is 19% less than the contour length of the DNA fragment in the absence of protein (Fig. 3E, hatched and open bars). This indicates that, within the resolution of our electron micrographs, 19% of the DNA fragment, or ≈90 bp, is associated with the protein mass. This suggests, in agreement with our photocrosslinking results, the existence of a long protein-DNA contact surface.
Third, and most important, the apparent overall DNA contour length in the complex—i.e., the protein-free DNA contour length plus the diameter of the protein mass—is 10% less than the contour length of the DNA fragment in the absence of protein (Fig. 3E, solid and open bars). This 10% difference, which is highly statistically significant (P < 0.0001), indicates that the DNA fragment is compacted by the equivalent of 10%, or ≈50 bp, by its association with the protein mass. Compaction of double-stranded DNA of this magnitude under physiological conditions is diagnostic of wrapping of DNA around a protein core (analogous to wrapping of thread around a spool; see ref. 35).
While this work was under review for publication, Forget et al. (32) reported electron microscopic observations of TBP-IIB-IIE-IIF-RNAPII-promoter complexes. Their electron microscopic observations are in complete agreement with ours.
DISCUSSION
Our photocrosslinking results establish that RNAPII interacts with ≈240 Å of promoter DNA, i.e., with a length of promoter DNA approximately twice the longest dimension of RNAPII (19). Our electron microscopy results establish that RNAPII compacts promoter DNA by the equivalent of ≈50 bp. Taken together, the results make a compelling case that RNAPII wraps promoter DNA around its circumference. Based on the length of the RNAP-DNA contact surface and the magnitude of the RNAPII-induced DNA compaction, we estimate that RNAPII wraps promoter DNA around fully two-thirds of its circumference.
Kornberg and coworkers (19) have determined the structure of RNAPII at 16 Å resolution and have proposed that the active center of RNAPII—and thus the transcription start in a preinitiation complex—is located between two prominent finger-like projections. In addition, Kornberg and coworkers have determined the location of IIB in the RNAP-IIB complex—and thus the likely location of the promoter −30 region and TATA element in a preinitiation complex (20). Based on these results, and on the assumption that RNAPII does not undergo large structural changes upon formation of a preinitiation complex, Kornberg and coworkers have proposed a model for the trajectory of the promoter DNA segment between the TATA element and the transcription start in a preinitiation complex (20, 36). However, Kornberg and coworkers were not able to define the rotational phasing of the DNA segment, and—because the location of IIB was determined only in x-y projection, and not in three dimensions (20)—they also were not able to define whether the DNA segment interacts with the “top” or “bottom” faces of RNAPII (20).
Our photocrosslinking results provide three constraints for efforts to model the trajectory of DNA in a preinitiation complex. First, the observation that RNAPII crosslinks to DNA upstream of the TATA element (Figs. 1D and 2B) indicates that the TBP-induced DNA bend at the TATA element (16–18) must be phased to maximize contacts to the DNA segment upstream of the TATA element. Second, the observation that RNAPII crosslinks to DNA well downstream of the transcription start (Figs. 1D and 2B) indicates that the complex must contain a second DNA bend (or set of DNA bends), centered at or near the transcription start and phased to maximize contacts to the DNA segment downstream of the transcription start. Third, the observation that RNAPII crosslinks to DNA at half of tested phosphates between positions −53 and −6 (26 of 48; Figs. 1D and 2B) but at nearly all tested phosphates between −5 and +19 (23 of 24; Figs. 1D and 2B) indicates that RNAPII interacts with positions −53 to −6 through a shallow, relatively open channel but interacts with positions −5 to +19 through a deep, nearly completely enclosed, channel.
