Abstract
Glutamate transporters play a crucial role in physiological glutamate homeostasis, neurotoxicity, and glutamatergic regulation of opioid tolerance. However, how the glutamate transporter turnover is regulated remains poorly understood. Here we show that chronic morphine exposure induced posttranscriptional down-regulation of the glutamate transporter EAAC1 in C6 glioma cells with a concurrent decrease in glutamate uptake and increase in proteasome activity, which were blocked by the selective proteasome inhibitor MG-132 or lactacystin but not the lysosomal inhibitor chloroquin. At the cellular level, chronic morphine induced the PTEN (phosphatase and tensin homolog deleted on chromosome Ten)-mediated up-regulation of the ubiquitin E3 ligase Nedd4 via cAMP/protein kinase A signaling, leading to EAAC1 ubiquitination and proteasomal degradation. Either Nedd4 or PTEN knockdown with small interfering RNA prevented the morphine-induced EAAC1 degradation and decreased glutamate uptake. These data indicate that cAMP/protein kinase A signaling serves as an intracellular regulator upstream to the activation of the PTEN/Nedd4-mediated ubiquitin-proteasome system activity that is critical for glutamate transporter turnover. Under an in vivo condition, chronic morphine exposure also induced posttranscriptional down-regulation of the glutamate transporter EAAC1, which was prevented by MG-132, and transcriptional up-regulation of PTEN and Nedd4 within the spinal cord dorsal horn. Thus, inhibition of the ubiquitin-proteasome-mediated glutamate transporter degradation may be an important mechanism for preventing glutamate overexcitation and may offer a new strategy for treating certain neurological disorders and improving opioid therapy in chronic pain management.
Glutamate transporters play a crucial role in physiological glutamate homeostasis, neurotoxicity, and glutamatergic regulation of opioid tolerance (1-5). However, how the glutamate transporter degradation is regulated remains unclear (6-8). The ubiquitin-proteasome system (UPS)2 is a major non-lysosomal proteolytic pathway that degrades cellular proteins including those with important roles in the regulation of cell growth and function (9-11). In addition, activation of UPS has been shown to regulate the PSD-95 degradation and α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor surface expression (12), suggesting a possible relationship between UPS and glutamatergic activities.
Ubiquitination is a process involving three enzymes: E1 (ubiquitin-activating enzyme), E2 (ubiquitin-conjugating enzyme), and E3 (ubiquitin ligase) (13, 14). Interactions between an E3 ligase and its target molecule are considered a key step in determining the selectivity of UPS for a target molecule and its subsequent proteasomal degradation, a process that is subject to intracellular modulation by various upstream regulators (14). PTEN (phosphatase and tensin homolog deleted on chromosome Ten) is a tumor suppressor and lipid phosphatase, which has been shown to regulate cell survival (15, 16), stem cell proliferation (17, 18), and neuronal function (19, 20). Recently, PTEN was shown to regulate E3 ligases (21), suggesting a potential regulatory role for PTEN in the UPS activity.
Antinociceptive tolerance induced by chronic morphine has been shown to be mediated, at least in part, through a central glutamatergic mechanism including an altered glutamate transporter expression (5, 22-25). Inhibition of glutamate transporter activity directly contributes to a heightened activity of N-methyl-d-aspartate receptors and the development of opioid tolerance (26). These data suggest that opioids may regulate the glutamate transporter turnover. Utilizing a C6 glioma cell line that endogenously expresses glutamate transporter EAAC1 (27, 28) and opioid receptors (29), we examined the hypothesis that chronic morphine exposure would induce the PTEN-mediated UPS activation resulting in an enhanced glutamate transporter degradation.
EXPERIMENTAL PROCEDURES
Cell Lines and Cell Culture—Rat glioma C6 cells, human glioma U87MG cells, and human embryonic kidney 293T cells were grown in Dulbecco's modified Eagle's medium plus 10% fetal bovine serum and 100 units/ml each of penicillin and streptomycin (Invitrogen) in a humidified atmosphere of 5% CO2 and 95% air at 37 °C. The C6 glioma cell line was used as a model system because it endogenously expresses EAAC1 (27, 28) and opioid receptors (29). This cell line has been extensively used to examine the biosynthesis, metabolism, and regulatory mechanisms of the EAAC1 glutamate transporter (30-32). Because C6 glioma cells co-express both opioid receptors and EAAC1, this cell line was chosen to examine the hypothesis of the current study. U87MG cells with the PTEN-deficient gene were extensively used to investigate PTEN functions in cell cycle regulation and tumorgenesis (33-36). We used PTEN-deficient U87MG cells to demonstrate that PTEN would be necessary for Nedd4 expression after chronic morphine exposure, although U87 cells do not express the same transporter of EAAC1 as C6 cells. Cells were passaged fewer than 30 times and used between 75 and 85% confluence. There was no evidence for passage-dependent changes in morphology or effects measured in the present study.
Immunoprecipitation (IP)—Antibodies used in the IP experiments included EAAC1 (Alpha Diagnostics International (ADI), San Antonio, TX, rabbit polyclonal, 1:100); PTEN (Cell Signaling Technology, Inc., Beverly, MA, rabbit polyclonal, 1:100); Nedd4, Parkin, and Cbl (Abcam, Cambridge, MA, rabbit polyclonal, 1:200); Mdm2 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, rabbit polyclonal, 1:200), and normal rabbit IgG (Upstate, Lake Placid, NY, 1:200). Whole lysates of either control or drug-treated C6, 293T, and U87 cells were prepared in a lysis buffer (20 mm Tris (pH 7.5), 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1% Triton X-100, 2.5 mm sodium pyrophosphate, 1 mm β-glycerophosphate, 1 mm Na3VO4, 10 μg/ml of each leupeptin, aprotinin, pepstatin, and 1 mm phenylmethylsulfonyl fluoride) pre-cleared with 50 μl of anti-rabbit IgG beads (eBioscience, San Diego, CA) at 4 °C for 2 h, incubated with primary antibodies at 4 °C overnight, and then followed by anti-rabbit IgG bead pulldown at 4 °C for 2 h. After four washes with the lysis buffer, immune complexes were released in 100 μl of 2× reducing sample buffer by boiling at 90-95 °C for 5 min, resolved by SDS-PAGE, transferred to nitrocellulose membranes (Amersham Biosciences), and then subjected to the Western blot analysis.
Western Blotting—Western blotting was carried out using the following antibodies: monoclonal ubiquitin and polyclonal PTEN (Cell Signaling, 1:1,000); monoclonal PTEN (Pharmingen, 1:1,000); polyclonal μ-opioid receptor (Chemicon International, Inc., Temecula, CA, 1:1,000); EAAC1 (ADI, 1:100); β-actin (Santa Cruz, 1:25,000); polyclonal Nedd4 (Abcam Inc., 1:2,500); and monoclonal α-tubulin (Sigma, 1:1,000). Cell and tissue lysates (30 μg of protein) were subjected to standard SDS-PAGE and then transferred to a nitrocellulose membrane. After being blocked at room temperature for 1 h in a blocking buffer (PBS(-), 5% skim milk, 0.1% Tween 20), the membranes were incubated with antibodies diluted (as described above) in blocking buffer at room temperature for 2 h or at 4 °C overnight. This was followed by three washes in washing buffer (PBS(-), 0.1% Tween 20) and incubation with horseradish peroxidase-linked anti-mouse IgG (1:5,000), horseradish peroxidase-linked anti-rabbit IgG (1:10,000) (Amersham Biosciences), or horseradish peroxidase-linked anti-guinea pig IgG (1:3000) (Sigma) at room temperature for 1 h. Horseradish peroxidase-linked anti-rabbit IgG TrueBlot (eBioscience, San Diego, CA) was used in the IP experiments. After three washes, the bands were visualized by the ECL Plus Western blotting Detection System (Amersham Biosciences). The intensity of each protein level was analyzed using ImageJ 1.34s software (National Institutes of Health).
