Abstract
Lack of functional hereditary hemochromatosis protein, HFE, causes iron overload predominantly in hepatocytes, the major site of HFE expression in the liver. In this study, we investigated the role of HFE in the regulation of both transferrin-bound iron (TBI) and non-transferrin-bound iron (NTBI) uptake in HepG2 cells, a human hepatoma cell line. Expression of HFE decreased both TBI and NTBI uptake. It also resulted in a decrease in the protein levels of Zip14 with no evident change in the mRNA level of Zip14. Zip14 (Slc39a14) is a metal transporter that mediates NTBI into cells (Liuzzi, J. P., Aydemir, F., Nam, H., Knutson, M. D., and Cousins, R. J. (2006) Proc. Natl. Acad. Sci. U. S. A. 103, 13612–13617). Knockdown of Zip14 with siRNA abolished the effect of HFE on NTBI uptake. To determine if HFE had a similar effect on Zip14 in another cell line, HeLa cells expressing HFE under the tetracycline-repressible promoter were transfected with Zip14. As in HepG2 cells, HFE expression inhibited NTBI uptake by ∼50% and decreased Zip14 protein levels. Further analysis of protein turnover indicated that the half-life of Zip14 is lower in cells that express HFE. These results suggest that HFE decreases the stability of Zip14 and therefore reduces the iron loading in HepG2 cells.
Iron absorbed from the intestine is transported directly to the liver, a key organ involved in iron homeostasis. Hepatocytes, making up about 80% of the liver in volume, play an important role in maintaining iron homeostasis and iron sensing. They take up both transferrin-bound iron (TBI)2 and non-transferrin-bound iron (NTBI). NTBI uptake requires both reduction by ferric reductase and transport by a ferrous transporter. Steap3 is the candidate reductase in the liver (1, 2), and DMT1 (divalent metal transporter 1) and Zip14 (Zrt- and Irt-like protein 14) are candidate transporters. DMT1 was the first iron transporter identified and is ubiquitously expressed (3, 4). Zip14, a member of the SLC39A metal ion transporter family, initially identified as zinc transporter (5, 6), was recently reported to be abundantly expressed in hepatocytes and involved in NTBI uptake (7). Patients with hereditary hemochromatosis have significant levels of NTBI in their serum (8, 9).
Mutation of a single base pair in the hereditary hemochromatosis gene (HFE) causes iron overload in the liver, heart, pancreas, and parathyroid and pituitary glands, leading to multiorgan dysfunction (10, 11). This mutation results in a substitution of tyrosine for cysteine in the hereditary hemochromatosis protein, HFE, which disrupts a disulfide bond required for proper folding, preventing it from binding to β2-microglobulin and trafficking to the cell surface (12). The importance of functional HFE in iron metabolism is also supported by the evidence that hepatocytes from Hfe-/- knock-out mice can take up more NTBI (9) and have an 8-fold higher iron accumulation in the liver than wild-type mice (13).
Several mechanisms have been proposed by which HFE regulates iron metabolism. HFE competes with transferrin (Tf) for binding to TfR1 (transferrin receptor 1), lowering iron uptake into cells (14–16). Alternately, Tf binding to TfR1 releases HFE to bind to TfR2 in hepatocytes to increase hepcidin transcription (17). Hepcidin is a hormone secreted by the liver that negatively regulates dietary iron uptake by the intestine. HFE can also lower NTBI uptake in isolated primary mouse hepatocytes (9) and in Chinese hamster ovary cells lacking endogenous TfR1 (18). Thus, the mechanism by which HFE regulates iron metabolism still remains elusive.
