Abstract
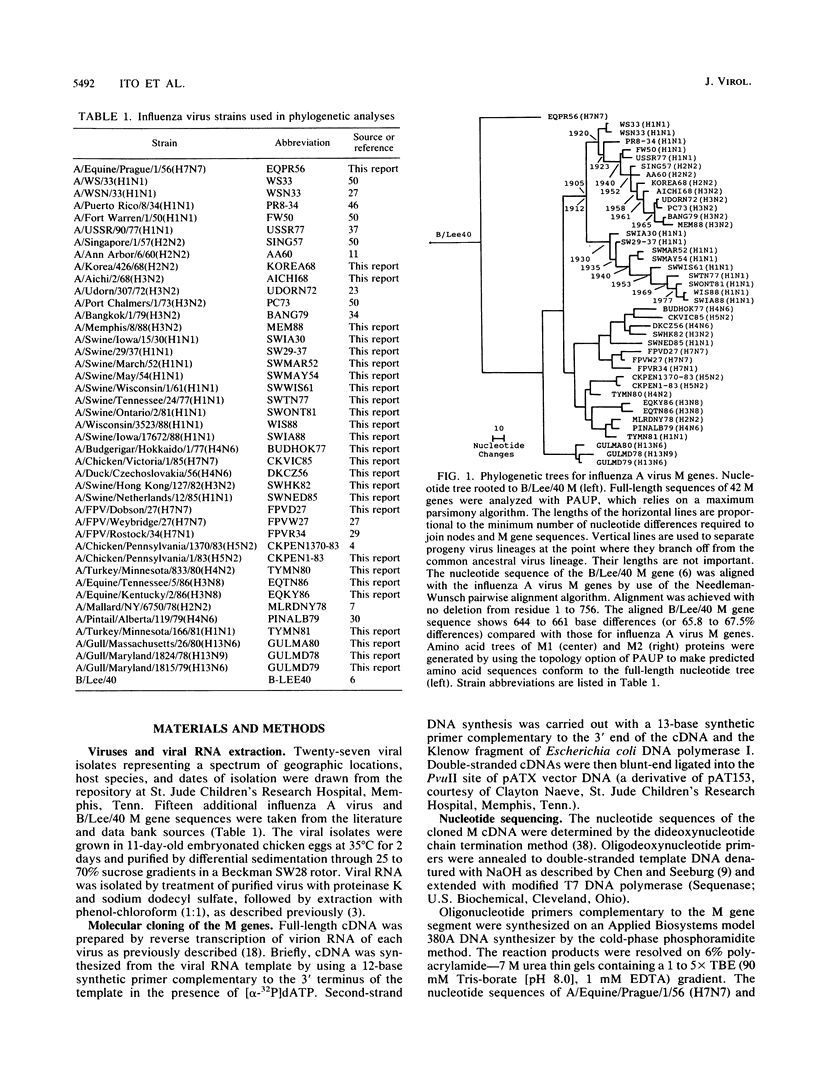
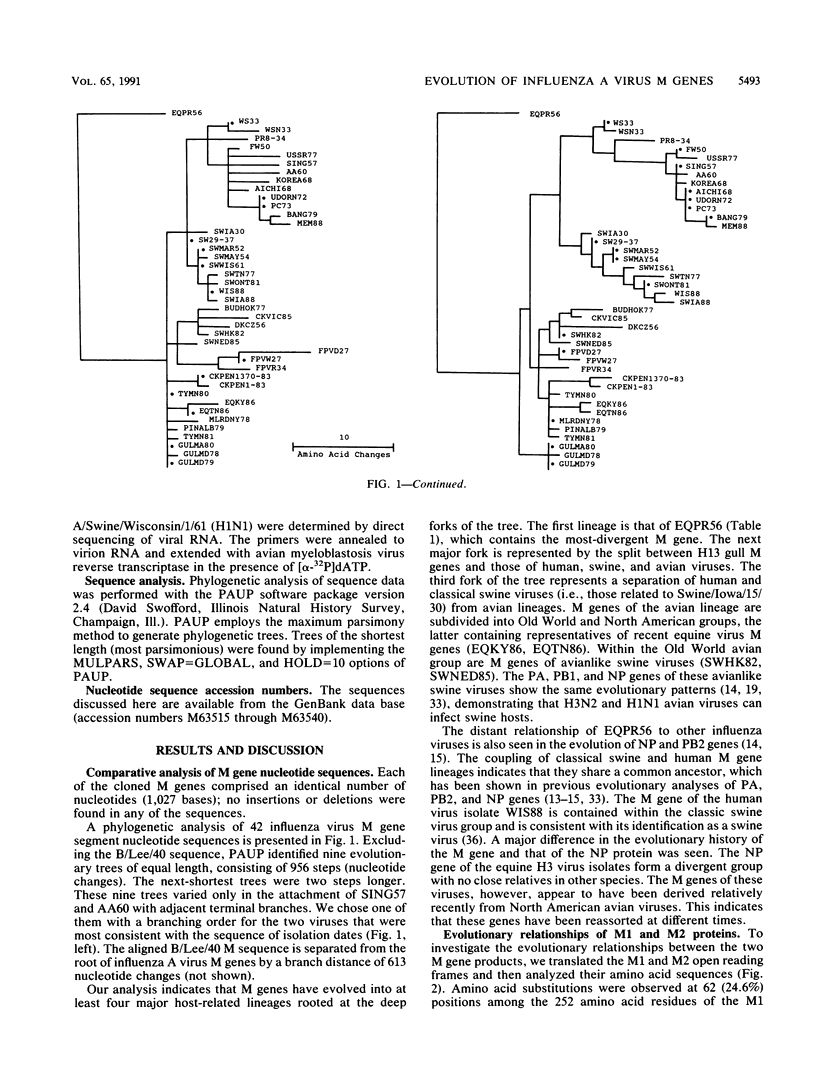
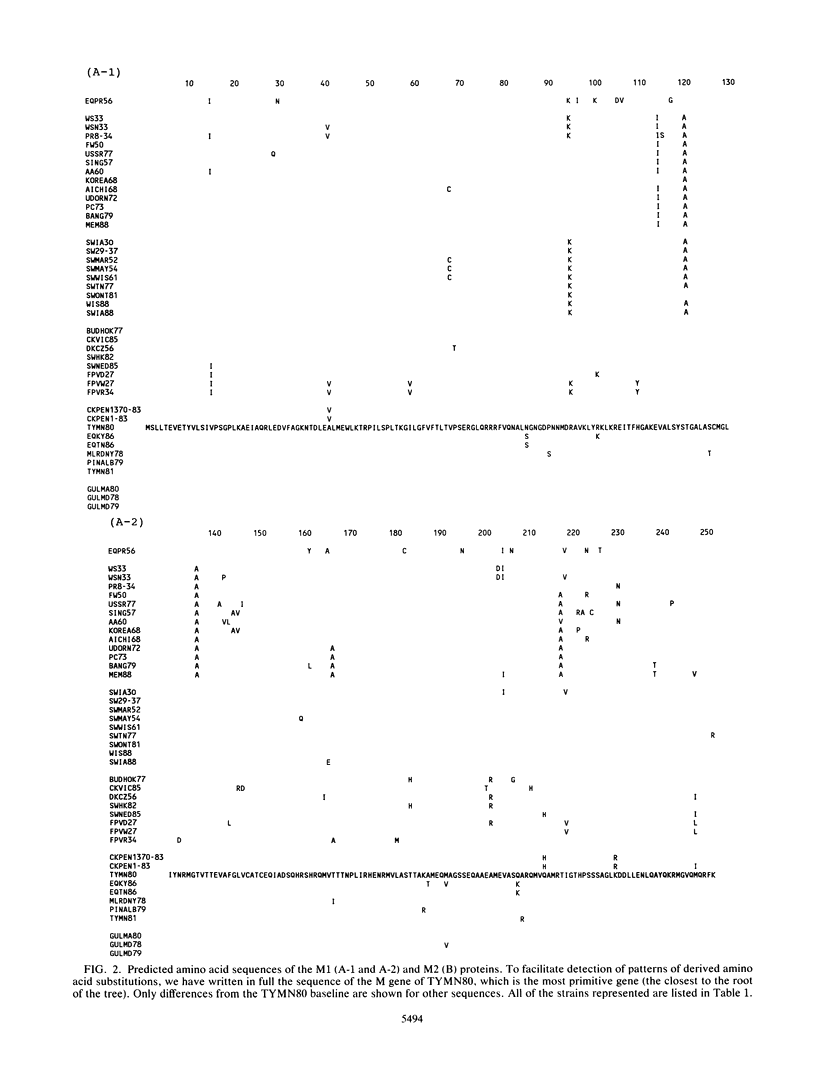
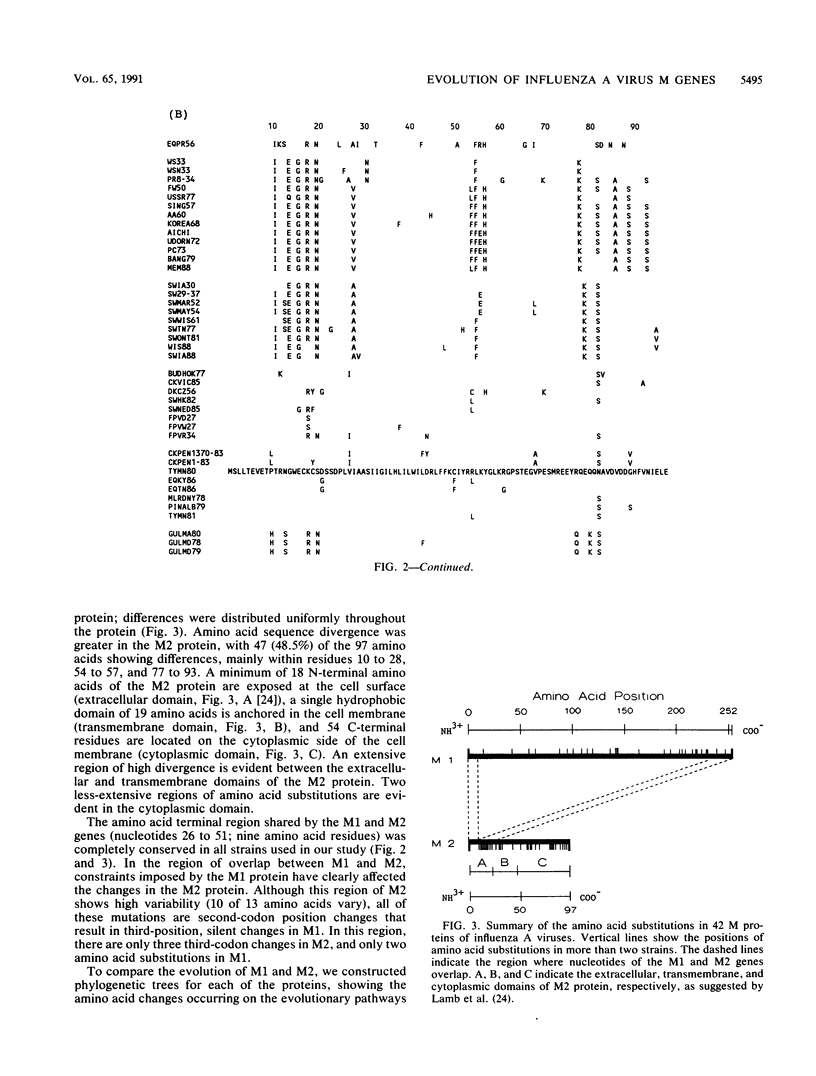
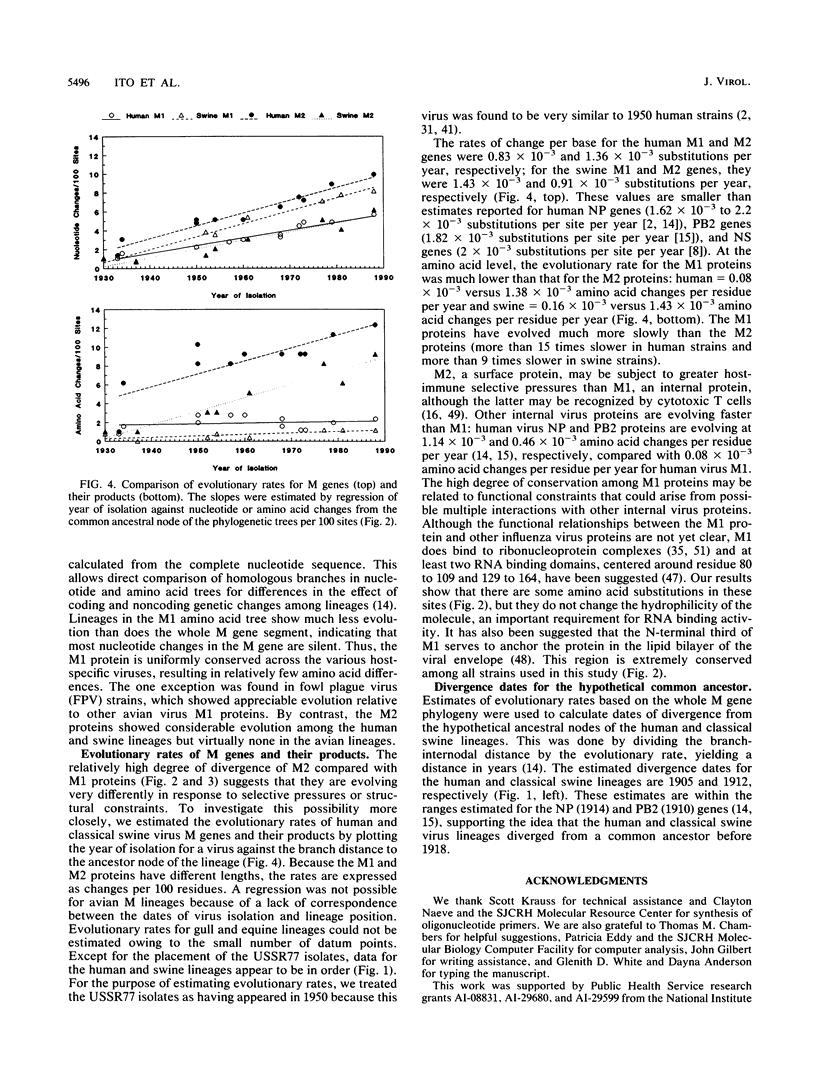
Phylogenetic analysis of 42 membrane protein (M) genes of influenza A viruses from a variety of hosts and geographic locations showed that these genes have evolved into at least four major host-related lineages: (i) A/Equine/prague/56, which has the most divergent M gene; (ii) a lineage containing only H13 gull viruses; (iii) a lineage containing both human and classical swine viruses; and (iv) an avian lineage subdivided into North American avian viruses (including recent equine viruses) and Old World avian viruses (including avianlike swine strains). The M gene evolutionary tree differs from those published for other influenza virus genes (e.g., PB1, PB2, PA, and NP) but shows the most similarity to the NP gene phylogeny. Separate analyses of the M1 and M2 genes and their products revealed very different patterns of evolution. Compared with other influenza virus genes (e.g., PB2 and NP), the M1 and M2 genes are evolving relatively slowly, especially the M1 gene. The M1 and M2 gene products, which are encoded in different but partially overlapping reading frames, revealed that the M1 protein is evolving very slowly in all lineages, whereas the M2 protein shows significant evolution in human and swine lineages but virtually none in avian lineages. The evolutionary rates of the M1 proteins were much lower than those of M2 proteins and other internal proteins of influenza viruses (e.g., PB2 and NP), while M2 proteins showed less rapid evolution compared with other surface proteins (e.g., H3HA). Our results also indicate that for influenza A viruses, the evolution of one protein of a bicistronic gene can affect the evolution of the other protein.(ABSTRACT TRUNCATED AT 400 WORDS)