In Vitro Glutamate Uptake Assay—An in vitro preparation was used to assess glutamate uptake activity according to a previously published method (5, 22, 37). In brief, C6 cell lysates were homogenized in 500 μl of ice-cold homogenization buffer (0.32 m sucrose, 2 mm EDTA, 2 mm EGTA, and 20 mm HEPES, pH 7.2, plus 1 mm phenylmethylsulfonyl fluoride and protease inhibitor mixture tablet (Roche Diagnostics GmbH, Mannheim, Germany)) and centrifuged at 1,500 × g for 10 min at 4 °C, and the supernatant was collected. Pellets were re-suspended in the same homogenization buffer, re-centrifuged as above. Both supernatants were combined and again centrifuged at 13,000 × g for 10 min at 4 °C. The so-obtained pellets were suspended in 1 ml of Locke's buffer (154 mm NaCl, 5.6 mm KCl, 2.3 mm CaCl2, 1.0 mm MgCl2, 3.6 mm NaHCO3, 5 mm glucose, 5 mm HEPES, pH 7.2, and saturated with 95% O2, 5% CO2). Glutamate uptake activity was determined by incubating the preparation (100 μg of protein content) with 0.4 μCi of l-[3H]glutamic acid (PerkinElmer Life Sciences) in a total volume of 1 ml of Locke's buffer for 5 min at 37 °C. The reaction was terminated by filtering the pellets through a Whatman (Maidstone, UK) GF/C 2.4-cm filter presoaked in Locke's buffer. The filter was then washed with 2 ml of ice-cold Locke's buffer three times, air-dried, and transferred into vials containing 10 ml of scintillation mixture (Fisher Scientific). The radioactivity was measured by Liquid Scintillation Analyzer Tri-Carb 2900TR (PerkinElmer). The basal uptake activity in counts per minute (cpm) was measured in the absence of any treatment. -Fold change in glutamate uptake activity was calculated with the following equation: (basal cpm without treatment - cpm with treatment)/(basal cpm without treatment).
26 S Proteasome Activity Assay—26 S proteasome activity was measured as described previously (38). Briefly, cells were washed with PBS followed with buffer I (50 mm Tris, pH 7.4, 2 mm dithiothreitol, 5 mm MgCl2, 2 mm ATP), and pelleted by centrifugation. Glass beads and homogenization buffer (50 mm Tris, pH 7.4, 1 mm dithiothreitol, 5 mm MgCl2, 2 mm ATP, 250 mm sucrose) were added and vortexed for 1 min. Beads and cell debris were removed by centrifugation at 1,000 × g for 5 min and 10,000 × g for 20 min. Protein concentration was determined by the BCA protocol (Pierce). One hundred μg of protein of each sample was diluted with buffer I to a final volume of 1,000 μl and the fluorogenic proteasome substrate SucLLVY-7-amido-4-methylcoumarin was added in a final concentration of 50 μm in 1% dimethyl sulfoxide. Cleavage activity was monitored continuously by the detection of free 7-amido-4-methylcoumarin using a Synergy HT Multi-detection Microplate Reader (Bio-Tek Instruments, Inc., Winooski, VT) at 380/460 nm and 37 °C. As controls, 7-amido-4-methylcoumarin (2 μm) was incubated with each agent in buffer I without cell extracts and measurements of proteasome activity were corrected when necessary.
Vector-based Short Hairpin RNA Oligomers—The DNA vectors for small interfering RNA (siRNA) constructs, pRNAT-H1.1/Neo/GFP (SD1216), were obtained from GenScript Corp. (Piscataway, NJ). Three short hairpin RNA insert sequences for the rat PTEN (GeneBank™ accession number NM_031606) and one scramble sequence for negative control were selected as follows by using the software siRNA Construct Builder (GenScript Corp.): GGATCCCGTTTATCTCTGGTCCTTACTTCTTCAAGAGAGAAGTAAGGACCAGAGATAAATTTTTTCCAAAAGCTT (75 bp, siPTEN construct 1), GGATCCCGTATACACCTTCAAGTCTTTCTTTCAAGAGAAGAAAGACTTGAAGGTGTATATTTTTTCCAAAAGCTT (75 bp, siPTEN construct 2), GGATCCCGTAATCCAGGTGATTCTTTAACTTCAAGAGAGTTAAAGAATCACCTGGATTATTTTTTCCAAAAGCTT (75 bp, siPTEN construct 3), GGATCCCGTCGCTTACCGATTCAGAATGGTTCAAGAGACCATTCTGAATCGGTAAGCGATTTTTTCCAAAAGCTT (75 bp, scrambled siPTEN construct 4). The four siRNA constructs were transfected into C6 cells using Lipofectamine 2000 (Invitrogen) transfection reagents according to the manufacturer's protocol. Stable cell clones were obtained with neomycin selection, based on GFP expression. PTEN expression following the transfection was assessed by Western blot and fluorescence microscopy.
RNA Interference—Custom SMARTpool siRNA to target the rat Nedd4 and negative control siCONTROL Non-targeting siRNA pool were designed and synthesized by Dharmacon (Lafayette, CO). Each siRNA (100 nm) was transfected into C6 cells using Lipofectamine 2000 (Invitrogen) transfection reagents according to the manufacturer's protocol. At 24 h after transfection, cells were treated with vehicle, morphine (10 μm, 48 h), morphine plus MG-132 (20 μm added during the last 12 h of the 48-h morphine exposure), or MG-132 alone for the indicated times, followed by relevant assays as described.
Immunohistochemistry and Fluorescence Microscopy—Stably transfected siPTEN or scramble siRNA glioma C6 cells (plated on glass cover-slips) were fixed with 4% paraformaldehyde at room temperature for 10 min and permeabilized with acetone at -20 °C for 3 min. After being blocked in a blocking solution (10% normal goat serum/PBS) at room temperature for 2 h, cell-plated coverslips were incubated with polyclonal anti-EAAC1 (ADI, 1:100) at 4 °C overnight, followed by incubation with Cy3-conjugated AffiniPure goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories, Inc., 1:500) in blocking solution at room temperature for 2 h, nuclei were counter-stained with Hoechst (0.5 μg/ml in PBS) for 10 min. Coverslips were washed with PBS between steps. Stained cells were mounted in Vectorshield (Vector Laboratories, Burlingame, CA) and observed under the OLYMPUS 1X71 fluorescence microscope.
RT-PCR—Total RNA was extracted from drug-treated C6 cells and spinal cord dorsal horns. Isolations were performed using the RNeasy Mini Kit (Qiagen) according to the manufacturer's protocol. RT-PCR were carried out using Titan One Tube RT-PCR System (Roche Diagnostics) according to the manufacturer's manual. All primers for rat glutamate transporters, PTEN, Nedd4, and a housekeeping gene were designed by Invitrogen OligoPerfect based on GenBank sequences (EAAC1, NM_013032; PTEN, NM_031606; Nedd4, XM_343427, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), XO 02231) and synthesized by Invitrogen: for EAAC1, forward, 5′-TGGTTCGAGGACACAGTGAG-3′ and reverse, 5′-GATCAGTGGCAGCACTACGA-3′, amplifying 800-base pair products; for PTEN, forward, 5′-ACACCGCCAAATTTAACTGC-3′ and reverse, 5′-AGGTTTCCTCTGGTCCTGGT-3′, amplifying 630-base pair products; for Nedd4, forward, 5′-GATCCTCGGATGCAGAATGT-3′ and reverse, 5′-ATCGCCACTGGATTACAAGG-3′, amplifying 715-base pair products; for GAPDH, forward, 5′-GGTGATGCTGGTGCTGAGTA-3′ and reverse, 5′-GGATGCAGGGATGATGTTCT-3′, amplifying 369-base pair products.
RT-PCR was started with incubating the sample (20 ng of total RNA) at 50 °C for 30 min for reverse transcription, followed by one cycle of denaturation at 94 °C for 2 min, 10 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, and elongation at 68 °C for 1 min and 25 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, and elongation at 68 °C for 1 min with cycle elongation of 5 s for each cycle. A 7-min extension at 68 °C was carried out at the end of the final cycle. The samples were then cooled to 4 °C. Ten μl of the RT-PCR product was loaded onto one lane and subjected to electrophoresis at 50 V through 1.8% (w/v) agarose gel containing 0.4 μg/ml ethidium bromide. The RT-PCR product bands and a 100-bp ladder molecular weight marker (New England BioLabs, Ipswich, MA) were visualized under UV light and imaged using the AlphaEaseFC system (Alpha Innotech Corporation, San Leandro, CA). The intensity of each transporter mRNA level was analyzed using ImageJ 1.34s software (NIH, Bethesda, MD). The GAPDH level was used for sample normalization.