In the present study, the regulation of both TBI and NTBI uptake by HFE was studied in HepG2 cells, a human hepatoma cell line. We found that expression of HFE decreased both TBI and NTBI uptake. Expression of HFE resulted in a decrease in the protein level of Zip14 with no evident change in the level of Zip14 mRNA. Knockdown of Zip14 with siRNA abolished the effect of HFE on NTBI uptake. HeLa cells had no detectable Zip14 protein. When HeLa cells expressing HFE were transfected with Zip14, NTBI uptake was decreased by comparison with the corresponding controls. HFE was also found to reduce NTBI uptake by ∼50% and to decrease Zip14 protein level. These results suggest that HFE controls the stability of Zip14, which consequently influences the iron loading of hepatocytes.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection—HepG2 cells were maintained in minimum essential medium Eagle (Invitrogen) supplemented with 1.0 mm sodium pyruvate, 0.1 mm nonessential amino acids (Invitrogen), and 10% fetal bovine serum. Cells (2 × 106) were transfected with 5 μg of pcDNA3.1/HFE-FLAG (HepG2/HFE) or pcDNA3.1 (HepG2/Con) using Nucleofector kit V (Amaxa Biosystems, Gaithersburg, MD) according to the manufacturer's instructions and plated into a 78-cm2 dish. Stable cell clones were obtained by selection with 800 μg/ml G418.
The HeLa/tTA-HFE-FLAG cell line, expressing FLAG epitope-tagged HFE under control of the tetracycline-responsive promoter (19), was maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 400 μg/ml G418, and 300 ng/ml puromycin with or without 2 μg/ml doxycyline (dox). HeLa/tTA-HFE-FLAG cells (70–80% confluent) in 78-cm2 dishes were transiently transfected using 100 μl of Lipofectamine (Invitrogen) and 10 μg of pCMV/Zip14-myc encoding mouse Zip14 tagged with the Myc epitope at the C terminus. Two days later, cells were split into a 12-well plate in the presence or absence of 2 μg/μl dox for 24 h.
Iron Treatment of Cells—Fe-NTA was freshly prepared by mixing 500 mm FeCl3 and 530 mm NTA at a volume ratio of 1:3.77 (18). HepG2 cells and HepG2/HFE cells were seeded in a 6-well plate at a concentration of 2 × 105 cells/well. After 2 days, cells were treated with 100 μm Fe-NTA for 24 h and harvested.
55Fe-NTA Uptake—Cells grown in a 6-well plate were washed twice and equilibrated in DMEM supplemented with 20 mm Hepes (pH 7.4) at 37 °C, 5% CO2 for 15 min. The medium was replaced with 1 ml of 100 nm 55Fe-NTA in DMEM supplemented with 20 mm Hepes (pH 7.4) and 2 mg/ml ovalbumin. After 5, 15, 30, or 60 min at 37 °C, 5% CO2, cells were washed twice with 2 ml of 5 mm EDTA, phosphate-buffered saline at 4 °C and solubilized with 1 ml of solubilization detergent (0.1% Triton X-100, 0.1% NaOH). Lysates were mixed with 6 ml of UniverSol scintillation fluid (CN Chemical Co., Costa Mesa, CA), and the radioactivity was counted for 10 min in a scintillation counter (20, 21).
55Fe-Tf Uptake—55Fe-Tf uptake was conducted as previously described (21). Briefly, cells grown in a 6-well plate were washed twice and equilibrated in DMEM with 20 mm Hepes (pH 7.4) for 15 min at 37 °C, 5% CO2. Wash medium was replaced with 1 ml of uptake medium containing 100 nm 55Fe-Tf in DMEM supplemented with 20 mm Hepes (pH 7.4) and 2 mg/ml ovalbumin, pH 7.4. After 1 h, cells were placed on ice, and externally bound 55Fe-Tf was stripped with an acidic buffer (0.2 n acetic acid, 500 mm NaCl, 1 mm FeCl3) for 3 min. Cells were solubilized in 1 ml of solubilization detergent (0.1% Triton X-100, 0.1% NaOH). Lysates were mixed with 6 ml of UniverSol scintillation fluid, and the radioactivity was counted for 10 min in a scintillation counter (20, 21).
