Abstract
In vitro DNA amplification methods, such as polymerase chain reaction (PCR), rely on synthetic oligonucleotide primers for initiation of the reaction. In vivo, primers are synthesized on-template by DNA primase. The bacteriophage T7 gene 4 protein (gp4) has both primase and helicase activities. In this study, we report the development of a primase-based Whole Genome Amplification (pWGA) method, which utilizes gp4 primase to synthesize primers, eliminating the requirement of adding synthetic primers. Typical yield of pWGA from 1 ng to 10 ng of human genomic DNA input is in the microgram range, reaching over a thousand-fold amplification after 1 h of incubation at 37°C. The amplification bias on human genomic DNA is 6.3-fold among 20 loci on different chromosomes. In addition to amplifying total genomic DNA, pWGA can also be used for detection and quantification of contaminant DNA in a sample when combined with a fluorescent reporter dye. When circular DNA is used as template in pWGA, 108-fold of amplification is observed from as low as 100 copies of input. The high efficiency of pWGA in amplifying circular DNA makes it a potential tool in diagnosis and genotyping of circular human DNA viruses such as human papillomavirus (HPV).
INTRODUCTION
Whole genome amplification (WGA) technologies are useful tools in many fields of studies, such as genomic research, disease diagnostics, as well as forensic analysis. Indeed, DNA samples used in these studies are often available in limited quantities. Amplifying the entire genome enables researchers to perform more tests on the samples that would otherwise be impossible. Two types of WGA are currently being commercialized for research applications: (1) methods derived from PCR and (2) multiple displacement amplification (MDA).
Examples of PCR-based WGA technologies include primer extension preamplification (PEP) (1) and degenerate oligonucleotide primed PCR (DOP-PCR) (2), which use random or degenerate primers, respectively, for PCR amplification of genomic DNA. These approaches and other methods developed based on them (3,4) are able to amplify DNA from a single cell (1–3,5). However, a major problem of these PCR-based methods is incomplete coverage of the genome due to amplification bias of PCR reactions over certain loci (6).
Recently, an alternative method of WGA, called OmniPlex™, has been developed. This method converts randomly fragmented genomic DNA into PCR-amplifiable units flanked by universal adaptors. This library of fragments is then amplified by PCR using universal primers (7). Compared with the aforementioned other PCR-based WGA methods, this approach gives significantly better genome coverage and lower amplification bias (8). On the other hand, the length of amplification products from this method is rather short, ranging from 75 to 1500 bp with a mean size of 400 bp, and it also requires considerably more steps in the reaction setup (OmniPlex™ product information, http://www.sigmaaldrich.com/sigma/bulletin/WGA1BUL.pdf).
Instead of PCR, MDA-based WGA relies on the strand displacement activity of phage Phi29 DNA polymerase (9). Random oligonucleotides of varying lengths, typically hexamers (10), are used in MDA to prime DNA synthesis by Phi29 DNA polymerase. Compared to the PCR-based WGA methods, MDA offers isothermal DNA amplification with less bias and no loss of essential sequences. It is therefore considered the first truly comprehensive WGA method (11–13). Despite the advantages of the Phi29 DNA polymerase based MDA system, there are still some limitations. The amplification time of the system is rather long, typically several hours (10). In addition, in this primer-based amplification system, an initial heat denaturation step of the input DNA is often performed before the isothermal amplification to facilitate primer annealing (14). This step involves extra labor and may introduce mutations to the template (10) and/or contaminations to the reaction.
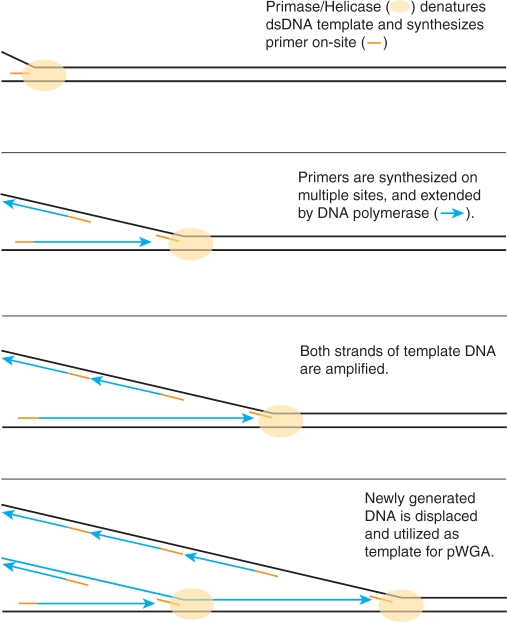
Both PCR and MDA methods require adding synthetic oligonucleotide primers to the amplification reactions. In this study, we report the development of a primase-based whole genome amplification (pWGA) method (Figure 1), which utilizes bacteriophage T7 gp4 primase to synthesize primers on-template, eliminating the requirement of adding synthetic primers. This technology is based on the in vitro reconstitution of the naturally existing cellular DNA replication machinery of bacteriophage T7 (15).
Figure 1.
Mechanism of pWGA reaction.
Bacteriophage T7 has one of the simplest DNA replication systems. Only four proteins are needed to replicate the entire 40-kb linear genome of T7 (16). The T7 gp4 is a bi-functional protein that serves as both a DNA helicase and a primase. The DNA helicase activity resides at the C-terminal half of the protein and it unwinds the duplex DNA template using the energy generated by deoxythymidine triphosphate (dTTP) hydrolysis (17–19). The primase activity resides on the N-terminal half of the protein and it recognizes nucleotide sequences 3′-CTGG(G/T)-5′ and 3′-CTGTG-5′, and generates short RNA primers for the lagging strand synthesis (20–22). T7 DNA polymerase holoenzyme is a heterodimer of two proteins, T7 gene 5 protein (gp5) and Escherichia coli (E. coli) thioredoxin, at 1:1 molar ratio (23–25). T7 gene 5 encodes the 5′–3′ DNA polymerase and the 3′–5′ exonuclease (26,27). E. coli thioredoxin binds to T7 gp5 with an affinity of 5 nM (28), and improves the polymerase processivity (29,30). The DNA polymerase holoenzyme is responsible for faithful and processive DNA synthesis from the short RNA primers generated by gp4 primase. Another protein needed for T7 DNA replication is a single-stranded DNA (ssDNA) binding protein encoded by T7 gene 2.5 (gp2.5). This protein binds single-stranded DNA template (31) and interacts with both DNA polymerase (32) and helicase/primase (33). It has been shown that these four proteins can generate replication products of up to 40 kb through rolling circle amplification mode on a primed DNA template (34,35).