Reagents—Morphine, naloxone, ddA, H89, RP-8-PCPT-CGMPS, SucLLVY-7-amido-4-methylcoumarin (chymotrypsin-like), and 7-amido-4-methylcoumarin were purchased from Sigma; Go6976, MG-132, lactacystin, and chloroquin were from Calbiochem (La Jolla, CA); LY294002 from Cell Signaling Technology, Inc. (Beverly, MA).
Experimental Animals and Intrathecal Drug Delivery—Adult male Sprague-Dawley rats (Charles River Laboratories, Wilmington, MA) weighing 300-350 g were housed in individual cages with water and food pellets available ad libitum. The animal room was artificially illuminated from 7:00 a.m. to 7:00 p.m. The protocol was approved through our Institutional Animal Care and Use Committee. A polyethylene-10 intrathecal catheter was implanted in each rat according to a previously described method (39). Rats exhibiting neurological deficits after the implantation were excluded. The experiment began at 24 h after the intrathecal catheter implantation. Morphine (15 nmol) plus vehicle, MG-132 (5 nmol in 10 μl of saline with 0.4% dimethyl sulfoxide) alone, morphine plus MG-132 (5 nmol), or vehicle (saline with 0.4% dimethyl sulfoxide) alone was administrated (n = 6 rats/group) twice daily for 7 days through the intrathecal catheter in a total volume of 20 μl followed by 10 μl of saline flush to purge the residual from the catheter dead space. Spinal cord tissue samples were taken at the end of the 7-day treatment.
RESULTS
Chronic Morphine Exposure Induces EAAC1 Down-regulation in C6 Glioma Cells
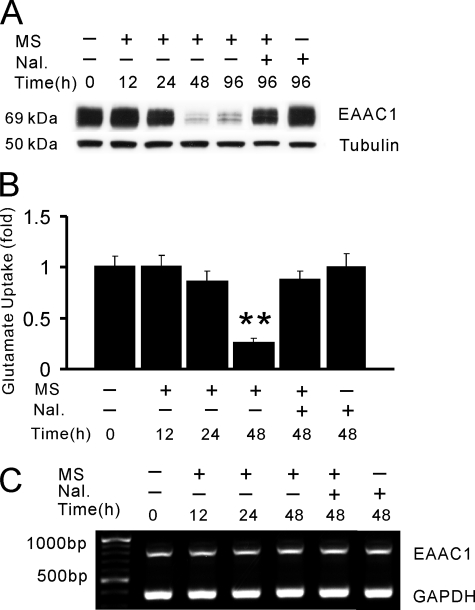
Expression of glutamate transporter EAAC1 was examined at 12-, 24-, 48-, and 96-h exposure of C6 glioma cells. Although the EAAC1 expression (Western blot) was not changed within the first 24 h of morphine exposure, the EAAC1 level was dramatically decreased by nearly 90% when examined at 48 h of morphine exposure (Fig. 1A), which lasted up to at least 96 h after morphine exposure. In contrast, no changes of opioid receptor levels were detected at these time points in C6 glioma cells under the same condition (data not shown), indicating that chronic morphine exposure induced a selective down-regulation of glutamate transporter EAAC1.
FIGURE 1.
Chronic morphine exposure down-regulates EAAC1 expression. C6 cells were exposed to morphine (MS, 10 μm) or morphine plus naloxone (Nal., 1 μm) for 0, 12, 24, 48, and 96 h. A, Western blotting of whole cell lysates. B, in vitro glutamate uptake activity. C, RT-PCR analysis. Results are representative of three independent experiments or the mean ± S.E. for at least three independent extract preparations. **, p < 0.01, compared with vehicle control using Student's t test. Tubulin, loading control.
Consistent with the down-regulation of EAAC1 expression, glutamate uptake activity in C6 glioma cells was decreased by nearly 70% at 48 h of morphine exposure (Fig. 1B, p < 0.01), as assessed using an in vitro glutamate uptake assay (5, 37). Co-treatment of morphine with the opioid receptor antagonist naloxone (1 μm) completely prevented the down-regulation of EAAC1 and the decrease in glutamate uptake activity, whereas naloxone alone did not alter the baseline EAAC1 expression or glutamate uptake activity (Fig. 1, A and B). To determine whether EAAC1 down-regulation occurred at the mRNA level, we examined the EAAC1 mRNA expression (RT-PCR) in C6 glioma cells under the same condition of morphine exposure. The EAAC1 mRNA level did not differ from the baseline when examined at 12, 24, or 48 h of morphine exposure (Fig. 1C). These results indicate that chronic morphine exposure induced the opioid receptor-mediated EAAC1 down-regulation, which occurred at the posttranscriptional level and critically regulated glutamate uptake activity.
EAAC1 Down-regulation Is Mediated through Ubiquitin-proteasomal Degradation
It has been shown that proteasome-mediated degradation of cellular proteins including oncoproteins and cell cycle proteins is a critical mechanism for the posttranscriptional regulation of protein turnover (40). Given that the EAAC1 down-regulation after chronic morphine exposure occurred at the posttranscriptional level, we examined whether proteasomal degradation would play a major role in this process. To our surprise, this chronic morphine-induced EAAC1 down-regulation in C6 glioma cells was completely blocked by co-treatment of morphine with the selective proteasome inhibitor MG-132 (20 μm) or lactacystin (10 μm), when MG-132 or lactacystin was added into the incubation for 12 h beginning at 36 h after the initiation of morphine exposure. In contrast, co-treatment of morphine with chloroquin (50 μm), a lysosome inhibitor, did not prevent morphine-induced EAAC1 down-regulation (Fig. 2A). The MG-132 or lactacystin treatment alone had no effect on the expression of EAAC1 or opioid receptors, nor did the treatment with 50 μm chloroquine alone have detectable cytotoxic effects on C6 cells (data not shown). These results indicate that morphine induced the proteasome-mediated EAAC1 degradation.
FIGURE 2.
Chronic morphine induces ubiquitin-proteasome-mediated degradation of EAAC1. C6 cells were treated with morphine (MS, 48 h) or morphine plus the selective proteasome inhibitor MG-132 (20 μm for 12 h) or lactacystin (Lac., 10 μm for 12 h) or the lysosome inhibitor chloroquin (Chl., 50 μm for 12 h). A, Western blotting (WB) of whole cell lysates. B, in vitro glutamate uptake activity. C, 26 S proteasome activity assay. D, C6 cell lysates were co-immunoprecipitated with an anti-EAAC1 antibody (IP), followed by Western blotting with an anti-ubiquitin antibody and anti-EAAC1 antibody (WB). Ubn, the migration position of polyubiquitin conjugates; +, the EAAC1 position. Results are representative of three independent experiments or the mean ± S.E. for at least three independent extract preparations. *, p < 0.05 and **, p < 0.01, compared with vehicle control using Student's t test.
Prevention of EAAC1 down-regulation by either MG-132 or lactacystin also reversed the decreased glutamate uptake activity following chronic morphine exposure, whereas the lysosomal inhibitor chloroquin had no effect (Fig. 2B). Because changes in EAAC1 expression and glutamate uptake are shown as a ratio between control and experimental groups, the exact difference in the magnitude of change between these two assays were unable to be determined. Nonetheless, changes in both EAAC1 and glutamate uptake after either MG-132 or lactacystin indicate a functional relationship between the proteasome-mediated EAAC1 degradation and glutamate uptake activity in C6 glioma cells. Interestingly, the basal level of EAAC1 turnover and glutamate uptake activity must also be regulated by the proteasomal system, because both the EAAC1 protein level and glutamate uptake activity were elevated above their basal level (before morphine exposure) after C6 glioma cells were co-exposed to morphine and MG-132 or lactacystin (Fig. 2, A and B, p < 0.05). Moreover, chronic morphine exposure resulted in an increase in proteasome activity in C6 glioma cells with a time course parallel to that of the EAAC1 degradation (Fig. 2C). This concurrent increase in proteasome activity was reversed by treatment of either naloxone or MG-132 (Fig. 2C, p < 0.05), indicating that chronic morphine exposure enhanced proteasome activity through activation of opioid receptors.