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altmüller A., Fitch W. M., Scholtissek C. Biological and genetic evolution of the nucleoprotein gene of human influenza A viruses. J Gen Virol. 1989 Aug;70(Pt 8):2111–2119. doi: 10.1099/0022-1317-70-8-2111. [DOI] [PubMed] [Google Scholar]
- Bean W. J., Jr, Sriram G., Webster R. G. Electrophoretic analysis of iodine-labeled influenza virus RNA segments. Anal Biochem. 1980 Feb;102(1):228–232. doi: 10.1016/0003-2697(80)90343-7. [DOI] [PubMed] [Google Scholar]
- Bean W. J., Threlkeld S. C., Webster R. G. Biologic potential of amantadine-resistant influenza A virus in an avian model. J Infect Dis. 1989 Jun;159(6):1050–1056. doi: 10.1093/infdis/159.6.1050. [DOI] [PubMed] [Google Scholar]
- Both G. W., Sleigh M. J., Cox N. J., Kendal A. P. Antigenic drift in influenza virus H3 hemagglutinin from 1968 to 1980: multiple evolutionary pathways and sequential amino acid changes at key antigenic sites. J Virol. 1983 Oct;48(1):52–60. doi: 10.1128/jvi.48.1.52-60.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briedis D. J., Lamb R. A., Choppin P. W. Sequence of RNA segment 7 of the influenza B virus genome: partial amino acid homology between the membrane proteins (M1) of influenza A and B viruses and conservation of a second open reading frame. Virology. 1982 Jan 30;116(2):581–588. doi: 10.1016/0042-6822(82)90150-7. [DOI] [PubMed] [Google Scholar]
- Buckler-White A. J., Naeve C. W., Murphy B. R. Characterization of a gene coding for M proteins which is involved in host range restriction of an avian influenza A virus in monkeys. J Virol. 1986 Feb;57(2):697–700. doi: 10.1128/jvi.57.2.697-700.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonagurio D. A., Nakada S., Parvin J. D., Krystal M., Palese P., Fitch W. M. Evolution of human influenza A viruses over 50 years: rapid, uniform rate of change in NS gene. Science. 