In addition to the four replicative proteins described above, three additional factors enhance the replication efficiency of the pWGA system in vitro; (i) a nucleoside diphosphokinase, (ii) an inorganic pyrophosphatase and (iii) an ATP regeneration system consisting of creatine kinase and phosphocreatine. As the helicase activity of T7 gp4 uses dTTP hydrolysis to drive the unwinding of the double stranded DNA (dsDNA) (18), it is necessary to regenerate dTTP as the reaction progresses to avoid bias in the ratio of the four deoxynucleoside triphosphates (dNTPs). Nucleoside diphosphokinase transfers the γ phosphate group from nucleoside triphosphates (NTPs) to nucleoside diphosphates (NDPs) (36), therefore equilibrates the ratio of dNDP and dNTPs, regardless of the base. This activity rebalances the pool of available dNTPs for DNA synthesis. The accumulation of inorganic pyrophosphate during DNA synthesis is eliminated by inorganic pyrophosphatase (37) in order to avoid product inhibition of the polymerase. Finally, creatine kinase transfers the high-energy phosphate from phosphocreatine to dNDPs to regenerate the dNTPs consumed by nucleoside diphosphokinase (38), and thus help increase the DNA yield.
The pWGA system performs fast isothermal DNA amplification without the need of thermocycling, prior heat-denaturation, or added primers. In this system, separation of DNA template strands is achieved by the T7 DNA helicase, and initiation of DNA synthesis is achieved by the T7 primase. Efficient amplification of template strands is then carried out by T7 DNA polymerase holoenzyme (Figure 1).
MATERIALS AND METHODS
Proteins
T7 gp4 helicase/primase and T7 gp2.5 ssDNA binding protein were purified as described previously (31,39). Native T7 DNA polymerase was purchased from New England BioLabs, Inc (NEB) (Ipswich, MA). T7 Sequenase was purchased from USB (Cleveland, OH). Nucleotide diphosphokinase, inorganic pyrophosphatase, and micrococcal nuclease were purchased from Sigma (St Louis, MO). Creatine kinase was purchased from Roche (Basel, Switzerland).
pWGA reaction
Twenty microliters of pWGA reaction master mix was assembled as described in (15) and it contains: 20 mM Tris–Glutamate pH 7.5, 6 mM DTT, 9 mM MgCl2, 100 mM potassium glutamate, 550 μM of each of the four dNTPs, 330 μM rATP, 440 μM rCTP, 0.05 mg/ml BSA, 3.6 μg T7 gp2.5, 0.047 μg native T7 DNA polymerase, 0.47 μg T7 Sequenase, 0.45 μg T7 gp4, 25 ng nucleotide diphosphokinase, 15 ng pyrophosphatase, 750 ng creatine kinase, and 10 mM creatine phosphate in a final reaction volume of 25 μl. Two forms of the T7 DNA polymerase were used in the pWGA master mix, the ‘native’ protein is the wild-type T7 DNA polymerase and ‘T7 Sequenase’ is a mutant enzyme which lacks the 3′–5′ exonuclease activity (40). A combination of these two forms of polymerases results in the most efficient DNA synthesis (15). The reaction master mix was treated with 0.05 unit of micrococcal nuclease in the presence of 0.5 mM CaCl2 at 30°C for 1 h to remove any DNA contamination. The nuclease treatment was stopped by addition of EGTA to a final concentration of 5 mM.
Amplification was carried out simply by adding DNA template to the master mix to reach the final volume of 25 μl and incubating the reactions at 37°C for 30–120 min. The reaction was stopped by inactivation of T7 DNA polymerase at 65°C for 20 min. The amplification product was analyzed on a 0.6% agarose gel after addition of 5 × DNA agarose gel loading buffer containing 15% Ficoll400, 0.3% orange G, 5 mM EDTA and 0.1% SDS.
Quantification of amplification product by PicoGreen® assay
pWGA amplification products were quantified using Quant-iT™ PicoGreen® dsDNA kit (Invitrogen, Carlsbad, CA), following the protocol provided by the manufacturer. Briefly, the Quant-iT™ PicoGreen® dsDNA reagent was diluted by 20-fold in TE buffer (10 mM Tris–HCl, 1 mM EDTA, pH 7.5). The un-amplified human genomic DNA (gDNA) (Promega, Madison, WI) was diluted to 50 ng/ml in TE buffer. Solutions containing 0, 2, 10, 20, 100, and 200 μl of the diluted DNA were added to TE buffer to make 1 ml solutions, each containing 0, 0.1, 0.5, 1, 5, and 10 ng of DNA, respectively. To these DNA solutions, 1 ml of the diluted Quant-iT™ PicoGreen® dsDNA reagent was added followed by incubation at room temperature for 3 min. Triplets of each DNA solution were prepared. After incubation, sample fluorescence was measured on a SpectraMax M5 microplate reader (Molecular Devices, Sunnyvale, CA) with excitation at 480 nm and emission at 520 nm. The average fluorescence for each DNA sample was plotted against the amount of DNA present in the sample and fitted to a linear equation, which served as the standard curve for quantification. Triplets of pWGA reactions were performed to amplify from 1 or 10 ng of input gDNA. The pWGA products were then diluted by 20-fold in TE buffer and 1 μl of the diluted product was added to the cuvette for fluorescence measurement. The average fluorescent intensity was then mapped onto the standard curve to determine the amount of DNA present in the cuvette. pWGA product yield was then calculated as (the amount of DNA present in the cuvette) × (20, dilution factor) × (25, pWGA reaction volume).
Using pWGA amplification product as template for DNA sequencing
One nanogram of each of the three plasmids, pTYB3 (NEB), pET28a(+), and pET15b (Novagen, Gibbstown, NJ), was amplified by pWGA at 37°C for 1 h and the reactions were stopped by heat inactivation of the enzymes at 65°C for 20 min. The pWGA products were purified by QIAquick PCR purification kit (Qiagen, Valecia, CA) to remove unincorporated nucleotides. A 10 μl aliquot of each purified pWGA product was used as template for DNA sequencing by Dye-Terminator sequencing method using AmpliTaq® DNA polymerase FS on a 3130xl Genetic analyzer (Applied Biosystems (ABI), Foster city, CA). For each pWGA product, two separate sequencing experiments were performed, each using a primer that is of the opposite direction to the other. The primer sequences for each sequencing experiment are given in Table 1.
Table 1.
