Abstract
Generation of replicative defective viruses is frequently observed during viral infections. We now report that encapsidation and reverse transcription of spliced viral RNA is an additional mechanism for synthesis of defective viral particles. We have investigated the in vivo synthesis of a spliced hepatitis B virus (HBV) RNA. By using the polymerase chain reaction with different sets of primers on DNA purified from infected livers and the HepG2 HBV cell line, we detected a subgenomic HBV DNA complementary to the spliced viral RNA. Its nucleotide sequence was found to be identical to that previously described for the spliced RNA. This HBV RNA is packaged and reverse transcribed in vivo, the cDNA being incorporated into circulating particles. This finding establishes the synthesis of spliced HBV RNA in vivo and indicates that its reverse transcription can give rise to defective viruses.
Full text
PDF
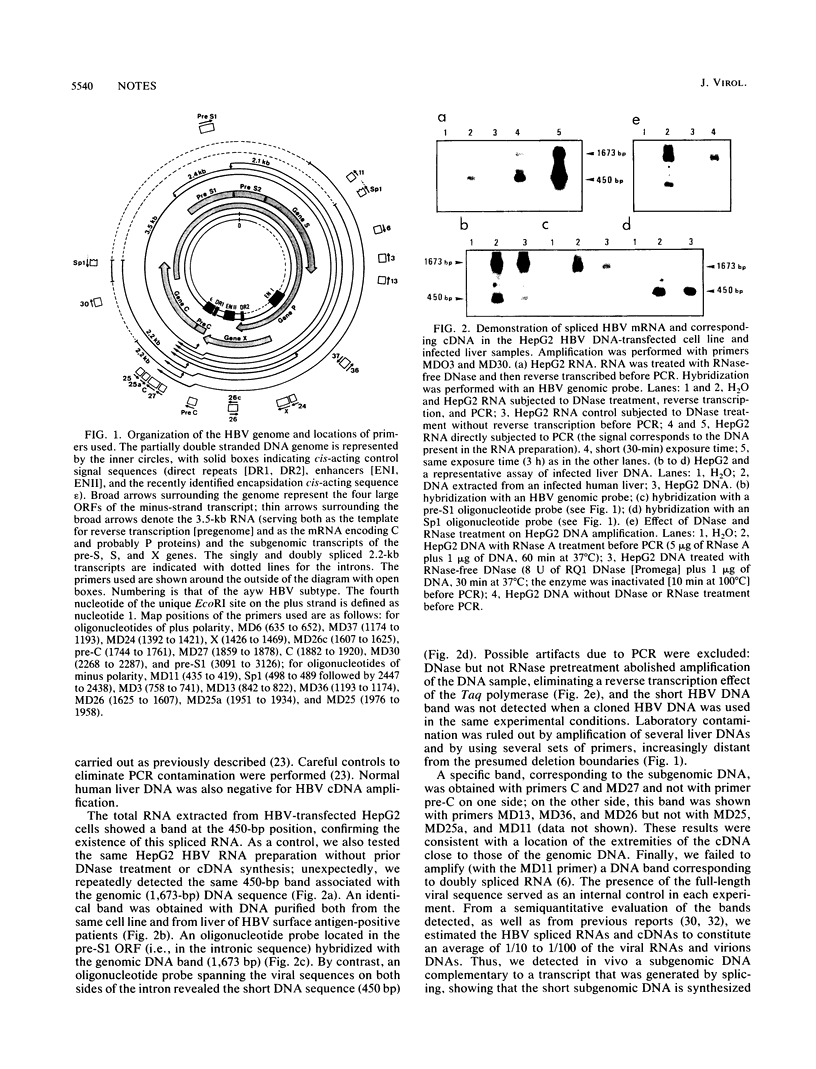
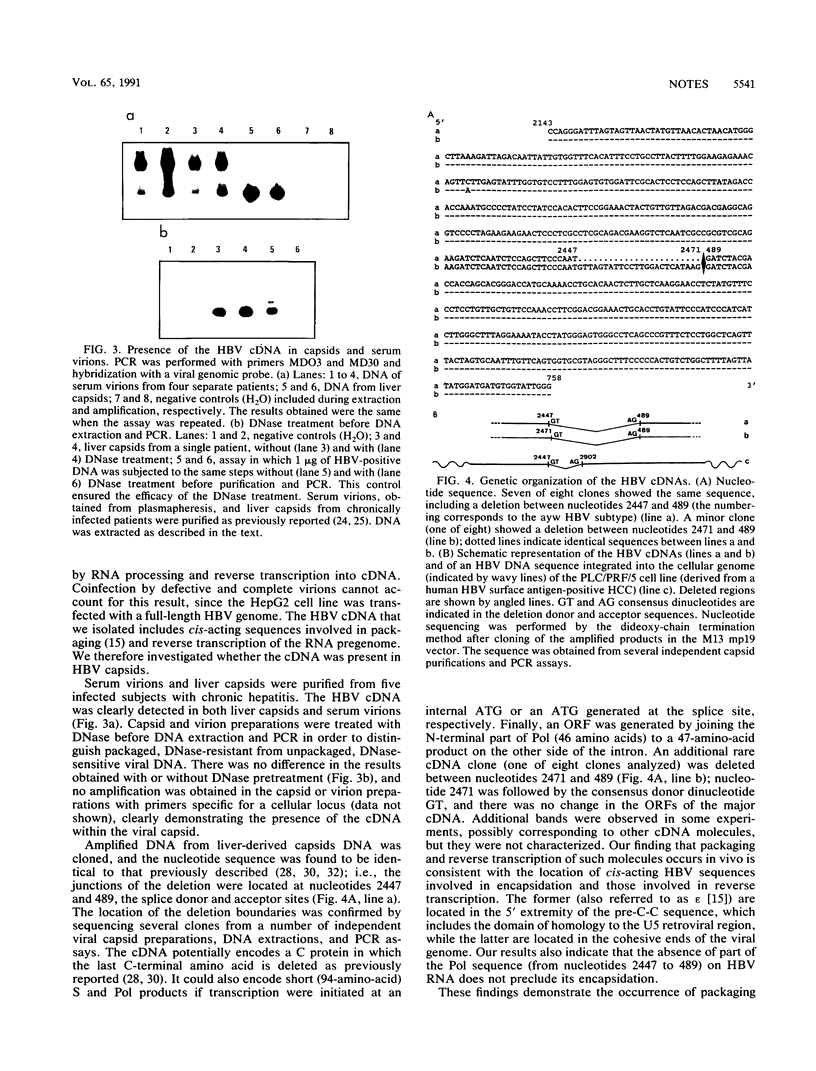


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adam M. A., Miller A. D. Identification of a signal in a murine retrovirus that is sufficient for packaging of nonretroviral RNA into virions. J Virol. 1988 Oct;62(10):3802–3806. doi: 10.1128/jvi.62.10.3802-3806.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz D. C., Hanna Z., Jolicoeur P. Severe immunodeficiency disease induced by a defective murine leukaemia virus. Nature. 1989 Apr 6;338(6215):505–508. doi: 10.1038/338505a0. [DOI] [PubMed] [Google Scholar]
- Bartenschlager R., Junker-Niepmann M., Schaller H. The P gene product of hepatitis B virus is required as a structural component for genomic RNA encapsidation. J Virol. 1990 Nov;64(11):5324–5332. doi: 10.1128/jvi.64.11.5324-5332.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodor J., Svoboda J. The LTR, v-src, LTR provirus generated in the mammalian genome by src mRNA reverse transcription and integration. J Virol. 1989 Feb;63(2):1015–1018. doi: 10.1128/jvi.63.2.1015-1018.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S. K., Morse H. C., 3rd, Makino M., Ruscetti S. K., Hartley J. W. Defective virus is associated with induction of murine retrovirus-induced immunodeficiency syndrome. Proc Natl Acad Sci U S A. 1989 May;86(10):3862–3866. doi: 10.1073/pnas.86.10.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P. J., Chen C. R., Sung J. L., Chen D. S. Identification of a doubly spliced viral transcript joining the separated domains for putative protease and reverse transcriptase of hepatitis B virus. J Virol. 1989 Oct;63(10):4165–4171. doi: 10.1128/jvi.63.10.4165-4171.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavel F., Orenstein J. M. A mutant of human immunodeficiency virus with reduced RNA packaging and abnormal particle morphology. J Virol. 1990 Oct;64(10):5230–5234. doi: 10.1128/jvi.64.10.5230-5234.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman A. D., Triezenberg S. J., McKnight S. L. Expression of a truncated viral trans-activator selectively impedes lytic infection by its cognate virus. Nature. 1988 Sep 29;335(6189):452–454. doi: 10.1038/335452a0. [DOI] [PubMed] [Google Scholar]