1986 May 23;232(4753):980–982. doi: 10.1126/science.2939560. [DOI] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Cox N. J., Kitame F., Kendal A. P., Maassab H. F., Naeve C. Identification of sequence changes in the cold-adapted, live attenuated influenza vaccine strain, A/Ann Arbor/6/60 (H2N2). Virology. 1988 Dec;167(2):554–567. [PubMed] [Google Scholar]
- Fang R., Min Jou W., Huylebroeck D., Devos R., Fiers W. Complete structure of A/duck/Ukraine/63 influenza hemagglutinin gene: animal virus as progenitor of human H3 Hong Kong 1968 influenza hemagglutinin. Cell. 1981 Aug;25(2):315–323. doi: 10.1016/0092-8674(81)90049-0. [DOI] [PubMed] [Google Scholar]
- Gammelin M., Altmüller A., Reinhardt U., Mandler J., Harley V. R., Hudson P. J., Fitch W. M., Scholtissek C. Phylogenetic analysis of nucleoproteins suggests that human influenza A viruses emerged from a 19th-century avian ancestor. Mol Biol Evol. 1990 Mar;7(2):194–200. doi: 10.1093/oxfordjournals.molbev.a040594. [DOI] [PubMed] [Google Scholar]
- Gorman O. T., Bean W. J., Kawaoka Y., Webster R. G. Evolution of the nucleoprotein gene of influenza A virus. J Virol. 1990 Apr;64(4):1487–1497. doi: 10.1128/jvi.64.4.1487-1497.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman O. T., Donis R. O., Kawaoka Y., Webster R. G. Evolution of influenza A virus PB2 genes: implications for evolution of the ribonucleoprotein complex and origin of human influenza A virus. J Virol. 1990 Oct;64(10):4893–4902. doi: 10.1128/jvi.64.10.4893-4902.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotch F., McMichael A., Smith G., Moss B. Identification of viral molecules recognized by influenza-specific human cytotoxic T lymphocytes. J Exp Med. 1987 Feb 1;165(2):408–416. doi: 10.1084/jem.165.2.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall R. M., Air G. M. Variation in nucleotide sequences coding for the N-terminal regions of the matrix and nonstructural proteins of influenza A viruses. J Virol. 