A model built by starting with the model of Kornberg and coworkers (20, 36) and incorporating the above constraints is presented in Fig. 4. The model invokes two DNA bends: (i) the ≈80° TBP-induced DNA bend (16–18), phased to maximize contacts to the DNA segment upstream of the TATA element, and (ii) a second ≈70° DNA bend, centered at the transcription start and phased to maximize contacts to the DNA segment at and downstream of the transcription start. The model places the DNA segment between the TATA element and the transcription start on the “top” face of RNAPII (“top” as defined in ref. 20; Fig. 4A) and places the DNA segment corresponding to positions −53 to −6 within a shallow, relatively open channel (Fig. 4A) and the DNA segment corresponding to positions −5 to +20 within a deep, nearly completely enclosed, channel (Fig. 4B). (An alternative model placing the DNA segment between the TATA element and the transcription start on the “bottom” face of RNAPII does not allow placement of the DNA segment corresponding to positions −5 to +20 within a deep, nearly completely enclosed, channel.) The model is consistent with the observed range of DNA bend angles (Fig. 3C), the observed DNA bend center (Fig. 3D), the observed degree of compaction of DNA (Fig. 3E; ref. 32), and the observed handedness of the DNA writhe, i.e., clockwise proceeding from upstream to downstream with the apex of the DNA bend at the top (cf., Figs. 3A and 4). The model is able to accommodate the crystallographic structures of the TBP-IIB-DNA (ref. 18; Fig. 4A) and TBP-IIA-DNA complexes (refs. 37 and 38; not shown) without steric hindrance and appears to have ample space to accommodate IIF RAP30 and IIF RAP74 (Fig. 4A).
Figure 4.
Model for the structure of the TBP-IIB-IIF-RNAPII-promoter complex (two views: upstream end at right in A, downstream end at right in B). TBP, white; IIB, blue; positions of IIF RAP30 and IIF RAP74, yellow; RNAPII, red; DNA template strand, green; DNA nontemplate strand, turquoise; the transcription start, yellow. NT, proposed binding site for the nontemplate strand of the transcription bubble in open and elongation complexes. RNA, proposed emergence point of nascent RNA in elongation complexes.
One important feature of the model in Fig. 4 is that it suggests a candidate for the binding site for the nontemplate strand of the transcription bubble in open and elongation complexes, i.e., a 40 Å long, 8 Å wide groove connecting positions −11 and +3 of the DNA binding channel and separated from the DNA binding channel by a 20 × 15 × 15 Å knob-like projection (Fig. 4A). A second important feature of the model is that it suggests a candidate for the binding site for nascent RNA in elongation complexes, i.e., a 50 Å long, 8–10 Å wide tunnel connecting the floor of the DNA binding channel near position −9 with the surface of RNAPII (Fig. 4A).
The model in Fig. 4 provides a framework for interpretation of genetic, biochemical, and structural data on RNAPII-dependent transcription initiation and regulation. Based on similarities between RNAPII and other multisubunit RNA polymerases (i.e., RNA polymerases I and III, E. coli RNA polymerase) in three-dimensional structure (39, 40), subunit primary structure (41, 42), DNA crosslinking (43–47), DNA bending (48–51), and DNA wrapping (51–54), we suggest that the model in Fig. 4 may apply generally to multisubunit RNA polymerases.
Acknowledgments
We thank S. Burley and S. Darst for structural data and G. Parkinson for assistance in reformatting electron-density files. This work was supported by a Howard Hughes Medical Institute investigatorship to D.R. and by National Institutes of Health Grants GM31819 to J.D.G., GM37120 to D.R., and GM51527 and GM53665 to R.H.E.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: RNAPII, RNA polymerase II; TBP, TATA-element binding protein.