Using pWGA product as template for DNA sequencing
| Plasmid |
Sequencing primer |
Continuous accurate sequence read |
|||||
|---|---|---|---|---|---|---|---|
| Name | Length | Direction | Name | Start | Primer sequence (5′–3′) | Start–End | Length |
| pTYB3 | 7477 | Forward | S1248 | 5637 | TAATACGACTCACTATAGGG | 5691–6317 | 627 |
| Reverse | S1261 | 5862 | ACCCATGACCTTATTACCAACCTC | 5824–5100 | 725 | ||
| pET28a(+) | 5369 | Forward | S1271 | 66 | TATGCTAGTTATTGCTCAG | 135–799 | 665 |
| Reverse | S1248 | 386 | TAATACGACTCACTATAGGG | 331–1, 5369–4968 | 733 | ||
| pET15b | 5708 | Forward | S1271 | 253 | TATGCTAGTTATTGCTCAG | 280–897 | 618 |
| Reverse | S1248 | 479 | TAATACGACTCACTATAGGG | 424–1, 5708–5490 | 643 | ||
Measurement of amplification bias by real-time PCR
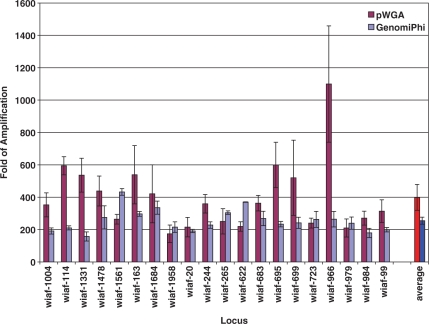
The amplification bias of pWGA reactions was evaluated, in comparison with MDA, by surveying the genome coverage of pWGA and MDA amplification over 20 SNP markers across from different chromosomes. The names of the SNP loci are indicated on the x-axis of Figure 4. The sequences of the primers are listed in Table S1 (Supplementary Data) and can be found in Hosono et al. (13).
Figure 4.
Fold of pWGA and GenomiPhi amplification over 20 SNP loci. The average fold of amplification of 10 ng input human genomic DNA for each locus from three pWGA (magenta) or GenomiPhi (light blue) experiments is plotted with standard deviation against the primer name used to amplify that locus. The average fold of amplification over the 20 loci among three pWGA (red) or GenomiPh (blue) reactions is also plotted.
For pWGA reactions 1, 10 and 50 ng of purified human genomic DNA were amplified at 37°C for 1 h. Each pWGA reaction was performed in triplets and was stopped by incubation at 65°C for 20 min to inactivate the proteins. The same amounts of gDNA were amplified in triplets by MDA using the GenomiPhi V2 DNA amplification kit (GE Healthcare, Piscataway, NJ) at 30°C for 1.5 h after heat denaturation at 95°C for 3 min. The reactions were stopped by incubation at 65°C for 10 min. The pWGA and GenomiPhi products were diluted by 25-fold prior to use as templates for subsequent real-time PCR assays. In these real-time PCR assays, 20 pairs of SNP primers, each of which maps to the p or q arms of different chromosomes, were randomly chosen from the Whitehead Institute-Affymetrix (WIAF) human SNP database (13). Those primers result in amplification products ranging from 100 to 250 bp. Detection of fluorescent signal in the real-time PCR reactions was achieved by binding of SYBR® green dye to the double stranded PCR amplification product. Real-time PCR reactions were performed using the DyNAmo™ SYBR® green qPCR kit (Finnzymes Oy, Finland) on an ABI7300 real-time PCR system. For each pair of primers, six reactions, each containing 0, 10 pg, 100 pg, 1 ng, 10 ng or 100 ng of un-amplified gDNA, were performed to serve as a standard curve for absolute quantification. One microliter of the diluted pWGA or GenomiPhi product was assayed for each pair of primers by real-time PCR using the standard curve generated for that primer set. The fold of amplification by pWGA or GenomiPhi for each SNP locus is calculated as the ratio between the average amount of triplicate pWGA or GenomiPhi reaction products and the amount of unamplified sample. Locus bias from amplification of a particular concentration of a sample is calculated as the highest fold of amplification over the lowest fold of amplification among the 20 loci. The yield of a pWGA or GenomiPhi reaction is estimated from this method as the average amount from 20 loci.
Amplification of circular DNA
The copy numbers of pCMV_GLuc (NEB) plasmid were calculated from the concentrations provided by the suppliers. Ten-fold serial dilutions in water were performed to get desired copy numbers. For pWGA amplification, 1 μl diluted DNA was mixed with 4 μl TE and added to 20 μl of pWGA master mix. The reaction proceeded at 37°C for 1 h before inactivation at 65°C for 20 min. Amplification by MDA was performed using the GenomiPhi V2 DNA amplification kit, following the protocol from manufacturer. Briefly, 1 μl diluted DNA with 9 μl sample buffer was heated at 95°C for 3 min, chilled on ice, and then mixed with 9 μl reaction buffer and 1 μl enzyme mix. The reaction was incubated at 30°C for 90 min and inactivated at 65°C for 10 min. These reaction products were then diluted by 200-fold, 5 μl of which were used as input substrate for quantification by real-time PCR using the DyNAmo™ SYBR® green qPCR kit. The primers used in the real-time PCR reactions are: Luc-3F (AAAGGGCTTGCCAACGTGCAGTGTTCT) and Luc-3R (TTGATCTTGTCCACCTGGCCCTGGATCTT) (NEB).
Universal DNA detection
Each 25 μl reaction contained 20 μl of pWGA master mix, 1 × ROX reference dye, 0.2 × EvaGreen® fluorescent dye (Biotium, Hayward, CA), and 0, 100 fg, 1 pg, 10 pg, 100 pg or 1 ng of purified human genomic DNA. Triplicates of reactions were assembled on ice. The reaction was carried out on the ABI7300 real-time PCR machine at 37°C constant temperature for 120 min.