55Fe-NTA Efflux—Cells grown in a 6-well plate were washed twice and equilibrated in DMEM with 20 mm Hepes (pH 7.4) for 15 min at 37 °C, 5% CO2. The medium was replaced with 1 ml of DMEM containing 100 nm 55Fe-NTA, 20 mm Hepes (pH 7.4), and 2 mg/ml ovalbumin. After 3 h, cells were immediately washed twice at room temperature with 2 ml of 5 mm EDTA in phosphate-buffered saline (pH 7.4) to remove nonspecific 55Fe from the cell surface, and 1 ml of efflux medium (DMEM, 20 mm Hepes (pH 7.4), 1 mm Fe-NTA) was added to the cells, and they were incubated at 37 °C for 0, 5, 15, 30, or 60 min. The plates were quickly washed twice with 2 ml of 5 mm EDTA in phosphate-buffered saline to remove any extracellular 55Fe, and the cells were solubilized in 1 ml of solubilization detergent (0.1% Triton X-100, 0.1% NaOH). Lysates were mixed with 6 ml of UniverSol scintillation fluid, and the radioactivity was counted for 10 min in a scintillation counter (20, 21).
Real Time Quantitative Reverse Transcription (qRT)-PCR—Total RNA was isolated from cells using the RNeasy RNA isolation kit (Qiagen, Valencia, CA) and treated with DNase (Roche Applied Science) to remove any contaminating genomic DNA. cDNA was synthesized using oligo(dT) primers and Superscript II reverse transcriptase according to the manufacturer's instructions. Primers specific for human HFE, TfR1, DMT1 (for both iron-responsive element and non-iron-responsive element DMT1 forms), Zip14, and GAPDH were designed using the Primer Express software package (PE Biosystems, Foster City, CA) (20). The primer sequences are listed in Table 1. The qRT-PCR was carried out in triplicate for each sample in at least three independent experiments using a SYBR Green detection system on an ABI PRISM 7900 machine (Applied Biosystems, Foster City, CA) (20, 22). The reaction volume was 15 μl. Forty cycles of PCR amplification were denatured at 95 °C for 15 s, were annealed at 55 °C for 30 s, and were extended at 72 °C for 30 s. PCR products were detected by measuring the increase of fluorescence from the binding of SYBR Green to double-stranded DNA. Melting curve experiments previously established that the fluorescent signal for each amplicon was derived from the products only and not from primer dimers. All primer sets used in these studies were validated against the reference primers (GAPDH) to ensure that they amplified equally across the range of template concentrations.
TABLE 1.
List of primers used for qRT-PCR
| Gene | Forward primer | Reverse primer |
|---|---|---|
| GAPDH (868F/968R)a | 5′-ACCCACTCCTCACCTTTGA-3′ | 5′-CTGTTGCTGTACCAAATTCGT-3′ |
| TfR1 (305F/435R) | 5′-CAGGAACCGATCTCCAGTGA-3′ | 5′-CTTGATGGTGCGGTGAAGT-3′ |
| DMT1b (1415F/1555R) | 5′-ATGGACTAGGTGGCGGATT-3′ | 5′-GATAAGCCACGTGACCACA-3′ |
| Zip14 (1381F/1484R) | 5′-GTCTGGCCTTTGGCATCCT-3′ | 5′-AGGGAACATATCAGCCAGAGAAAT-3′ |
F indicates forward; R indicates reverse
DMT1 primers can amplify both iron-responsive element and non-iron-responsive element forms of DMT1 cDNA
Knockdown of Zip14 Using siRNA—Lipofectamine RNAiMAX transfection reagent (Invitrogen) was used to transfect siRNA specific for human Zip14 (Dharmacon, Lafayette, CO) or negative control siRNA into cells at a final concentration of 10 nm following the manufacturer's instructions (7, 23). Briefly, 2 μl of Lipofectamine RNAiMAX and 12 pmol of RNAi duplex were mixed in 200 μl of Opti-MEM medium and added into each well of a 12-well plate. After incubation at room temperature for 15 min, ∼2 × 105 cells in 1 ml of minimum essential medium Eagle supplemented with 1.0 mm sodium pyruvate, 0.1 mm nonessential amino acids, and 10% fetal bovine serum were added to each well. Three days later, Zip14 mRNA levels were detected using qRT-PCR, and protein levels were detected using immunoblots to determine the efficiency of knockdown.