References
- 1.Orphanides G, Lagrange T, Reinberg D. Genes Dev. 1996;8:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 2.Roeder R. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 3.Nikolov D, Burley S. Proc Natl Acad Sci USA. 1997;94:15–22. doi: 10.1073/pnas.94.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conaway R, Garrett P, Hanley J, Conaway J. Proc Natl Acad Sci USA. 1991;88:6205–6209. doi: 10.1073/pnas.88.14.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flores O, Lu H, Killeen M, Greenblatt J, Burton Z, Reinberg D. Proc Natl Acad Sci USA. 1991;88:9999–10003. doi: 10.1073/pnas.88.22.9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parvin J, Sharp P. Cell. 1993;73:533–540. doi: 10.1016/0092-8674(93)90140-l. [DOI] [PubMed] [Google Scholar]
- 7.Timmers H T M. EMBO J. 1994;13:391–399. doi: 10.1002/j.1460-2075.1994.tb06273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parvin J, Shykind B, Meyers R, Kim J, Sharp P. J Biol Chem. 1994;269:18414–18421. [PubMed] [Google Scholar]
- 9.Pan G, Greenblatt J. J Biol Chem. 1994;269:30101–30104. [PubMed] [Google Scholar]
- 10.Holstege F, Tantin D, Carey M, van der Vliet P, Timmers H T M. EMBO J. 1995;14:810–819. doi: 10.1002/j.1460-2075.1995.tb07059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holstege F, van der Vliet P, Timmers H T M. EMBO J. 1996;15:1666–1677. [PMC free article] [PubMed] [Google Scholar]
- 12.Spassky A, Kirkegaard K, Buc H. Biochemistry. 1985;24:2723–2731. doi: 10.1021/bi00332a019. [DOI] [PubMed] [Google Scholar]
- 13.Cowing D, Mecsas J, Record M T, Gross C. J Mol Biol. 1989;210:521–530. doi: 10.1016/0022-2836(89)90128-9. [DOI] [PubMed] [Google Scholar]
- 14.Schickor P, Metzger W, Werel W, Lederer H, Heumann H. EMBO J. 1990;19:2215–2220. doi: 10.1002/j.1460-2075.1990.tb07391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mecsas J, Cowing D, Gross C. J Mol Biol. 1991;220:585–597. doi: 10.1016/0022-2836(91)90102-c. [DOI] [PubMed] [Google Scholar]
- 16.Kim Y, Geiger J, Hahn S, Sigler P. Nature (London) 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 17.Kim J, Nikolov D, Burley S. Nature (London) 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 18.Nikolov D, Chen H, Halay E, Usheva A, Hisatake K, Lee D K, Roeder R, Burley S. Nature (London) 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- 19.Darst S, Edwards A, Kubalek E, Kornberg R. Cell. 1991;66:121–128. doi: 10.1016/0092-8674(91)90144-n. [DOI] [PubMed] [Google Scholar]
- 20.Leuther K, Bushnell D, Kornberg R. Cell. 1996;85:773–779. doi: 10.1016/s0092-8674(00)81242-8. [DOI] [PubMed] [Google Scholar]
- 21.Lagrange T, Kim T-K, Orphanides G, Ebright Y, Ebright R, Reinberg D. Proc Natl Acad Sci USA. 1996;93:10620–10625. doi: 10.1073/pnas.93.20.10620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayer A, Barany F. Gene. 1995;153:1–8. doi: 10.1016/0378-1119(94)00752-e. [DOI] [PubMed] [Google Scholar]
- 23.Maldonado E, Ha I, Cortes P, Weis L, Reinberg D. Mol Cell Biol. 1990;10:6335–6347. doi: 10.1128/mcb.10.12.6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maldonado E, Drapkin R, Reinberg D. Methods Enzymol. 1996;274:72–100. doi: 10.1016/s0076-6879(96)74009-0. [DOI] [PubMed] [Google Scholar]
- 25.Bartholomew B, Kassavetis G, Geiduschek E P. Mol Cell Biol. 1991;11:5181–5189. doi: 10.1128/mcb.11.10.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan S, Conaway R, Conaway J. BioTechniques. 1994;16:824–828. [PubMed] [Google Scholar]
- 27.Wang B, Burton Z. J Biol Chem. 1995;270:27035–27044. doi: 10.1074/jbc.270.45.27035. [DOI] [PubMed] [Google Scholar]