A necessary step in the proteasomal degradation pathway is the formation of an ubiquitin-protein conjugate (11). Subsequently, the covalent binding of ubiquitin ligase and its target molecule leads to the recognition and degradation of a target molecule by the 26 S proteasome (41). To further confirm the findings indicating a role for the proteasomal pathway in morphine-induced EAAC1 degradation, we examined whether morphine exposure would lead to ubiquitination of EAAC1 proteins in C6 glioma cells by using an IP assay. If morphine-induced EAAC1 ubiquitination and subsequent proteasomal degradation, the state of EAAC1 ubiquitination should be displayed after inhibition of the proteasomal activity. Indeed, a smear of ubiquitin-conjugated EAAC1 protein bands, which migrated slowly in gels, was clearly shown in C6 glioma cells co-treated with morphine and either MG-132 or lactacystin, but not the lysosomal inhibitor chloroquin, vehicle, morphine, MG-132, or lactacystin alone (Fig. 2D).
Collectively, the preventive effect of proteasome inhibitors on EAAC1 degradation, increased proteasome activity, and a functional relationship between EAAC1 degradation and glutamate uptake activity indicate that chronic morphine exposure induced polyubiquitination of EAAC1 proteins with their subsequent proteasomal degradation in C6 glioma cells.
Nedd4 Is the E3 Ligase Associated with EAAC1
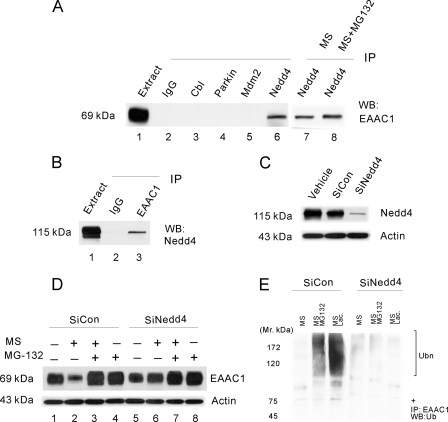
The specificity of ubiquitination is determined by E3 ligases, which interact with their selective protein substrates. In the next step, we sought to identify the E3 ligase partner of EAAC1 responsible for the EAAC1 ubiquitination and its proteasomal degradation. We first used the IP assay to examine possible EAAC1 conjugation with several candidate E3 ligases, including Cb1, Parkin, Mdm2, and Nedd4, in C6 glioma cells and each of these immune complexes was screened for copurification of EAAC1 by immunoblot. EAAC1 was detected only in Nedd4 immunoprecipitates (Fig. 3A, lane 6) but not in the control IgG, Cb1, Parkin, or Mdm2 complex (Fig. 3A, lanes 2-5). Morphine or morphine plus MG-132 did not disturb EAAC1-Nedd4 association (Fig. 3A, lanes 7 and 8) and we did not detect associations between EAAC1 and other tested ligases. Additional control experiments confirmed that each antibody did indeed immunoprecipitate its appropriate antigen (data not shown). In another experiment using a reciprocal approach, Nedd4 copurified with EAAC1 (Fig. 3B, lane 3) but not with a control IgG (Fig. 3B, lane 2). Similarly, neither Cb1 nor Parkin and Mdm2 copurified with EAAC1 in the same experiment (data not shown). These data provide strong evidence that EAAC1 and Nedd4 interact in C6 glioma cells, which is consistent with previous reports suggesting a possible role of a Nedd4 family member (Nedd4-2) in the regulation of EAAC1 function (42, 43) and dopamine transporter ubiquitination (44).
FIGURE 3.
The ubiquitin E3 ligase Nedd4 interacts with EAAC1. A, EAAC1 copurified with Nedd4. Extracts of C6 cells were subjected to immunoprecipitation using antibodies against several ubiquitin E3 ligases (lanes 2-6). EAAC1 co-purified with Nedd4 (lane 6) but not with a control IgG, Cbl, Parkin, or Mdm2. Morphine or morphine plus MG-132 did not disturb the interaction between EAAC1 and Nedd4 (lane 7 and 8). EAAC1 extract (lane 1) represents 4% of the amount used in the immunoprecipitation. B, Nedd4 co-purified with EAAC1. Extracts of C6 cells were immunoprecipitated with antibodies against EAAC1. Nedd4 was specifically copurified in EAAC1 precipitates (lane 3) but not in control IgG precipitates (lane 2). Extract (lane 1) represents 1% of the amount used in the immunoprecipitation. C, Nedd4 was markedly knocked down with siRNA. C6 cells were transfected with negative control siCONTROL non-targeting siRNA pool, Nedd4 siRNA pool, or vehicle, the Nedd4 expression level was examined at 48 h after each transfection. D, Nedd4 knockdown abolished the morphine-induced EAAC1 down-regulation. C6 cells transiently transfected with siCONTROL or siNedd4 were treated with vehicle, morphine (10 μm for 48 h), morphine plus MG-132, or MG-132 (MG-132 was added during the final 12 h of the 48-h morphine exposure), the EAAC1 expression level was examined in each group. E, C6 cells transiently transfected with siCONTROL or siNedd4 were treated with vehicle, morphine (10 μm for 48 h), morphine plus MG-132, or MG-132 (MG-132 was added during the final 12 h of the 48-h morphine exposure), cell lysates were co-immunoprecipitated with an anti-EAAC1 antibody (IP), followed by Western blotting with an anti-ubiquitin antibody (WB, Western blot). Ubn, the migration position of polyubiquitin conjugates; +, the EAAC1 position. Results are representative of three independent experiments. Actin, a loading control.
To determine a causal relationship between the roles of Nedd4 E3 ligase and EAAC1 degradation, we examined whether knockdown of the Nedd4 E3 ligase using siRNA would prevent the UPS-mediated EAAC1 degradation after chronic morphine exposure. Of C6 glioma cells transfected with a vehicle, negative control (siCONTROL) non-targeting siRNA pool, or the Nedd4 siRNA pool, the Nedd4 expression level was reduced by nearly 90% only in those cells transfected with the Nedd4 siRNA pool when examined at 48 h after the transfection (Fig. 3C). The siRNA knockdown of Nedd4 successfully abolished chronic morphine-induced EAAC1 down-regulation as compared with the Nedd4 siCONTROL transfectants (Fig. 3D, lanes 1 and 2 versus lanes 5 and 6). The effect of Nedd4 siRNA knockdown on the prevention of EAAC1 down-regulation was quantitatively similar to that after co-treatment of morphine with the proteasome inhibitor MG-132. Of significance is that Nedd4 knockdown with siRNA prevented the appearance of the high molecular weight bands as detected in Fig. 3E. Taken together, these results indicate that Nedd4 played a critical role as an E3 ligase in this UPS-mediated EAAC1 degradation.
PTEN Regulates the E3 Ligase Nedd4 via the cAMP/PKA Signaling
Because the Nedd4 E3 ligase is critically involved in UPS-mediated EAAC1 degradation, how does chronic morphine exposure regulate this process? PTEN is a tumor suppressor and lipid phosphatase. Recently, there has been increasing interest in the role of PTEN for the normal development of neuronal tissues and potential regulation of E3 ligases (21). Accordingly, we asked whether PTEN would have a role in the regulation of Nedd4 expression after chronic morphine exposure in the following experiments.