RESULTS
Reaction time, amplicon length and product yield of pWGA
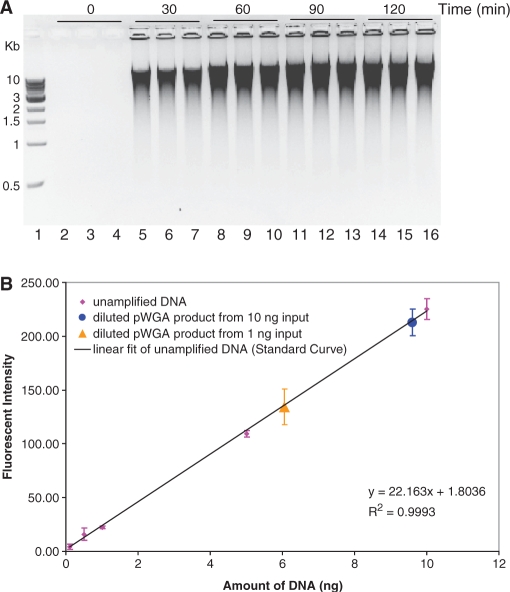
In order to understand the capability of the pWGA system, performance of the pWGA reactions was studied in detail. First, the speed of pWGA in DNA amplification was evaluated using purified human genomic DNA as input template. A total amount of 10 ng of DNA was added to each 25 μl of pWGA reaction mix. Reactions proceeded at a constant temperature of 37°C. The reactions were terminated after 0, 30, 60, 90 or 120 min of incubation, respectively, by heat inactivation of the proteins at 65°C for 20 min. Five microliters of each of the reactions were analyzed on a 0.6% agarose gel. Each reaction was performed in triplicate. As seen from the gel (Figure 2A), a significant amount of amplification product was observed at 30 min of reaction (Figure 2A, lanes 5–7). The amount of amplification product reached maximum between 60 and 90 min (Figure 2A, lanes 8–13). Incubation longer than 90 min did not give higher amount of product (Figure 2A, lanes 14–16).
Figure 2.
pWGA reaction time, product length, and product yield. (A) pWGA reactions containing 10 ng human genomic DNA were terminated after a specified incubation time [0 (lanes 2–4), 30 (lanes 5–7), 60 (lanes 8–10), 90 (lanes 11–13) or 120 (lanes 14–16) minutes], as indicated at the top of the gel, and 5 µl of each 25 µl reaction was analyzed on a 0.6% agarose gel. The reactions were performed in triplicate, and lane 1 shows 500 ng of 1-kb DNA ladder (NEB) with the sizes of bands indicated on the left of the gel. The lane numbers are indicated at the bottom of the gel. (B) Quantification of pWGA product yield by PicoGreen® assay. Fluorescent intensity of 0.1, 0.5, 1, 5 and 10 ng of un-amplified DNA (magenta diamonds) is plotted against the amount of DNA and is fitted to the linear equation shown at the bottom right of the plot (black line) as a standard curve for quantification. Triplets of pWGA product from 1 ng (orange triangle) and 10 ng (blue circle) were diluted 20 times. One microliter of the diluted products were assayed for PicoGreen® fluorescence and mapped on the standard curve according to their intensity. The ± SD over three experiments are shown as error bars on each data point.
The products of pWGA amplification ran as a smear on the agarose gel, indicating the presence of branched DNA or mixed lengths of amplification products. The size of the pWGA product was mostly larger than 3 kb, with a significant amount larger than 10 kb (Figure 2A; compare the DNA ladder in lane 1 with the smear of pWGA products in lanes 5–16).
The yield of pWGA reactions from 1 ng or 10 ng of input human genomic DNA was quantified by measuring PicoGreen® fluorescence (41). A standard curve was generated with triplicates of 0.1, 0.5, 1, 5 and 10 ng of un-amplified human genomic DNA (Figure 2B, magenta diamonds). A linear fit of these data points resulted in a slope of 22.16 units of fluorescent intensity per ng of DNA. Triplicates of pWGA products from 1 ng or 10 ng input DNA were diluted by 20-fold and 1 μl of the diluted sample was added to each PicoGreen® assay tube. The fluorescent intensities were measured, which corresponded to about 6 (from 1 ng input) or 10 (from 10 ng input) ng of DNA (Figure 2B, orange triangle and blue circle). Based on the dilution factors, the total yield of pWGA product in a 25 μl reaction was about 3 μg from 1 ng of input and 5 μg from 10 ng of input [(6 or 10 ng) × (20, fold of dilution) × (25 μl, reaction volume)], reaching up to 3000-fold of amplification.
Using pWGA amplification product as template for DNA sequencing
Three commonly used cloning plasmids, pTYB3 (NEB), pET28a(+) and pET15b (Novagen), were amplified by pWGA. The sizes of these plasmids range from 5.3 kb to 7.4 kb (Table 1). After amplification, the pWGA products were purified by QIAquick PCR purification kit (Qiagen) to remove unincorporated nucleotides. These purified products were used as templates for subsequent DNA sequencing experiments.
For each pWGA product, two separate sequencing experiments were performed, using primers that are of opposite direction to each other. The commercially available sequencing primers that are widely used in cloning experiments were chosen for this study. For the pET28a(+) and pET15b plasmids, T7 promoter primer (S1248, NEB) and T7 terminator primer (S1271, NEB) were used as sequencing primers. For pTYB3, T7 promoter primer and intein reverse sequencing primer (S1261, NEB) were selected (Table 1).
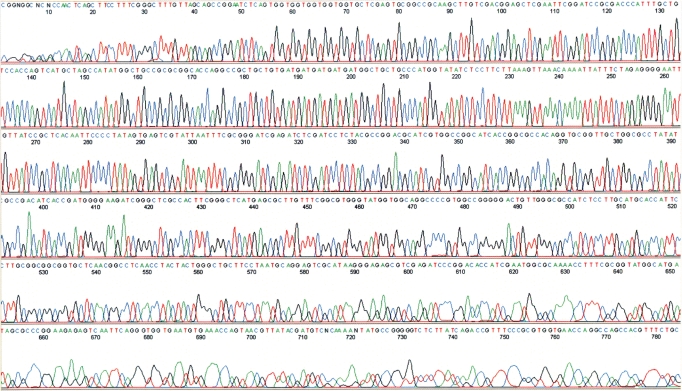
The results from the sequencing experiments showed typical patterns of sequencing peaks (Figure 3), undistinguishable from those using un-amplified plasmids (data not shown). Among the six sequencing experiments, the lengths of sequences that could be clearly read were about 750 bp (Figure 3). The lengths of sequences that were continuous and accurate were between 618 bp and 733 bp (Table 1), similar to a typical sequencing results using a plasmid as template. Sequencing experiments using either forward or reverse primers yielded similar results, indicating that both strands of the plasmid template were amplified in similar ways by pWGA.
Figure 3.
Representative sequencing results using the pWGA amplification product as template. The template for sequencing was the pWGA amplification product from plasmid pET28a(+). The primer used for sequencing was the T7 terminator primer (S1271, NEB) that covers residues 66–84 on the plasmid and it is in the reverse direction of the plasmid. The sequencing experiment was performed by Dye-Terminator Sequencing method using AmpliTaq® DNA polymerase FS on an ABI 3130xl Genetic analyzer.