- Hirochika H., Takatsuji H., Ubasawa A., Ikeda J. E. Site-specific deletion in cauliflower mosaic virus DNA: possible involvement of RNA splicing and reverse transcription. EMBO J. 1985 Jul;4(7):1673–1680. doi: 10.1002/j.1460-2075.1985.tb03836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch R. C., Lavine J. E., Chang L. J., Varmus H. E., Ganem D. Polymerase gene products of hepatitis B viruses are required for genomic RNA packaging as wel as for reverse transcription. Nature. 1990 Apr 5;344(6266):552–555. doi: 10.1038/344552a0. [DOI] [PubMed] [Google Scholar]
- Horwich A. L., Furtak K., Pugh J., Summers J. Synthesis of hepadnavirus particles that contain replication-defective duck hepatitis B virus genomes in cultured HuH7 cells. J Virol. 1990 Feb;64(2):642–650. doi: 10.1128/jvi.64.2.642-650.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A. S., Baltimore D. Defective viral particles and viral disease processes. Nature. 1970 Apr 25;226(5243):325–327. doi: 10.1038/226325a0. [DOI] [PubMed] [Google Scholar]
- Junker-Niepmann M., Bartenschlager R., Schaller H. A short cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of foreign RNA. EMBO J. 1990 Oct;9(10):3389–3396. doi: 10.1002/j.1460-2075.1990.tb07540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever A., Gottlinger H., Haseltine W., Sodroski J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J Virol. 1989 Sep;63(9):4085–4087. doi: 10.1128/jvi.63.9.4085-4087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majors J. E., Varmus H. E. Nucleotide sequencing of an apparent proviral copy of env mRNA defines determinants of expression of the mouse mammary tumor virus env gene. J Virol. 1983 Sep;47(3):495–504. doi: 10.1128/jvi.47.3.495-504.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann R., Baltimore D. Varying the position of a retrovirus packaging sequence results in the encapsidation of both unspliced and spliced RNAs. J Virol. 1985 May;54(2):401–407. doi: 10.1128/jvi.54.2.401-407.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann R., Mulligan R. C., Baltimore D. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell. 1983 May;33(1):153–159. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- Miller R. H., Girones R., Cote P. J., Hornbuckle W. E., Chestnut T., Baldwin B. H., Korba B. E., Tennant B. C., Gerin J. L., Purcell R. H. Evidence against a requisite role for defective virus in the establishment of persistent hepadnavirus infections. Proc Natl Acad Sci U S A. 1990 Dec;87(23):9329–9332. doi: 10.1073/pnas.87.23.9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto H., Tsuda F., Mayumi M. Defective mutants of hepatitis B virus in the circulation of symptom-free carriers. Jpn J Exp Med. 1987 Aug;57(4):217–221. [PubMed] [Google Scholar]
- Overbaugh J., Donahue P. R., Quackenbush S. L., Hoover E. A., Mullins J. I. Molecular cloning of a feline leukemia virus that induces fatal immunodeficiency disease in cats. Science. 1988 Feb 19;239(4842):906–910. doi: 10.1126/science.2893454. [DOI] [PubMed] [Google Scholar]