1981 Apr;38(1):1–7. doi: 10.1128/jvi.38.1.1-7.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huddleston J. A., Brownlee G. G. The sequence of the nucleoprotein gene of human influenza A virus, strain A/NT/60/68. Nucleic Acids Res. 1982 Feb 11;10(3):1029–1038. doi: 10.1093/nar/10.3.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka Y., Krauss S., Webster R. G. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J Virol. 1989 Nov;63(11):4603–4608. doi: 10.1128/jvi.63.11.4603-4608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida H., Kawaoka Y., Naeve C. W., Webster R. G. Antigenic and genetic conservation of H3 influenza virus in wild ducks. Virology. 1987 Jul;159(1):109–119. doi: 10.1016/0042-6822(87)90353-9. [DOI] [PubMed] [Google Scholar]
- Kida H., Shortridge K. F., Webster R. G. Origin of the hemagglutinin gene of H3N2 influenza viruses from pigs in China. Virology. 1988 Jan;162(1):160–166. doi: 10.1016/0042-6822(88)90405-9. [DOI] [PubMed] [Google Scholar]
- Lamb R. A., Lai C. J. Conservation of the influenza virus membrane protein (M1) amino acid sequence and an open reading frame of RNA segment 7 encoding a second protein (M2) in H1N1 and H3N2 strains. Virology. 1981 Jul 30;112(2):746–751. doi: 10.1016/0042-6822(81)90319-6. [DOI] [PubMed] [Google Scholar]
- Lamb R. A., Zebedee S. L., Richardson C. D. Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell. 1985 Mar;40(3):627–633. doi: 10.1016/0092-8674(85)90211-9. [DOI] [PubMed] [Google Scholar]
- Laver W. G., Webster R. G. Ecology of influenza viruses in lower mammals and birds. Br Med Bull. 1979 Jan;35(1):29–33. doi: 10.1093/oxfordjournals.bmb.a071537. [DOI] [PubMed] [Google Scholar]