Amplification bias over 20 SNP loci
To evaluate the genome coverage and amplification bias of pWGA reaction products, we analyzed 20 genetic markers on different chromosomes. Namely, 20 pairs of primers that cover single nucleotide polymorphism (SNP) loci on different chromosomes (13) were selected and real-time PCR reactions were performed using these primers. Using different amounts of un-amplified human genomic DNA as input template, a standard curve was generated for each primer set. These standard curves were then used to quantify the amount and thus calculate the fold of amplification of each locus in a pWGA product. Triplicates of pWGA reactions were performed using 10 ng of purified human genomic DNA as input template. To serve as a reference for comparison, the same experiments were performed for MDA using the GenomiPhi V2 DNA amplification kit. The amplification products from pWGA and GenomiPhi were then diluted and used as template in the following qPCR reaction. Fold of amplification for each locus was calculated based on the standard curves and averaged over the three individual pWGA or GenomiPhi experiments (Figure 4). Locus bias from 10 ng input over 20 loci for pWGA is 6.3 with locus wiaf-966 having the highest fold of amplification (1099-fold) and locus wiaf-1958 having the lowest fold of amplification (173-fold). The average fold of amplification for pWGA over the 20 selected loci is about 400 from 10 ng of input (Figure 4, magenta and red), giving a final yield of about 4 μg, consistent with the results from PicoGreen® assays (Figure 2B). In comparison, locus bias from 10 ng input over the same 20 loci for GenomiPhi is 2.7 with locus wiaf-1561 having the highest fold of amplification (432.5-fold) and locus wiaf-1331 having the lowest fold of amplification (157.5-fold). The average fold of amplification for GenomiPhi over these 20 loci is about 250 (Figure 4, light blue and blue).
pWGA reactions were also performed with different amounts of input human genomic DNA template and the product yield and locus bias were tested, the results of which are listed in Table 2. Consistent with the PicoGreen® measurement, we found that pWGA product yield was about 2 μg when the input template was 1 ng and reaches about 5 μg when larger amount of input was used (Table 2). Locus bias increases when the amount of input was limited. However, it was not greater than 11 when only 1 ng of human DNA was used as input template for pWGA amplification. Overall, in comparison with the GenomiPhi V2 kit, pWGA has slightly higher amplification yield and amplification bias.
Table 2.
Locus bias of pWGA and GenomiPhi from different amounts of input DNA template
| Amount of input DNA (ng) | 1 |
10 |
50 |
|||
|---|---|---|---|---|---|---|
| pWGA | MDA | pWGA | MDA | pWGA | MDA | |
| Fold of amplificationa | 1829 | 2084 | 399 | 254.9 | 93 | 54.7 |
| SDa | 188 | 49 | 79.8 | 21.5 | 25 | 8.4 |
| Product yield (μg) | 1.8 | 2.1 | 4.0 | 2.5 | 4.6 | 2.7 |
| Locus bias | 10.9 | 3.2 | 6.3 | 2.7 | 4.9 | 2.4 |
aFold of amplification and SD are the average from triplicates of pWGA reactions.
Amplification of circular DNA
In addition to amplifying human genomic DNA, we have examined the performance of pWGA system in amplifying circular DNA templates. We used the 5.8 kb pCMV_GLuc control plasmid (NEB) as input template. pWGA reactions containing 100, 1000, 10 000, 100 000 and 1 000 000 copies of plasmid pCMV_Gluc, respectively, were performed at 37°C for 1 h. In parallel, the same plasmids were amplified by MDA using GenomiPhi V2 DNA amplification kit. The products of amplification were quantified by real-time PCR using primers specific to pCMV_GLuc.
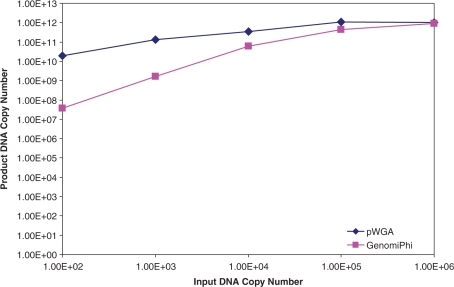
We found that pWGA was able to efficiently amplify the plasmid from as low as 100 copies. The yield was 1.9 × 1010 copies of amplification product from 100 copies of input (Figure 5, blue diamonds), reaching 1.9 × 108-fold of amplification. In comparison, amplification by MDA from the same amount of input plasmid resulted in lower yield, which was only 3.8 × 107 copies of product DNA (Figure 5, magenta squares), reaching 3.8 × 105-fold of amplification. Overall, in our hands, product yield of pWGA over the range of 102–106 copies of input plasmid was higher than that of MDA, especially when very small amount (100–1000 copies) of DNA was used as input template. The difference of amplification between pWGA and GenomiPhi from 100 copies of input plasmid was 5 × 102-fold, and that from 1000 copies of input was 81-fold. When more plasmid (106 copies) was added as input template, pWGA and GenomiPhi performed similarly in terms of fold of amplification.
Figure 5.
Amplification of circular DNA. Various amounts of pCMV_GLuc plasmid were used as substrates for side-by-side pWGA (blue diamonds) and MDA reactions (magenta squares). The copy numbers of amplified plasmids DNA were determined by comparing with standard curves generated from templates of known copy numbers through real-time PCR using primers Luc-3F and Luc-3R.
Real-time pWGA and universal DNA detection
The ability of pWGA system to amplify DNA in real-time was examined by performing the reaction in the presence of SYBR® green (42) or EvaGreen® (43) fluorescent dyes. Real-time fluorescent signal was detected within 1 h of 37°C reaction when 0.1 × to 0.2 × of either dye was present in pWGA reaction. The fluorescent signal over time displayed a typical shape similar to that of real-time PCR reaction (44), with an initial background phase, followed by an exponential amplification phase, a linear phase and then the plateau phase (data not shown). The real-time signal was specific for pWGA amplification of template DNA. If the DNA template or DNA polymerase in the reaction was omitted, no real-time signal was detected (data not shown).