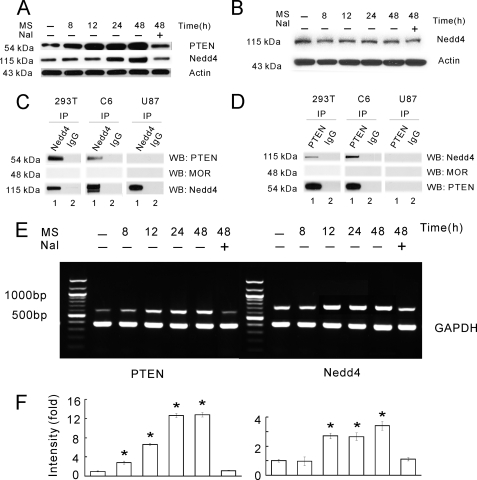
PTEN Is Up-regulated after Chronic Morphine—Chronic morphine exposure induced a time-dependent up-regulation of PTEN in C6 glioma cells starting at 12 h, which was followed by the up-regulation of Nedd4 beginning at 24 h after morphine exposure and the up-regulation of both PTEN and Nedd4 after morphine exposure were blocked by naloxone (Fig. 4A). These findings are consistent with morphine-induced EAAC1 down-regulation, which occurred at 48 h after morphine exposure (Fig. 1A), indicating a temporal correlation among these cellular changes. Morphine treatment did not induce Nedd4 up-regulation in the PTEN-null U87 cell line (Fig. 4B), indicating that Nedd4 regulation is PTEN-dependent. In addition, the RT-PCR method showed that this up-regulation occurred at the transcriptional level. mRNA levels of PTEN started increasing significantly after 8 h of morphine exposure and reached 13-fold at 24 h as compared with the level at 0 h; mRNA levels of Nedd4 started increasing significantly after 12 h of morphine exposure and reached 3.5-fold at 48 h as compared with the level at 0 h; the up-regulation of both PTEN and Nedd4 after morphine exposure was blocked by naloxone (Fig. 4, E and F). Taken together, these data suggest that morphine-induced EAAC1 down-regulation may be mediated through inducing PTEN and Nedd4 to initiate the ubiquitin-proteasomeal degradation pathway.
FIGURE 4.
PTEN regulates the Nedd4 expression. A, C6 cells were treated with morphine (MS, 10 μm) or morphine plus naloxone (Nal, 1 μm) for 0, 8, 12, 24, and 48 h. Western blotting of PTEN or Nedd4 was performed using whole cell lysates. B, time course effect of morphine on the level of Nedd4 protein in PTEN-null U87 cells. C, PTEN copurified with Nedd4. Lysates from the PTEN-expressing 293T and C6 cell lines and the PTEN-null U87 cell line were IP with Nedd4 (lane 1) or normal rabbit IgG (lane 2) and were Western blotted (WB) with PTEN, μ-opioid receptor (MOR), or Nedd4. PTEN, but not μ-opioid receptor, copurified with Nedd4 (lane 1), but not control IgG (lane 2) in 293T and C6 cells. D, Nedd4 copurified with PTEN. Extracts of 293T, C6, and U87 cells were immunoprecipitated with antibodies against PTEN (lane 1) or normal rabbit IgG (lane 2) and Western blotted (WB) with Nedd4, μ-opioid receptor, and PTEN. Nedd4 but not μ-opioid receptor specifically copurified in PTEN precipitates (lane 1) but not in control IgG precipitates (lane 2) in 293T and C6 cells. PTEN-null U87 cells were used as control. E, time course PTEN and Need4 mRNA level (RT-PCR) after each treatment in C6 cells. F, intensity (-fold) quantification and statistical analysis of the RT-PCR results in E. Results are the mean ± S.E. for at least three independent extract preparations. *, p < 0.05, as compared with the vehicle group using Student's t test.
PTEN Interacted with the Nedd4 E3 Ligase—To examine the relationship between PTEN and the Nedd4 E3 ligase, we first used two PTEN-expressing cell lines (C6 and 293T) and a PTEN-null cell line (U87) to examine whether PTEN would physically interact with Nedd4 using the IP assay. Specific interactions between PTEN and Nedd4 were detected in the IP experiments: 1) PTEN conjugated with Nedd4 (Fig. 4C, lane 1 in 293T and C6 cells) but not with a control IgG (Fig. 4C, lane 2 in all three cell lines); 2) Nedd4 also conjugated with PTEN (Fig. 4D, lane 1 in 293T and C6 cells) but not with a control IgG in the reciprocal IP experiment (Fig. 4D, lane 2 in all three cell lines); 3) μ-opioid receptor, as a control, did not copurify with either PTEN or Nedd4 (Fig. 4, C and D); and 4) copurification of PTEN and Nedd4 was present only in PTEN-expressing cell lines (C6 and 293T) but not in the PTEN-null cell line U87 (Fig. 4, C and D).
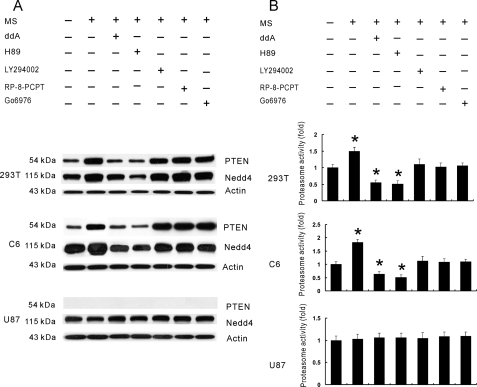
cAMP/PKA Signaling Regulated the PTEN and Nedd4 Expression—Up-regulation of the cAMP/PKA signal pathway has long been associated with the molecular adaptation to long-term opioid administration, which is thought to play a key role in the development of opioid tolerance and dependence (45). Thus, it is likely that cAMP/PKA signaling may be an initial cellular step after chronic morphine exposure that leads to the up-regulation and functional recruitment of both PTEN and Nedd4. To test this hypothesis, we again used two PTEN-expressing cell lines (C6 and 293T) and a PTEN-null cell line (U87). Each of these cell lines was pretreated for 30 min with the adenylyl cyclase inhibitor ddA, the PKA inhibitor H89, the phosphatidylinositol 3-kinase inhibitor LY294002, the cGMP-dependent protein kinase inhibitor RP-8-PCPT, the PKC inhibitor Go6976, or a vehicle (see Fig. 5 legend for doses), followed by co-treatment of each of these agents with morphine for 48 h. The up-regulation of both PTEN and Nedd4 after chronic morphine exposure was blocked only by ddA or H89 but not by other agents (Fig. 5A), and ddA and H89 alone had no effect on the expression of PTEN and Nedd4 (data not shown). Moreover, chronic morphine-induced Nedd4 up-regulation in two PTEN-expressing cell lines (C6 and 293T) was absent in the PTEN-null U87 cell line (Fig. 5A), indicating that PTEN is necessary for Nedd4 expression after chronic morphine exposure.
FIGURE 5.
Morphine recruited PTEN through the cAMP/PKA signaling. A, 293T, C6, and U87 cells were pretreated with the adenylyl cyclase inhibitor ddA (30 μm), PKA inhibitor H89 (30 μm), phosphatidylinositol 3-kinase inhibitor LY294002 (10 μm), cGMP-dependent protein kinase inhibitor RP-8-PCPT (5 μm), or PKC inhibitor Go6976 (1 μm) for 30 min and then co-treated with morphine (10 μm) for 48 h. Western blotting of whole lysate was made. B, 26 S proteasome activity assay of these same samples from panel A. Results are representative of three independent experiments or the mean ± S.E. for at least three independent extract preparations. *, p < 0.05, as compared with the vehicle group using Student's t test.
Collectively, these findings indicate that cAMP/PKA signaling is upstream to the morphine-induced up-regulation of PTEN and Nedd4. Of interest to note is that this morphine-induced PTEN up-regulation through the cAMP/PKA signaling also regulated proteasome activity, because the adenylyl cyclase inhibitor ddA or the PKA inhibitor H89 blocked the morphine-induced increase in proteasome activity only in the PTEN-expressing 293T and C6 cells (Fig. 5B, p < 0.05) even below the baseline and ddA or H89 alone had no effect on proteasome activity (data not shown). Thus, chronic morphine exposure induced a cAMP/PKA-mediated up-regulation of PTEN, which in turn regulated the up-regulation of the Nedd4 E3 ligase and proteasome activity.