- Laver W. G., Webster R. G. Studies on the origin of pandemic influenza. II. Peptide maps of the light and heavy polypeptide chains from the hemagglutinin subunits of A 2 influenza viruses isolated before and after the appearance of Hong Kong influenza. Virology. 1972 May;48(2):445–455. doi: 10.1016/0042-6822(72)90055-4. [DOI] [PubMed] [Google Scholar]
- Markushin S., Ghiasi H., Sokolov N., Shilov A., Sinitsin B., Brown D., Klimov A., Nayak D. Nucleotide sequence of RNA segment 7 and the predicted amino sequence of M1 and M2 proteins of FPV/Weybridge (H7N7) and WSN (H1N1) influenza viruses. Virus Res. 1988 May;10(2-3):263–271. doi: 10.1016/0168-1702(88)90021-4. [DOI] [PubMed] [Google Scholar]
- Martínez C., del Rio L., Portela A., Domingo E., Ortín J. Evolution of the influenza virus neuraminidase gene during drift of the N2 subtype. Virology. 1983 Oct 30;130(2):539–545. doi: 10.1016/0042-6822(83)90108-3. [DOI] [PubMed] [Google Scholar]
- McCauley J. W., Mahy B. W., Inglis S. C. Nucleotide sequence of fowl plague virus RNA segment 7. J Gen Virol. 1982 Jan;58(Pt 1):211–215. doi: 10.1099/0022-1317-58-1-211. [DOI] [PubMed] [Google Scholar]
- Murphy B. R., Buckler-White A. J., London W. T., Snyder M. H. Characterization of the M protein and nucleoprotein genes of an avian influenza A virus which are involved in host range restriction in monkeys. Vaccine. 1989 Dec;7(6):557–561. doi: 10.1016/0264-410x(89)90283-1. [DOI] [PubMed] [Google Scholar]
- Nakajima K., Desselberger U., Palese P. Recent human influenza A (H1N1) viruses are closely related genetically to strains isolated in 1950. Nature. 1978 Jul 27;274(5669):334–339. doi: 10.1038/274334a0. [DOI] [PubMed] [Google Scholar]
- Nakajima K., Nobusawa E., Nakajima S. Evolution of the NS genes of the influenza A viruses. II. Characteristics of the amino acid changes in the NS1 proteins of the influenza A viruses. Virus Genes. 1990 Jun;4(1):15–26. doi: 10.1007/BF00308562. [DOI] [PubMed] [Google Scholar]