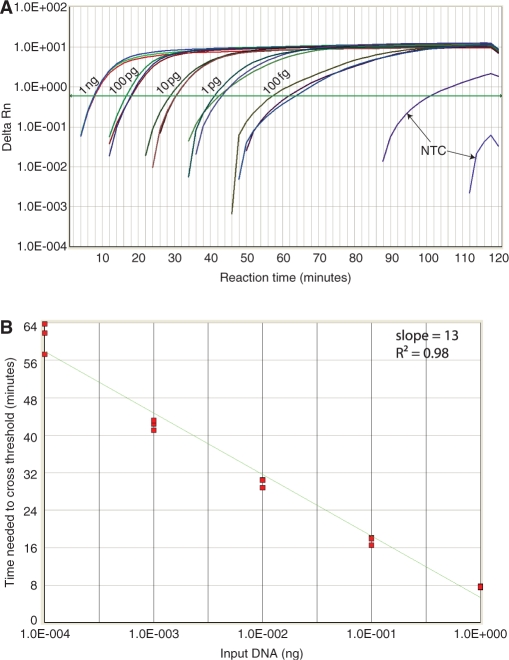
Furthermore, real-time pWGA reactions were performed with different amount of input template. Triplicates of reactions containing 100 fg, 1 pg, 10 pg, 100 pg and 1 ng of purified human genomic DNA were carried out at 37°C on an ABI7300 real-time PCR machine (Figure 6A). The real-time pWGA signal indicates that detection of 1 ng of input DNA took less than 10 min. Detection of as low as 100 fg of DNA was consistently observed at around 1 h of incubation (Figure 6A) and was significantly distinguished from background non-specific amplification, which occurred in only one of the triplets after 90 min of incubation (Figure 6A, NTC). The cycle numbers at which the fluorescent signal crossed the threshold (Ct) are plotted against the amount of input template and fitted to a linear equation (Figure 6B). This plot shows good linearality (R2 = 0.98) over 4 orders of magnitude of input template, from 100 fg to 1 ng, indicating an exponential amplification of the input template by pWGA over this range of input. The time it took for the product to increase 10 times was 13 min (Figure 6B, slope).
Figure 6.
Real-time quantitative pWGA. (A) Real-time amplification signal (Delta Rn) from triplets of 0 (NTC), 100 fg, 1 pg, 10 pg, 100 pg and 1 ng of input DNA is plotted against reaction time. The threshold is drawn as a green line. (B) Ct number from different amount of input template is plotted against the amount of input template with a linear fit shown as a green line. The slope and the goodness of fit (R2-value) are shown at the top right of the plot. The template amount on the x-axis is distributed on a log scale.
DISCUSSION
Sufficient quantity of genomic DNA is crucial in many aspects of genomic research, clinical diagnostics, and forensic studies. The DNA source often becomes a limiting factor in these studies. A method that can amplify a minimal amount of DNA from different sources to large quantity without significant loss of genome coverage is instrumental. In this study, we describe a new method of rapid, isothermal, whole genome amplification technology, pWGA. This method is based on the naturally existing bacteriophage T7 DNA replication system, in which the duplex template is separated by T7 DNA helicase. Single-stranded DNA is protected by T7 ssDNA-binding protein. Amplification is dependent on T7 primase to generate short primers for extension by DNA polymerase. These four proteins are sufficient to amplify the 40-kb T7 linear genome in vivo (16). When purified and assembled in vitro, together with the accessory proteins to re-generate and re-balance nucleotide substrates, we have shown that the T7 DNA replication proteins can amplify a small amount of input human genomic DNA (1 ng) to reach microgram quantity.
One of the advantages of the pWGA system is that it is truly isothermal. No thermal cycling is needed and heat denaturation of the template prior to amplification is not necessary in pWGA. The reaction is started and proceeds by incubation of the template with the reaction master mix at 37°C. The true isothermal nature of the system simplifies the reaction setup. In addition, it eliminates the exposure of template DNA to high temperature, thus reducing the chance of introducing unwanted mutations. Another advantage of the pWGA system is the reaction speed: microgram quantity of amplification product from as low as 1 ng of input DNA can be obtained within 1 h of incubation. The fast reaction speed, together with the simple isothermal reaction scheme, makes the system potentially useful in a point of care diagnostic setting where fast results are needed and laboratory instruments are limited.
pWGA reaction product is suitable for a variety of downstream assays. After a simple clean up step to remove excess nucleotides and proteins, pWGA product can be used for DNA sequencing (Figure 3). pWGA products can also be used directly as template for PCR reactions without purification. The T7 DNA polymerase in pWGA system efficiently incorporates modified nucleotides such as biotin-dNTP and the labeled products can then be applied to microarrays for genomic studies (data not shown).
Faithful representation of the input template is an essential requirement for any kind of WGA technology. The product quality of pWGA was evaluated by agarose gel electrophoresis and by surveying the genome coverage and amplification bias using real-time PCR. The high molecular weight product of pWGA observed on the gel is a close representation of the input template and is a desirable feature in assays such as enzymatic digestion, cloning and DNA sequencing. Amplification bias is a severe problem in PCR-based WGA technologies due to the possibility of uneven annealing of primers to the template. Instead of using added primers, the primase based WGA technology utilizes a primase to synthesize primers. The ability of the system to synthesis primers eliminates the primer annealing preference and could potentially reduce amplification bias. In this study, we surveyed 20 genetic markers on different chromosomes for the coverage and amplification bias of pWGA. We showed that from 10 ng of input DNA, all the 20 loci were amplified by pWGA by 173–1099-fold. There was no loss of the genome coverage among the 20 loci and the bias was less than 7-fold when input DNA was more than 10 ng. Even from as low as 1 ng of input (about 300 copies of the genome), the bias was only 11-fold. The amplification bias of our pWGA system is similar to that of MDA (Figure 4 and Table 2) and is much lower than those of PCR-based WGA (11). Low amplification bias is a prerequisite if the product is used in subsequent genome researches, such as comparative genomic hybridization (CGH), where genome copy number changes are investigated (45).
The ability of pWGA to efficiently amplify circular DNA enables the use of this system in detecting and subsequently identifying and genotyping DNA virus samples. The other usage of circular DNA amplification is to restore an archived DNA specimen, which is often fragmented and poses a problem for direct WGA amplification. Recently, Wang et al. described a strategy to recover archived DNA by restriction and circularization-aided rolling circle amplification (RCA-RCA) (46). In this strategy, fragmented DNA is circularized first and then amplified by rolling circle amplification. The pWGA system is particularly suited for use in this method because of its high efficiency in amplifying circular DNA.