PTEN Negatively Regulated the EAAC1 Expression
To determine the functional role of PTEN in the EAAC1 regulation in C6 glioma cells, we first generated, using siRNA, a series of stable clones with the PTEN knockdown (C1-C3) of more than 85% as shown by fluorescence microscopy and Western blotting (Fig. 6A). We then examined the effect of PTEN on EAAC1 expression using these stable PTEN knockdown cells. In a stable PTEN knock-out cell line (Fig. 6B, bottom), there was a marked increase in intracellular labeling of EAAC1 with large vesicle-like structures and clusters accumulated in the perinuclear area. By visual examination, about 90% of the PTEN knocked down C6 cells showed enhanced EAAC1 immunoreactivity. Among these cells, about 80% exhibited the pattern shown in the figure. In contrast, the scramble siPTEN cell exhibited dispersed, punctuating, and weak intracellular labeling of EAAC1 (Fig. 6B, top). Consistently, PTEN knockdown increased the basal glutamate uptake activity and completely reversed the morphine-induced decrease in glutamate uptake as compared with wild type and scramble siPTEN controls (Fig. 6C, p < 0.05). These findings indicate that PTEN serves as a negative intracellular regulator of the EAAC1 expression and glutamate uptake activity with and without chronic morphine exposure. Taken together with the data presented in Figs. 4 and 5, these findings indicate that PTEN plays a critical role in two key steps of the UPS-mediated EAAC1 degradation through the regulation of the Nedd4 E3 ligase and proteasomal activity.
FIGURE 6.
PTEN is a negative regulator in EAAC1 expression in C6 cells. C6 cells were stably transfected with vector-based short hairpin RNA constructs: PTEN/pRNAT-H1.1/Neo/GFP or scramble pRNAT-H1.1/Neo/GFP. A, fluorescence microscopy (top row) and Western blotting of the wild type (Con), scramble (Scr), and 3 siPTEN clones (C1-3) with anti-PTEN (middle row) or tubulin (bottom row). B, immunocytochemistry of scramble siPTEN (ssiPTEN) and siPTEN (C1) with anti-EAAC1 (red) and Hoechst (blue) nuclear staining. There was a marked increase in intracellular labeling of EAAC1 with large vesicle-like structures and clusters accumulated in the perinuclear area (bottom row). Ninety percent of the PTEN knocked down C6 cells represented enhanced EAAC1 immunoreactivities. Among them, 80% exhibits this pattern. In contrast, the scramble siPTEN cell exhibited dispersed, punctuating, and weak intracellular labeling of EAAC1 (top row). These images are representative of multiple fields examined for each treatment from two independent immunofluorescence experiments. C, glutamate uptake activity assay in wild type, SsiPTEN, and siPTEN-transfected cells. *, p < 0.05 as compared with the wide type (WT) control using Student's t test.
MG-132 Blocked Morphine-induced EAAC1 Down-regulation in the Spinal Cord Dorsal Horn of the Rat
To further examine the role of UPS activity in morphine-induced EAAC1 degradation under an in vivo condition, we analyzed the protein and mRNA expressions of EAAC1, PTEN, and Nedd4 in the spinal cord dorsal horn of the rats exposed to vehicle, morphine (15 nmol), morphine plus MG-132 (5 nmol), or MG132 alone (5 nmol) for seven consecutive days. Consistent with the findings observed in C6 glioma cells (Figs. 1A, 2A, and 4A), chronic morphine exposure induced down-regulation of EAAC1 and up-regulation of PTEN and Nedd4 within the spinal cord dorsal horn (Fig. 7A). Co-administration of morphine with MG-132 effectively prevented the down-regulation of EAAC1 and the up-regulation of PTEN and Nedd4 at the protein level, whereas MG-132 itself had no effect on the expression of EAAC1, PTEN, or Nedd4 (Fig. 7A). Moreover, morphine did not alter the baseline mRNA level of EAAC1 but increased the mRNA level of PTEN and Nedd4 (Fig. 7, B and C) by ∼9- and 3-fold, respectively, which are consistent with the results in C6 cells (Fig. 4E). MG-132 itself did not have effects on the mRNA levels of EAAC1, PTEN, or Nedd4. These in vivo results further demonstrate that chronic morphine exposure induced the UPS-mediated down-regulation of spinal glutamate transporter EAAC1 possibly through a PTEN and Nedd4-mediated cellular pathway.
FIGURE 7.
The proteasome inhibitor MG-132 prevented spinal cord EAAC1 down-regulation. Rats (n = 6) were administrated intrathecally (twice daily) with vehicle, morphine (MS, 15 nmol), morphine plus MG-132 (5 nmol), or MG132 (5 nmol) alone for seven consecutive days. A, Western blotting of glutamate transporter EAAC1, PTEN, and Need4 within the spinal cord dorsal horn. B, the mRNA level of EAAC1, PTEN, and Nedd4 examined using RT-PCR after each treatment. C, densitometric quantification of RT-PCR results are shown in B. Analysis of variance, *, p < 0.05, as compared with the vehicle control. Lane 1, vehicle; lane 2, morphine (15 nmol); lane 3, morphine (15 nmol) plus MG-132 (5 nmol); lane 4, MG-132 (5 nmol) alone. Results are representative of three independent experiments or the mean ± S.E. from at least three independent extract preparations. Tubulin, loading control.
DISCUSSION
The present results demonstrate a UPS-mediated cellular mechanism critical for the posttranscriptional regulation of the glutamate transporter turnover. Our data suggest that the UPS-mediated mechanism of glutamate transporter turnover has a significant functional role in the regulation of glutamate homeostasis and glutamatergic activities. In C6 glioma cells, the UPS-mediated glutamate transporter degradation contributed to the physiological glutamate transporter turnover because inhibition of the proteasome activity improved the basal level of endogenous glutamate transporter content. The UPS-mediated glutamate transporter turnover also was substantially enhanced following chronic morphine exposure resulting in an altered glutamate uptake activity in C6 glioma cells as well as the spinal cord dorsal horn.3 Because the glutamatergic system plays an important role in a variety of cellular events such as morphine tolerance and neurotoxicity (5, 23-25, 46, 47), preventing an excessive glutamate transporter turnover under such circumstances may be an important mechanism to maintain glutamate uptake activity and regional glutamate homeostasis.
In the current in vitro experiments, we used the C6 glioma cell line that expresses both EAAC1 (27, 28), a neuronal glutamate transporter, and opioid receptors (29). Although C6 glioma cells have been extensively used for studying the biosynthesis, metabolism, and regulatory mechanisms of EAAC1 glutamate transporter (30-32), C6 glioma cells are tumor-based cell lines with mixed neuronal and astroglial properties. Given that tumor cells may not possess the same cellular regulatory mechanisms as those of normal neuronal and glial cells, we examined the role of UPS activity in the down-regulation of EAAC1 (mainly a neuronal glutamate transporter) using spinal cord tissues obtained from rats repeatedly exposed to morphine for seven consecutive days. Our data demonstrate that MG-132 prevented the down-regulation of spinal cord EAAC1 at the protein but not mRNA level. Moreover, repeated morphine exposure also induced the up-regulation of PTEN and Nedd4 at both and also protein levels, similar to that observed in C6 glioma cells. These results indicate that the degradation of EAAC1 may be subject to the proteasomal regulation through a PTEN-Nedd4-mediated cellular pathway under in vivo conditions as well.
Although the present study does not differentiate the role of various opioid receptor subtypes in the UPS-mediated glutamate transporter degradation, the effectiveness of morphine, a prototypic μ-opioid receptor agonist with a much weaker affinity to other opioid receptors, in our experiments suggests a major contribution of μ-opioid receptor activation to this process. Of significance is that chronic opioid exposure induced a PTEN-dependent up-regulation of the ubiquitin E3 ligase Nedd4 and enhanced proteasome activity, leading to EAAC1 ubiquitination and its subsequent proteasomal degradation. The role of proteasome activity in this process is strongly supported by the data showing that two structurally distinct and selective proteasome inhibitors (MG-132 and lactacystin), but not the lysosome inhibitor chloroquin, both blocked EAAC1 degradation induced by chronic morphine exposure. In addition, the data suggest a cAMP/PKA-dependent mechanism contributing to the increased proteasome activity after chronic morphine exposure. In future studies, we will examine whether this altered proteasome activity is due to a cAMP/PKA-dependent increase in proteasome subunit levels and/or phosphorylation.