Immunoblot—Cells were washed with cold phosphate-buffered saline twice and lysed on ice in NET-Triton buffer (150 mm NaCl, 5 mm EDTA, 10 mm Tris, 1% Triton X-100, pH 7.4) with 1× Complete Mini Protease Inhibitor Mixture (Roche Applied Science) and 1 mm phenylmethylsulfonyl fluoride. The cell lysate was centrifuged at 16,000 × g for 5 min, and the supernatant was kept. Protein concentrations of the cell extracts were measured using the BCA Protein Assay (Pierce). The cell extracts were reduced and denatured with Laemmli buffer (24) for 5 min at 95 °C and subjected to SDS-PAGE on 12% gels. Protein was transferred to nitrocellulose. Immunoblot analysis was carried out using rabbit anti-ferritin (1:4,000; DAKO, Carpinteria, CA), M2 anti-FLAG (1:10,000; Sigma), mouse anti-Myc (1:5,000; Invitrogen), rabbit anti-Zip14 (1:2,000), and mouse anti-actin (1:10,000; Sigma) followed by goat anti-rabbit or anti-mouse secondary antibodies conjugated to horseradish peroxidase (1:10,000; Chemicon, Temecula, CA). Bands were detected by enhanced chemiluminescence (SuperSignal West-Pico; Pierce). To quantify the amount of Zip14 and actin on the blot, Alexa 680 goat anti-rabbit (1:5,000; Molecular Probes, Carlsbad, CA) and IRDye 800 donkey anti-mouse (1:5,000; Rockland Immunochemicals, Gilbertsville, PA) fluorescent secondary antibodies were used, respectively. The intensity of the band was quantified by fluorescence imaging (Odyssey Infrared Imaging System; Li-Cor, Lincoln, NE).
RESULTS
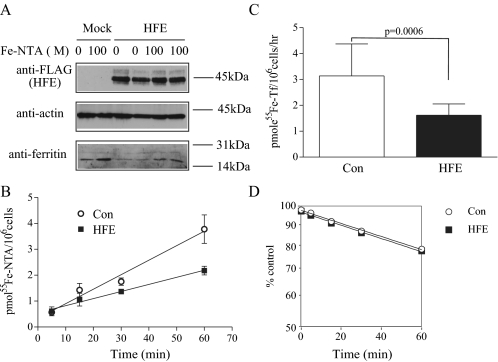
HFE Decreases Intracellular Iron Uptake in HepG2 Cells—HepG2 cells were used to investigate the role of HFE in iron uptake. Intracellular iron status was assessed initially by measuring levels of ferritin, an iron storage protein. When the intracellular iron concentration is low, iron-regulatory proteins 1 and 2 (IRP1 and -2) bind to a conserved iron-responsive element located in the 5′-untranslated regions of ferritin mRNAs to inhibit its translation. Conversely, increased iron concentration leads to decreased binding of IRP1 and -2 to the iron-responsive element, resulting in increased ferritin synthesis (25–27). We found that HFE expression in HepG2 cells decreased ferritin levels (Fig. 1A), indicating lower cellular iron concentrations. Reduced ferritin levels may result from a decrease in iron uptake and/or an increase in iron export. Iron uptake into cells is controlled by Tf and non-Tf-mediated pathways. Cells loaded with Fe-NTA (NTBI) showed increased ferritin levels, and cells expressing HFE showed decreased iron loading both with and without Fe-NTA treatment (Fig. 1A). To test how much NTBI uptake was inhibited by HFE expression, a direct assay of iron uptake using 55Fe-NTA was employed. HFE expression inhibited NTBI uptake by about 42% in HepG2 cells after 60 min (Fig. 1B). Furthermore, HFE expression decreased the TBI uptake through Tf-mediated iron transport pathway by ∼48% in HepG2 cells (Fig. 1C), indicating that the inhibition of cellular iron loading by HFE could be due to a decrease of both TBI and NTBI uptake. Iron efflux measurements were made to examine the possibility that the lower basal intracellular iron levels in HFE-HepG2 cells could result from an increase in iron export. HFE had no effect on iron export in HepG2 cells (Fig. 1D). These results suggest that HFE decreases intracellular iron through inhibition of NTBI and TBI uptake rather than iron export.