- Paterlini P., Gerken G., Nakajima E., Terre S., D'Errico A., Grigioni W., Nalpas B., Franco D., Wands J., Kew M. Polymerase chain reaction to detect hepatitis B virus DNA and RNA sequences in primary liver cancers from patients negative for hepatitis B surface antigen. N Engl J Med. 1990 Jul 12;323(2):80–85. doi: 10.1056/NEJM199007123230202. [DOI] [PubMed] [Google Scholar]
- Petit M. A., Capel F., Pillot J. Demonstration of a firm association between hepatitis B surface antigen proteins bearing polymerized human albumin binding sites and core-specific determinants in serum hepatitis B viral particles. Mol Immunol. 1985 Nov;22(11):1279–1287. doi: 10.1016/0161-5890(85)90047-1. [DOI] [PubMed] [Google Scholar]
- Petit M. A., Pillot J. HBc and HBe antigenicity and DNA-binding activity of major core protein P22 in hepatitis B virus core particles isolated from the cytoplasm of human liver cells. J Virol. 1985 Feb;53(2):543–551. doi: 10.1128/jvi.53.2.543-551.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schranz P., Zentgraf H., Loncarević I. F., Niepmann M., Schröder C. H. Defective replication units of hepatitis B virus. J Virol. 1990 Apr;64(4):1851–1854. doi: 10.1128/jvi.64.4.1851-1854.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey D. W. Expression of a subgenomic retroviral messenger RNA. Cell. 1980 Oct;21(3):811–820. doi: 10.1016/0092-8674(80)90444-4. [DOI] [PubMed] [Google Scholar]
- Su T. S., Lai C. J., Huang J. L., Lin L. H., Yauk Y. K., Chang C. M., Lo S. J., Han S. H. Hepatitis B virus transcript produced by RNA splicing. J Virol. 1989 Sep;63(9):4011–4018. doi: 10.1128/jvi.63.9.4011-4018.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers J., Mason W. S. Replication of the genome of a hepatitis B--like virus by reverse transcription of an RNA intermediate. Cell. 1982 Jun;29(2):403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Masui N., Kajino K., Saito I., Miyamura T. Detection and mapping of spliced RNA from a human hepatoma cell line transfected with the hepatitis B virus genome. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8422–8426. doi: 10.1073/pnas.86.21.8422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran A., Kremsdorf D., Capel F., Housset C., Dauguet C., Petit M. A., Brechot C. Emergence of and takeover by hepatitis B virus (HBV) with rearrangements in the pre-S/S and pre-C/C genes during chronic HBV infection. J Virol. 1991 Jul;65(7):3566–3574. doi: 10.1128/jvi.65.7.3566-3574.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. L., Chen P. J., Tu S. J., Lin M. H., Lai M. Y., Chen D. S. Characterization and genetic analysis of alternatively spliced transcripts of hepatitis B virus in infected human liver tissues and transfected HepG2 cells. J Virol. 1991 Apr;65(4):1680–1686. doi: 10.1128/jvi.65.4.1680-1686.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemer M., Garcia P., Shaul Y., Rutter W. J. Sequence of hepatitis B virus DNA incorporated into the genome of a human hepatoma cell line. J Virol. 1985 Mar;53(3):885–892. doi: 10.1128/jvi.53.3.885-892.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]