- Okazaki K., Kawaoka Y., Webster R. G. Evolutionary pathways of the PA genes of influenza A viruses. Virology. 1989 Oct;172(2):601–608. doi: 10.1016/0042-6822(89)90202-x. [DOI] [PubMed] [Google Scholar]
- Ortín J., Martínez C., del Río L., Dávila M., López-Galíndez C., Villanueva N., Domingo E. Evolution of the nucleotide sequence of influenza virus RNA segment 7 during drift of the H3N2 subtype. Gene. 1983 Aug;23(2):233–239. doi: 10.1016/0378-1119(83)90055-0. [DOI] [PubMed] [Google Scholar]
- Rees P. J., Dimmock N. J. Electrophoretic separation of influenza virus ribonucleoproteins. J Gen Virol. 1981 Mar;53(Pt 1):125–132. doi: 10.1099/0022-1317-53-1-125. [DOI] [PubMed] [Google Scholar]
- Rota P. A., Rocha E. P., Harmon M. W., Hinshaw V. S., Sheerar M. G., Kawaoka Y., Cox N. J., Smith T. F. Laboratory characterization of a swine influenza virus isolated from a fatal case of human influenza. J Clin Microbiol. 1989 Jun;27(6):1413–1416. doi: 10.1128/jcm.27.6.1413-1416.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samokhvalov E. I., Karginov V. A., Chizhikov V. E., Blinov V. M., Iuferov V. P. Pervichnaia struktura segmenta 7 RNK virusa grippa A/USSR/90/77(H1N1). Bioorg Khim. 1985 Aug;11(8):1080–1086. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild G. C. Evidence for a new type-specific structural antigen of the influenza virus particle. J Gen Virol. 1972 Apr;15(1):99–103. doi: 10.1099/0022-1317-15-1-99. [DOI] [PubMed] [Google Scholar]
- Scholtissek C., Rohde W., Von Hoyningen V., Rott R. On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology. 1978 Jun 1;87(1):13–20. doi: 10.1016/0042-6822(78)90153-8. [DOI] [PubMed] [Google Scholar]
- Smith M., Brown N. L., Air G. M., Barrell B. G., Coulson A. R., Hutchison C. A., 3rd, Sanger F. DNA sequence at the C termini of the overlapping genes A and B in bacteriophage phi X174. Nature. 1977 Feb 24;265(5596):702–705. doi: 10.1038/265702a0. [DOI] [PubMed] [Google Scholar]