FIGURE 1.
HFE expression results in decreased intracellular iron levels and in decreased iron uptake. A, HFE reduces ferritin level in HepG2 cells. Cell lysates (50 μg) from pcDNA3 or pcDNA3/wtHFE-FLAG stably transfected HepG2 cells (HepG2 cells and HepG2/HFE cells, respectively) were analyzed by immunoblot for HFE, ferritin, and actin levels. After cells were treated with or without 100 μm Fe-NTA for 24 h, chemiluminescence was used to detect ∼45 kDa, ∼42 kDa, and ∼19/21 kDa bands representing HFE, actin, and ferritin, respectively. The results are representative of one of three experiments without significant variation between experiments. HFE expression significantly inhibited the increase of ferritin (the marker of the intracellular Fe2+ level) stimulated by Fe-NTA. B, HFE reduces NTBI uptake in HepG2 cells. NTBI uptake was measured in HepG2 cells and HepG2/HFE cells by treatment with 100 nm 55Fe-NTA for 5, 15, 30, and 60 min in DMEM plus 20 mm Hepes (pH 7.4) supplemented with 2 mg/ml ovalbumin. The cell surface of excess 55Fe-NTA was washed twice with 2 ml of 5 mm EDTA plus phosphate-buffered saline, and the internal 55Fe was counted. The data shown represent one of two independent experiments performed with six replicates per time point with minimal variation between experiments. The rates of 55Fe uptake in pmol/106 cells for this experiment were in control (Con) (circles) and HFE-expressing (squares) HepG2 cells, respectively. The rate of 55Fe uptake was significantly decreased in HFE expressing HepG2 cells (p < 0.05). C, HFE decreases TBI uptake in HepG2 cells. TBI uptake was measured in HepG2 cells and HepG2/HFE cells by treatment with 100 nm 55Fe-Tf for 1 h in DMEM plus 20 mm Hepes (pH 7.4) supplemented with 2 mg/ml ovalbumin. Externally bound 55Fe-Tf was stripped with acidic buffer, and the internal 55Fe was counted. Results were expressed as average ± S.D. in pmol of 55Fe/106 cells/h for six samples per cell line. This graph was representative of one of three independent experiments. HFE expression significantly decreased TBI uptake in HepG2 cells (p < 0.05). D, HFE does not affect iron export in HepG2 cells. Iron export was measured in HepG2 cells and HepG2/HFE cells loaded with 100 nm 55Fe-NTA in DMEM plus 20 mm Hepes (pH 7.4) supplemented with 2 mg/ml ovalbumin for 3 h. Cells were washed with 5 mm EDTA plus phosphate-buffered saline to remove the excess 55Fe-NTA on the cell surface and incubated with 1 mm nonradioactive Fe-NTA in DMEM plus 20 mm Hepes (pH 7.4) for 0, 5, 15, 30, or 60 min. 55Fe radioactivity from the solubilized cells was counted. Iron export was expressed as the percentage of 55Fe in efflux medium to total 55Fe. The results shown are representative of one of three experiments with quadruplicate samples without significant variation between experiments. HFE had no effect on iron export in HepG2 cells.