The pWGA system can be used to quantitatively detect trace amount of DNA in a universal real-time assay without the need of adding target-specific primers. The pWGA assay can be used to detect and quantify residual DNA level in a given biological sample or in a pharmaceutical product. The detection sensitivity is 100 fg of DNA and the dynamic range for quantification is 104, from 100 fg to 1 ng. The reaction is very fast and detection of 100 fg of DNA occurs in only one hour. This fast and sensitive WGA-based universal DNA detection assay can be performed in a general laboratory setting using a real-time PCR machine. This is the first report of the use of WGA technology as a molecular tool for detecting trace amounts of contaminant DNA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was funded by the National Institute of Health grant HG004522-01 and the Department of Homeland Security Science and Technology Directorate, award NBCHC070097. We are grateful to Jared Rice and Jamie Wytiaz for their support during the project and Dr Richard Roberts for helpful discussions. The sequencing experiments were performed at the DNA sequencing facility at New England BioLabs. We also thank New England BioLabs for providing the enzymes, reagents, as well as the oligonucleotides used in this study. Funding to pay the Open Access publication charges for this article was provided by BioHelix Corporation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Zhang L, Cui X, Schmitt K, Hubert R, Navidi W, Arnheim N. Whole genome amplification from a single cell: implications for genetic analysis. Proc. Natl Acad. Sci. USA. 1992;89:5847–5851. doi: 10.1073/pnas.89.13.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Telenius H, Carter NP, Bebb CE, Nordenskjold M, Ponder BA, Tunnacliffe A. Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics. 1992;13:718–725. doi: 10.1016/0888-7543(92)90147-k. [DOI] [PubMed] [Google Scholar]
- 3.Dietmaier W, Hartmann A, Wallinger S, Heinmoller E, Kerner T, Endl E, Jauch KW, Hofstadter F, Ruschoff J. Multiple mutation analyses in single tumor cells with improved whole genome amplification. Am. J. Pathol. 1999;154:83–95. doi: 10.1016/S0002-9440(10)65254-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kittler R, Stoneking M, Kayser M. A whole genome amplification method to generate long fragments from low quantities of genomic DNA. Anal. Biochem. 2002;300:237–244. doi: 10.1006/abio.2001.5460. [DOI] [PubMed] [Google Scholar]
- 5.Wells D, Delhanty JD. Comprehensive chromosomal analysis of human preimplantation embryos using whole genome amplification and single cell comparative genomic hybridization. Mol. Hum. Reprod. 2000;6:1055–1062. doi: 10.1093/molehr/6.11.1055. [DOI] [PubMed] [Google Scholar]
- 6.Hawkins TL, Detter JC, Richardson PM. Whole genome amplification–applications and advances. Curr. Opin. Biotechnol. 2002;13:65–67. doi: 10.1016/s0958-1669(02)00286-0. [DOI] [PubMed] [Google Scholar]
- 7.Langmore JP. Rubicon Genomics, Inc. Pharmacogenomics. 2002;3:557–560. doi: 10.1517/14622416.3.4.557. [DOI] [PubMed] [Google Scholar]
- 8.Barker DL, Hansen MS, Faruqi AF, Giannola D, Irsula OR, Lasken RS, Latterich M, Makarov V, Oliphant A, Pinter JH, et al. Two methods of whole-genome amplification enable accurate genotyping across a 2320-SNP linkage panel. Genome Res. 2004;14:901–907. doi: 10.1101/gr.1949704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanco L, Bernad A, Lazaro JM, Martin G, Garmendia C, Salas M. Highly efficient DNA synthesis by the phage phi 29 DNA polymerase. Symmetrical mode of DNA replication. J. Biol. Chem. 1989;264:8935–8940. [PubMed] [Google Scholar]
- 10.Dean FB, Hosono S, Fang L, Wu X, Faruqi AF, Bray-Ward P, Sun Z, Zong Q, Du Y, Du J, et al. Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl Acad. Sci. USA. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lasken RS, Egholm M. Whole genome amplification: abundant supplies of DNA from precious samples or clinical specimens. Trends Biotechnol. 2003;21:531–535. doi: 10.1016/j.tibtech.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 12.Paez JG, Lin M, Beroukhim R, Lee JC, Zhao X, Richter DJ, Gabriel S, Herman P, Sasaki H, Altshuler D, et al. Genome coverage and sequence fidelity of phi29 polymerase-based multiple strand displacement whole genome amplification. Nucleic Acids Res. 2004;32:e71. doi: 10.1093/nar/gnh069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hosono S, Faruqi AF, Dean FB, Du Y, Sun Z, Wu X, Du J, Kingsmore SF, Egholm M, Lasken RS. Unbiased whole-genome amplification directly from clinical samples. Genome Res. 2003;13:954–964. doi: 10.1101/gr.816903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dean FB, Nelson JR, Giesler TL, Lasken RS. Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001;11:1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tabor S, Richardson CC. US Patent Application 20050164213. 2005. Isothermal amplification of DNA. [Google Scholar]
- 16.Richardson CC. Bacteriophage T7: minimal requirements for the replication of a duplex DNA molecule. Cell. 1983;33:315–317. doi: 10.1016/0092-8674(83)90411-7. [DOI] [PubMed] [Google Scholar]
- 17.Matson SW, Richardson CC. DNA-dependent nucleoside 5'-triphosphatase activity of the gene 4 protein of bacteriophage T7. J. Biol. Chem. 1983;258:14009–14016. [PubMed] [Google Scholar]
- 18.Matson SW, Tabor S, Richardson CC. The gene 4 protein of bacteriophage T7. Characterization of helicase activity. J. Biol. Chem. 1983;258:14017–14024. [PubMed] [Google Scholar]
- 19.Guo S, Tabor S, Richardson CC. The linker region between the helicase and primase domains of the bacteriophage T7 gene 4 protein is critical for hexamer formation. J. Biol. Chem. 1999;274:30303–30309. doi: 10.1074/jbc.274.42.30303. [DOI] [PubMed] [Google Scholar]
- 20.Romano LJ, Richardson CC. Characterization of the ribonucleic acid primers and the deoxyribonucleic acid product synthesized by the DNA polymerase and gene 4 protein of bacteriophage T7. J. Biol. Chem. 1979;254:10483–10489. [PubMed] [Google Scholar]