- Strauss J. H., Strauss E. G. Evolution of RNA viruses. Annu Rev Microbiol. 1988;42:657–683. doi: 10.1146/annurev.mi.42.100188.003301. [DOI] [PubMed] [Google Scholar]
- Sugrue R. J., Bahadur G., Zambon M. C., Hall-Smith M., Douglas A. R., Hay A. J. Specific structural alteration of the influenza haemagglutinin by amantadine. EMBO J. 1990 Nov;9(11):3469–3476. doi: 10.1002/j.1460-2075.1990.tb07555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster R. G. On the origin of pandemic influenza viruses. Curr Top Microbiol Immunol. 1972;59:75–105. doi: 10.1007/978-3-642-65444-2_3. [DOI] [PubMed] [Google Scholar]
- Winter G., Fields S. Cloning of influenza cDNA ino M13: the sequence of the RNA segment encoding the A/PR/8/34 matrix protein. Nucleic Acids Res. 1980 May 10;8(9):1965–1974. doi: 10.1093/nar/8.9.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z. P., Baylor N. W., Wagner R. R. Transcription-inhibition and RNA-binding domains of influenza A virus matrix protein mapped with anti-idiotypic antibodies and synthetic peptides. J Virol. 1989 Sep;63(9):3586–3594. doi: 10.1128/jvi.63.9.3586-3594.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z. P., Pal R., Fox J. W., Wagner R. R. Functional and antigenic domains of the matrix (M1) protein of influenza A virus. J Virol. 1987 Feb;61(2):239–246. doi: 10.1128/jvi.61.2.239-246.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebedee S. L., Lamb R. A. Nucleotide sequences of influenza A virus RNA segment 7: a comparison of five isolates. Nucleic Acids Res. 1989 Apr 11;17(7):2870–2870. doi: 10.1093/nar/17.7.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zvonarjev A. Y., Ghendon Y. Z. Influence of membrane (M) protein on influenza A virus virion transcriptase activity in vitro and its susceptibility to rimantadine. J Virol. 1980 Feb;33(2):583–586. doi: 10.1128/jvi.33.2.583-586.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]