There are two important caveats with regard to these findings. One is that demonstration of 26 S proteasome-mediated and ubiquitin-dependent degradation of EAAC1 under our experimental condition does not exclude the possibility that ubiquitin-independent degradation of EAAC1 may also play a role. This may be particularly relevant because the method used to detect proteasomal activity in the experiment may also detect certain ubiquitin-independent protein degradations, such as the Rb family of tumor suppressor (48, 49). It remains to be seen whether a ubiquitin-independent mechanism would contribute to the turnover of EAAC1 following chronic morphine exposure. Another caveat is that our data do not exclusively rule out the possibility that the lysosome activity may be involved in the regulation of glutamate transporter turnover, although we did not detect any significant effect of chloroquin on the EAAC1 degradation under our experimental condition.
An interesting finding from this study is that PTEN played a significant role in the regulation of Need4 E3 ligase and proteasome activity. With regard to the relationship between PTEN and Need4, our data indicate that 1) there was a temporal correlation between the change in PTEN and Need4 expression after chronic morphine exposure; 2) the increase in the Nedd4 protein level after morphine exposure was absent in the PTEN null U87 cells; and 3) chronic morphine exposure increased the PTEN and Nedd4 mRNA levels in C6 cells as detected using RT-PCR. These data indicate that chronic morphine exposure induced the PTEN and Nedd4 expression at the transcriptional level and that the morphine-induced increase in the Nedd4 protein level was PTEN-dependent. Of note, our data do not rule out the possibility that morphine may have an effect on the protein stability leading to the changed expression of PTEN and Nedd4. This issue can be addressed in future studies.
PTEN has been shown to play a role in cell survival (15, 16), stem cell proliferation (17, 18), and neuronal function (19, 20). Our findings indicate that PTEN also acted as an upstream regulator for the expression and function of Nedd4 E3 ligase and proteasome activity, two critical steps in the UPS-mediated glutamate transporter turnover. Moreover, cAMP/PKA signaling regulated the expression of PTEN after chronic morphine exposure, supporting a link between a known cellular mechanism of morphine tolerance and the UPS-mediated degradation of EAAC1. That modulation of several critical steps of UPS after chronic morphine exposure, including PTEN/Nedd4 knockdown and inhibition of proteasome activity, which each prevented the UPS-mediated glutamate transporter down-regulation indicates that the PTEN/Nedd4-mediated UPS activity plays a critical and sufficient role in the glutamate transporter turnover in C6 glioma cells. At the molecular level, PTEN may regulate the Nedd4 function by directly interacting with Nedd4 through its phosphatase-dependent and/or phosphatase-independent mechanisms. PTEN may also modulate protein expression at the transcriptional level through its lipid phosphatase function (50). The exact cellular mechanisms of PTEN in regulating Nedd4 E3 ligase expression and function will be an important step in our future studies. In addition, future studies may also examine the role of additional Nedd4 family members, such as Nedd4-2, as well as other intracellular regulators of UPS in the glutamate transporter turnover with and without chronic morphine exposure.
Our findings may have a significant functional implication in the cellular mechanisms of morphine tolerance. Recent studies have shed light on the neurobiology of opioid tolerance including those related to β-arrestin (51-53), G-protein signaling (54, 55), μ-opioid receptor oligomerization (53, 56, 57), and the central glutamatergic system (5, 22-25). The present data suggest a possible link between regulation of morphine-induced glutamate turnover after chronic morphine exposure and the development of morphine tolerance. Their exact relationship should be investigated in an in vivo system. The present findings may also have potential clinical implications. For example, using proteasome inhibitor and/or targeting key elements of the UPS-mediated glutamate transporter turnover (e.g. PTEN and Nedd4) could offer a new strategy for treating certain neurological disorders (1-5, 22, 58) and improving clinical opioid therapy in chronic and cancer pain treatment (59). These possibilities are supported by a recent study showing that inhibition of proteasomal activity attenuated neuropathic pain behaviors in rats (60), which is also regulated by the glutamatergic system including glutamate transporters (22).
Acknowledgments
We thank Dr. Nina Irwin, Department of Neurosurgery, Children's Hospital, for providing the 293T cell line.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 DA08835, NS42661, and NS45681. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: UPS, ubiquitin-proteasome system; PTEN, phosphatase and tensin homolog deleted on chromosome Ten; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IP, immunoprecipitation; PBS, phosphate-buffered saline; siRNA, small interfering RNA; RT, reverse transcriptase; PKA, protein kinase A; RP-8-PCPT, Rp-8-[(4-chlorophenyl)thio]-cGMPS triethylamine.
L. Yang, S. Wang, B. Sung, G. Lim, and J. Mao, unpublished data.
References
- 1.Lievens, J. C., Bernal, F., Forni, C., Mahy, N., and Kerkerian-Le Goff, L. (2000) Glia 29 222-232 [DOI] [PubMed] [Google Scholar]
- 2.Mennerick, S., Shen, W., Xu, W., Benz, A., Tanaka, K., Shimamoto, K., Isenberg, K. E., Krause, J. E., and Zorumski, C. F. (1999) J. Neurosci. 19 9242-9251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trotti, D., Aoki, M., Pasinelli, P., Berger, U. V., Danbolt, N. C., Brown, R. H., Jr., and Hediger, M. A. (2001) J. Biol. Chem. 276 576-582 [DOI] [PubMed] [Google Scholar]
- 4.Vorwerk, C. K., Naskar, R., Schuettauf, F., Quinto, K., Zurakowski, D., Gochenauer, G., Robinson, M. B., Mackler, S. A., and Dreyer, E. B. (2000) Investig. Ophthalmol. Vis. Sci. 41 3615-3621 [PubMed] [Google Scholar]
- 5.Mao, J., Sung, B., Ji, R. R., and Lim, G. (2002) J. Neurosci. 22 8312-8323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trotti, D. (2002) Adv. Exp. Med. Biol. 513 225-248 [DOI] [PubMed] [Google Scholar]
- 7.Gegelashvili, G., Robinson, M. B., Trotti, D., and Rauen, T. (2001) Prog. Brain Res. 132 267-286 [DOI] [PubMed] [Google Scholar]
- 8.Boehmer, C., Palmada, M., Rajamanickam, J., Schniepp, R., Amara, S., and Lang, F. (2006) J. Neurochem. 97 911-921 [DOI] [PubMed] [Google Scholar]
- 9.Coux, O., Tanaka, K., and Goldberg, A. L. (1996) Annu. Rev. Biochem. 65 801-847 [DOI] [PubMed] [Google Scholar]
- 10.Rolfe, M., Chiu, M. I., and Pagano, M. (1997) J. Mol. Med. 75 5-17 [DOI] [PubMed] [Google Scholar]