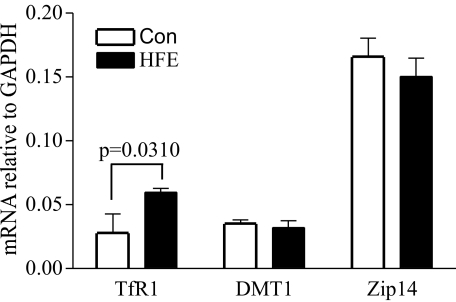
HFE Expression Increases TfR1 but Has No Effect on Zip14 or DMT1 mRNA Levels in HepG2 Cells—TfR1, DMT1, and Zip14 are iron transporters important in iron uptake. TfR1 mediates TBI uptake. DMT1 and Zip14 mediate NTBI uptake. The mRNA levels of TfR1, DMT1, and Zip14 were measured by qRT-PCR to investigate whether the inhibitory effect of HFE on iron uptake in HepG2 cells was caused by the change in expression of these mRNAs. HFE expression increased TfR1 mRNA levels in HepG2 cells (Fig. 2). These results are consistent with increased stability of TfR1 mRNA under low iron conditions (28). Thus, increased TfR1 expression in HFE-expressing HepG2 cells can be attributed to lower intracellular iron (Fig. 1A). DMT1 and Zip14 mRNA levels were not affected by HFE (Fig. 2). Notably, the relative level of Zip14 mRNA was high in HepG2 cells.
FIGURE 2.
HFE expression does not affect DMT1 and Zip14 mRNA levels. Total RNA was isolated from HepG2 cells and HepG2/HFE cells. The expression of each gene was measured by qRT-PCR using primers specific for TfR1, DMT1 (for both iron response element and non-iron-responsive element DMT1 forms), and Zip14. All were normalized to internal GAPDH control (Con). All samples were run in triplicate in three independent experiments. Data are shown as average ± S.D. HFE expression increased TfR1 mRNA abundance but has no effect on mRNA levels of DMT1 and Zip14.
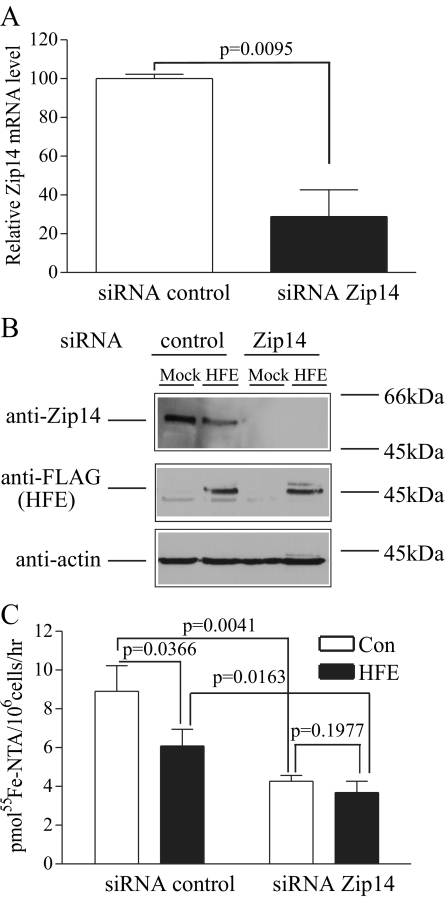
Knockdown of Zip14 Abolishes the Inhibitory Effect of HFE on NTBI Uptake in HepG2 Cells—Since the level of Zip14 mRNA was high in HepG2 cells and HFE expression inhibited NTBI uptake, we used siRNA knockdown of Zip14 in HepG2 cells to determine whether NTBI uptake was mediated by Zip14 and whether the inhibitory effect of HFE on NTBI uptake was mediated through Zip14. Knockdown of Zip14 resulted in a 71% decrease in mRNA level detected by qRT-PCR (Fig. 3A). Immunoblot analysis of cells treated with Zip14 siRNA indicated that the level of Zip14 protein was undetectable in HepG2 cells transfected with Zip14 siRNA compared with control cells (Fig. 3, A and B). In addition, knockdown of Zip14 decreased NTBI uptake by about 52% (Fig. 3C), implying that Zip14 is responsible for approximately half of the NTBI uptake in HepG2 cells.