- 28.Hansen J, Pheiffer B, Boehnert J. Anal Biochem. 1980;105:192–201. doi: 10.1016/0003-2697(80)90445-5. [DOI] [PubMed] [Google Scholar]
- 29.Griffith J, Makhov A, Zawel L, Reinberg D. J Mol Biol. 1995;246:576–584. doi: 10.1016/s0022-2836(05)80107-x. [DOI] [PubMed] [Google Scholar]
- 30.Sawadogo M, Roeder R. Cell. 1985;43:165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- 31.Robert F, Forget D, Li J, Greenblatt J, Coulombe B. J Biol Chem. 1996;271:8517–8520. doi: 10.1074/jbc.271.15.8517. [DOI] [PubMed] [Google Scholar]
- 32.Forget D, Robert F, Grondin G, Burton Z, Greenblatt J, Coulombe B. Proc Natl Acad Sci USA. 1997;94:7150–7155. doi: 10.1073/pnas.94.14.7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Dyke M, Roeder R, Sawadogo M. Science. 1988;241:1335–1338. doi: 10.1126/science.3413495. [DOI] [PubMed] [Google Scholar]
- 34.Buratowski S, Hahn S, Guarante L, Sharp P. Cell. 1989;56:549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- 35.Wolffe A. Chromatin, Structure and Function. London: Academic; 1995. [Google Scholar]
- 36.Kornberg R. Trends Biochem Sci. 1996;21:325–326. [PubMed] [Google Scholar]
- 37.Geiger J, Hahn S, Lee S, Sigler P. Science. 1996;272:830–836. doi: 10.1126/science.272.5263.830. [DOI] [PubMed] [Google Scholar]
- 38.Tan S, Hunziker Y, Sargent D, Richmond T. Nature (London) 1996;381:127–134. doi: 10.1038/381127a0. [DOI] [PubMed] [Google Scholar]
- 39.Schultz P, Celia H, Riva M, Sentenac A, Oudet P. EMBO J. 1993;12:2601–2607. doi: 10.1002/j.1460-2075.1993.tb05920.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Polyakov A, Severinova E, Darst S. Cell. 1995;83:365–373. doi: 10.1016/0092-8674(95)90114-0. [DOI] [PubMed] [Google Scholar]
- 41.Young R. Annu Rev Biochem. 1991;60:689–715. doi: 10.1146/annurev.bi.60.070191.003353. [DOI] [PubMed] [Google Scholar]
- 42.Sentenac A, Riva M, Thuriaux P, Buhler J-M, Treich I, Carles C, Werner M, Ruet A, Huet J, Mann C, Chiannilkulchai N, Stettler S, Mariotte S. In: Transcriptional Regulation. McKnight S, Yamamoto K, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 27–54. [Google Scholar]
- 43.Simpson R. Cell. 1979;18:277–285. doi: 10.1016/0092-8674(79)90047-3. [DOI] [PubMed] [Google Scholar]
- 44.Chenchick A, Beabealashvilli R, Mirzabekov A. FEBS Lett. 1981;128:46–50. doi: 10.1016/0014-5793(81)81076-9. [DOI] [PubMed] [Google Scholar]
- 45.Bartholomew B, Durkovich D, Kassavetis G, Geiduschek E P. Mol Cell Biol. 1993;13:942–952. doi: 10.1128/mcb.13.2.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bartholomew B, Braun B, Kassavetis G, Geiduschek E P. J Biol Chem. 1994;269:18090–18095. [PubMed] [Google Scholar]
- 47.Nudler E, Avetissova E, Markovtsov V, Goldfarb A. Science. 1996;273:211–217. doi: 10.1126/science.273.5272.211. [DOI] [PubMed] [Google Scholar]
- 48.Heumann H, Ricchetti M, Werel W. EMBO J. 1988;7:4379–4381. doi: 10.1002/j.1460-2075.1988.tb03336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rees W, Keller R, Vesenka J, Yang G, Bustamante C. Science. 1993;260:1646–1649. doi: 10.1126/science.8503010. [DOI] [PubMed] [Google Scholar]
- 50.Meyers-Almes F, Heumann H, Porschke D. J Mol Biol. 1994;236:1–6. doi: 10.1006/jmbi.1994.1112. [DOI] [PubMed] [Google Scholar]
- 51.Rippe K, Guthold M, von Hippel P, Bustamante C. J Mol Biol. 1997;270:125–138. doi: 10.1006/jmbi.1997.1079. [DOI] [PubMed] [Google Scholar]
- 52.Buc H. Biochem Soc Trans. 1986;14:196–199. doi: 10.1042/bst0140196. [DOI] [PubMed] [Google Scholar]
- 53.Amouyal H, Buc H. J Mol Biol. 1987;195:795–808. doi: 10.1016/0022-2836(87)90485-2. [DOI] [PubMed] [Google Scholar]
- 54.Craig M, Suh W, Record M T. Biochemistry. 1995;34:15624–15632. doi: 10.1021/bi00048a004. [DOI] [PubMed] [Google Scholar]