- 11.Hershko, A., and Ciechanover, A. (1998) Annu. Rev. Biochem. 67 425-479 [DOI] [PubMed] [Google Scholar]
- 12.Colledge, M., Snyder, E. M., Crozier, R. A., Soderling, J. A., Jin, Y., Langeberg, L. K., Lu, H., Bear, M. F., and Scott, J. D. (2003) Neuron 40 595-607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joazeiro, C. A., and Hunter, T. (2000) Science 289 2061-2062 [DOI] [PubMed] [Google Scholar]
- 14.Wojcik, C., and Di Napoli, M. (2004) Stroke 35 1506-1518 [DOI] [PubMed] [Google Scholar]
- 15.Backman, S., Stambolic, V., and Mak, T. (2002) Curr. Opin. Neurobiol. 12 516-522 [DOI] [PubMed] [Google Scholar]
- 16.Lian, Z., and Di Cristofano, A. (2005) Oncogene 24 7394-7400 [DOI] [PubMed] [Google Scholar]
- 17.Groszer, M., Erickson, R., Scripture-Adams, D. D., Lesche, R., Trumpp, A., Zack, J. A., Kornblum, H. I., Liu, X., and Wu, H. (2001) Science 294 2186-2189 [DOI] [PubMed] [Google Scholar]
- 18.Morrison, S. J. (2002) Nat. Med. 8 16-18 [DOI] [PubMed] [Google Scholar]
- 19.Ji, S. P., Zhang, Y., Van Cleemput, J., Jiang, W., Liao, M., Li, L., Wan, Q., Backstrom, J. R., and Zhang, X. (2006) Nat. Med. 12 324-329 [DOI] [PubMed] [Google Scholar]
- 20.Musatov, S., Roberts, J., Brooks, A. I., Pena, J., Betchen, S., Pfaff, D. W., and Kaplitt, M. G. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 3627-3631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mamillapalli, R., Gavrilova, N., Mihaylova, V. T., Tsvetkov, L. M., Wu, H., Zhang, H., and Sun, H. (2001) Curr. Biol. 11 263-267 [DOI] [PubMed] [Google Scholar]
- 22.Sung, B., Lim, G., and Mao, J. (2003) J. Neurosci. 23 2899-2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao, J., Price, D. D., and Mayer, D. J. (1994) J. Neurosci. 14 2301-2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tiseo, P. J., and Inturrisi, C. E. (1993) J. Pharmacol. Exp. Ther. 264 1090-1096 [PubMed] [Google Scholar]
- 25.Trujillo, K. A., and Akil, H. (1991) Science 251 85-87 [DOI] [PubMed] [Google Scholar]
- 26.Thomson, L. M., Zeng, J., and Terman, G. W. (2006) Neurosci. Lett. 407 64-69 [DOI] [PubMed] [Google Scholar]
- 27.Dowd, L. A., and Robinson, M. B. (1996) J. Neurochem. 67 508-516 [DOI] [PubMed] [Google Scholar]
- 28.Palos, T. P., Ramachandran, B., Boado, R., and Howard, B. D. (1996) Brain Res. Mol. Brain Res. 37 297-303 [DOI] [PubMed] [Google Scholar]
- 29.Bohn, L. M., Belcheva, M. M., and Coscia, C. J. (1998) J. Neurochem. 70 1819-1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang, W., and Kilberg, M. S. (2002) J. Biol. Chem. 277 38350-38357 [DOI] [PubMed] [Google Scholar]
- 31.Sims, K. D., Straff, D. J., and Robinson, M. B. (2000) J. Biol. Chem. 275 5228-5237 [DOI] [PubMed] [Google Scholar]
- 32.Davis, K. E., Straff, D. J., Weinstein, E. A., Bannerman, P. G., Correale, D. M., Rothstein, J. D., and Robinson, M. B. (1998) J. Neurosci. 18 2475-2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, J., Yen, C., Liaw, D., Podsypanina, K., Bose, S., Wang, S. I., Puc, J., Miliaresis, C., Rodgers, L., McCombie, R., Bigner, S. H., Giovanella, B. C., Ittmann, M., Tycko, B., Hibshoosh, H., Wigler, M. H., and Parsons, R. (1997) Science 275 1943-1947 [DOI] [PubMed] [Google Scholar]
- 34.Li, D. M., and Sun, H. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 15406-15411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park, M. J., Kim, M. S., Park, I. C., Kang, H. S., Yoo, H., Park, S. H., Rhee, C. H., Hong, S. I., and Lee, S. H. (2002) Cancer Res. 62 6318-6322 [PubMed] [Google Scholar]
- 36.Denning, G., Jean-Joseph, B., Prince, C., Durden, D. L., and Vogt, P. K. (2007) Oncogene 26 3930-3940 [DOI] [PubMed] [Google Scholar]
- 37.Mitrovic, A. D., Maddison, J. E., and Johnston, G. A. (1999) Neurochem. Int. 34 101-108 [DOI] [PubMed] [Google Scholar]
- 38.Glas, R., Bogyo, M., McMaster, J. S., Gaczynska, M., and Ploegh, H. L. (1998) Nature 392 618-622 [DOI] [PubMed] [Google Scholar]
- 39.Yaksh, T. L., and Rudy, T. A. (1976) Physiol. Behav. 17 1031-1036 [DOI] [PubMed] [Google Scholar]
- 40.Lee, D. H., and Goldberg, A. L. (1998) Trends Cell Biol. 8 397-403 [DOI] [PubMed] [Google Scholar]
- 41.Tolkacheva, T., Boddapati, M., Sanfiz, A., Tsuchida, K., Kimmelman, A. C., and Chan, A. M. (2001) Cancer Res. 61 4985-4989 [PubMed] [Google Scholar]
- 42.Boehmer, C., Henke, G., Schniepp, R., Palmada, M., Rothstein, J. D., Broer, S., and Lang, F. (2003) J. Neurochem. 86 1181-1188 [DOI] [PubMed] [Google Scholar]
- 43.Pham, N., and Rotin, D. (2001) J. Biol. Chem. 276 46995-47003 [DOI] [PubMed] [Google Scholar]
- 44.Sorkina, T., Miranda, M., Dionne, K. R., Hoover, B. R., Zahniser, N. R., and Sorkin, A. (2006) J. Neurosci. 26 8195-8205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nestler, E. J., and Aghajanian, G. K. (1997) Science 278 58-63 [DOI] [PubMed] [Google Scholar]
- 46.Mao, J., Sung, B., Ji, R. R., and Lim, G. (2002) J. Neurosci. 22 7650-7661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakagawa, T., Ozawa, T., Shige, K., Yamamoto, R., Minami, M., and Satoh, M. (2001) Eur. J. Pharmacol. 419 39-45 [DOI] [PubMed] [Google Scholar]
- 48.Kalejta, R. F., and Shenk, T. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 3263-3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Orlowski, M., and Wilk, S. (2003) Arch. Biochem. Biophys. 415 1-5 [DOI] [PubMed] [Google Scholar]
- 50.Chang, C. J., Freeman, D. J., and Wu, H. (2004) J. Biol. Chem. 279 29841-29848 [DOI] [PubMed] [Google Scholar]
- 51.Bohn, L. M., Gainetdinov, R. R., Lin, F. T., Lefkowitz, R. J., and Caron, M. G. (2000) Nature 408 720-723 [DOI] [PubMed] [Google Scholar]
- 52.Bohn, L. M., Lefkowitz, R. J., Gainetdinov, R. R., Peppel, K., Caron, M. G., and Lin, F. T. (1999) Science 286 2495-2498 [DOI] [PubMed] [Google Scholar]
- 53.Whistler, J. L., and von Zastrow, M. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 9914-9919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gintzler, A. R., and Chakrabarti, S. (2000) Mol. Neurobiol. 21 21-33 [DOI] [PubMed] [Google Scholar]
- 55.Ingram, S. L., Vaughan, C. W., Bagley, E. E., Connor, M., and Christie, M. J. (1998) J. Neurosci. 18 10269-10276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Finn, A. K., and Whistler, J. L. (2001) Neuron 32 829-839 [DOI] [PubMed] [Google Scholar]
- 57.He, L., Fong, J., von Zastrow, M., and Whistler, J. L. (2002) Cell 108 271-282 [DOI] [PubMed] [Google Scholar]
- 58.Rothstein, J. D., Patel, S., Regan, M. R., Haenggeli, C., Huang, Y. H., Bergles, D. E., Jin, L., Dykes, H. M., Vidensky, S., Chung, D. S., Toan, S. V., Bruijn, L. I., Su, Z. Z., Gupta, P., and Fisher, P. B. (2005) Nature 433 73-77 [DOI] [PubMed] [Google Scholar]
- 59.Mao, J. (2002) Pain 100 213-217 [DOI] [PubMed] [Google Scholar]
- 60.Moss, A., Blackburn-Munro, G., Garry, E. M., Blakemore, J. A., Dickinson, T., Rosie, R., Mitchell, R., and Fleetwood-Walker, S. M. (2002) J. Neurosci. 22 1363-1372 [DOI] [PMC free article] [PubMed] [Google Scholar]