- 21.Tabor S, Richardson CC. Template recognition sequence for RNA primer synthesis by gene 4 protein of bacteriophage T7. Proc. Natl Acad. Sci. USA. 1981;78:205–209. doi: 10.1073/pnas.78.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frick DN, Baradaran K, Richardson CC. An N-terminal fragment of the gene 4 helicase/primase of bacteriophage T7 retains primase activity in the absence of helicase activity. Proc. Natl Acad. Sci. USA. 1998;95:7957–7962. doi: 10.1073/pnas.95.14.7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Modrich P, Richardson CC. Bacteriophage T7 Deoxyribonucleic acid replication in vitro. A protein of Escherichia coli required for bacteriophage T7 DNA polymerase activity. J. Biol. Chem. 1975;250:5508–5514. [PubMed] [Google Scholar]
- 24.Modrich P, Richardson CC. Bacteriophage T7 deoxyribonucleic acid replication invitro. Bacteriophage T7 DNA polymerase: an an emzyme composed of phage- and host-specific subunits. J. Biol. Chem. 1975;250:5515–5522. [PubMed] [Google Scholar]
- 25.Mark DF, Richardson CC. Escherichia coli thioredoxin: a subunit of bacteriophage T7 DNA polymerase. Proc. Natl Acad. Sci. USA. 1976;73:780–784. doi: 10.1073/pnas.73.3.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hori K, Mark DF, Richardson CC. Deoxyribonucleic acid polymerase of bacteriophage T7. Purification and properties of the phage-encoded subunit, the gene 5 protein. J. Biol. Chem. 1979;254:11591–11597. [PubMed] [Google Scholar]
- 27.Hori K, Mark DF, Richardson CC. Deoxyribonucleic acid polymerase of bacteriophage T7. Characterization of the exonuclease activities of the gene 5 protein and the reconstituted polymerase. J. Biol. Chem. 1979;254:11598–11604. [PubMed] [Google Scholar]
- 28.Huber HE, Russel M, Model P, Richardson CC. Interaction of mutant thioredoxins of Escherichia coli with the gene 5 protein of phage T7. The redox capacity of thioredoxin is not required for stimulation of DNA polymerase activity. J. Biol. Chem. 1986;261:15006–15012. [PubMed] [Google Scholar]
- 29.Huber HE, Tabor S, Richardson CC. Escherichia coli thioredoxin stabilizes complexes of bacteriophage T7 DNA polymerase and primed templates. J. Biol. Chem. 1987;262:16224–16232. [PubMed] [Google Scholar]
- 30.Tabor S, Huber HE, Richardson CC. Escherichia coli thioredoxin confers processivity on the DNA polymerase activity of the gene 5 protein of bacteriophage T7. J. Biol. Chem. 1987;262:16212–16223. [PubMed] [Google Scholar]
- 31.Kim YT, Tabor S, Bortner C, Griffith JD, Richardson CC. Purification and characterization of the bacteriophage T7 gene 2.5 protein. A single-stranded DNA-binding protein. J. Biol. Chem. 1992;267:15022–15031. [PubMed] [Google Scholar]
- 32.Kim YT, Tabor S, Churchich JE, Richardson CC. Interactions of gene 2.5 protein and DNA polymerase of bacteriophage T7. J. Biol. Chem. 1992;267:15032–15040. [PubMed] [Google Scholar]
- 33.Kim YT, Richardson CC. Acidic carboxyl-terminal domain of gene 2.5 protein of bacteriophage T7 is essential for protein-protein interactions. J. Biol. Chem. 1994;269:5270–5278. [PubMed] [Google Scholar]
- 34.Debyser Z, Tabor S, Richardson CC. Coordination of leading and lagging strand DNA synthesis at the replication fork of bacteriophage T7. Cell. 1994;77:157–166. doi: 10.1016/0092-8674(94)90243-7. [DOI] [PubMed] [Google Scholar]
- 35.Lee J, Chastain PD, 2nd, Kusakabe T, Griffith JD, Richardson CC. Coordinated leading and lagging strand DNA synthesis on a minicircular template. Mol. Cell. 1998;1:1001–1010. doi: 10.1016/s1097-2765(00)80100-8. [DOI] [PubMed] [Google Scholar]
- 36.Jong AY, Ma JJ. Saccharomyces cerevisiae nucleoside-diphosphate kinase: purification, characterization, and substrate specificity. Arch. Biochem. Biophys. 1991;291:241–246. doi: 10.1016/0003-9861(91)90129-7. [DOI] [PubMed] [Google Scholar]
- 37.Cooperman BS, Chiu NY, Bruckmann RH, Bunick GJ, McKenna GP. Yeast inorganic pyrophosphatase. I. New methods of purification, assay, and crystallization. Biochemistry. 1973;12:1665–1669. doi: 10.1021/bi00733a001. [DOI] [PubMed] [Google Scholar]
- 38.McLeish MJ, Kenyon GL. Relating structure to mechanism in creatine kinase. Crit. Rev. Biochem. Mol. Biol. 2005;40:1–20. doi: 10.1080/10409230590918577. [DOI] [PubMed] [Google Scholar]
- 39.Notarnicola SM, Park K, Griffith JD, Richardson CC. A domain of the gene 4 helicase/primase of bacteriophage T7 required for the formation of an active hexamer. J. Biol. Chem. 1995;270:20215–20224. doi: 10.1074/jbc.270.34.20215. [DOI] [PubMed] [Google Scholar]
- 40.Tabor S, Richardson CC. Selective inactivation of the exonuclease activity of bacteriophage T7 DNA polymerase by in vitro mutagenesis. J. Biol. Chem. 1989;264:6447–6458. [PubMed] [Google Scholar]
- 41.Sambrook J, Maniatis T, Fritsch EF. Molecular cloning: a laboratory manual, 3 v. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 42.Giglio S, Monis PT, Saint CP. Demonstration of preferential binding of SYBR Green I to specific DNA fragments in real-time multiplex PCR. Nucleic Acids Res. 2003;31:e136. doi: 10.1093/nar/gng135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang W, Chen K, Xu C. DNA quantification using EvaGreen and a real-time PCR instrument. Anal. Biochem. 2006;356:303–305. doi: 10.1016/j.ab.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 44.Walker NJ. Real-time and quantitative PCR: applications to mechanism-based toxicology. J. Biochem. Mol. Toxicol. 2001;15:121–127. doi: 10.1002/jbt.8. [DOI] [PubMed] [Google Scholar]
- 45.Houldsworth J, Chaganti RS. Comparative genomic hybridization: an overview. Am. J. Pathol. 1994;145:1253–1260. [PMC free article] [PubMed] [Google Scholar]
- 46.Wang F, Wang L, Briggs C, Sicinska E, Gaston SM, Mamon H, Kulke MH, Zamponi R, Loda M, Maher E, et al. DNA degradation test predicts success in whole-genome amplification from diverse clinical samples. J. Mol. Diagn. 2007;9:441–451. doi: 10.2353/jmoldx.2007.070004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.