FIGURE 3.
Knockdown of Zip14 abolishes HFE inhibitory effect on NTBI uptake in HepG2 cells. A, knockdown of endogenous Zip14 mRNA in HepG2 cells using specific siRNA to Zip14. HepG2 cells were transfected with specific human Zip14 siRNA or with control siRNA. Total RNA from Zip14 siRNA or control siRNA-transfected HepG2 cells was used to measure Zip14 mRNA expression by qRT-PCR normalized to internal GAPDH control. All samples were run in triplicate in three independent experiments. Data are shown as average ± S.D. Human Zip14 siRNA significantly reduces endogenous Zip14 mRNA levels in HepG2 cells. B, knockdown of endogenous Zip14 protein in HepG2 cells and HepG2/HFE cells using specific siRNA to Zip14. HepG2 cells and HepG2/HFE cells were transfected with specific human Zip14 siRNA or with control siRNA. Cell lysates (50 μg) from Zip14 siRNA or control siRNA-transfected HepG2 cells and HepG2/HFE cells were analyzed by immunoblot for Zip14 and HFE expression normalized to actin control. Bands at ∼55, ∼45, and ∼42 kDa represent Zip14, HFE, and actin, respectively. These results were representative of one of three experiments without significant variation between experiments. Human Zip14 siRNA significantly inhibited endogenous Zip14 protein level in both HepG2 cells and HepG2/HFE cells. Also, HFE expression decreased endogenous Zip14 protein. C, knockdown of Zip14 abolishes HFE inhibitory effect on NTBI uptake in HepG2 cells. NTBI uptake was measured in HepG2 cells and HepG2/HFE cells transfected with specific human Zip14 siRNA or control (Con) siRNA. Cells were treated with 100 nm 55Fe-NTA in DMEM, supplemented with 20 mm Hepes (pH 7.4) and 2 mg/ml ovalbumin for 1 h. Cells were washed twice with 1 ml of 5 mm EDTA plus phosphate-buffered saline to strip the cell surface of excess 55Fe-NTA, and the internal 55Fe was counted. Results were expressed as average ± S.D. in pmol of 55Fe/106 cells/h for six samples per cell line. The data shown represent one of three independent experiments with minimal variation between experiments. HFE expression inhibited NTBI uptake in HepG2 cells, and this inhibition was abolished by knockdown of Zip14 expression.
HFE expression decreased endogenous Zip14 protein levels (Fig. 3B) and inhibited NTBI uptake in HepG2 cells. This inhibition of HFE on NTBI uptake was abolished by Zip14 knockdown (Fig. 3C). These results suggest that in HepG2 cells, the inhibitory effect of HFE on NTBI uptake is mainly mediated by the down-regulation of endogenous Zip14 protein.
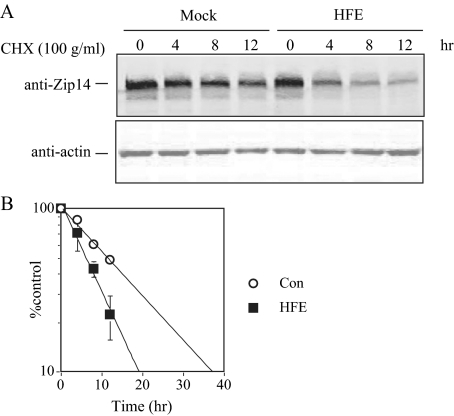
Expression of HFE Promotes Zip14 Degradation in HepG2 Cells—The observation that HFE expression reduced Zip14 protein while not changing the level of Zip14 mRNA suggested that HFE might affect the stability of the Zip14 protein. We measured Zip14 half-life by immunoblot using cycloheximide to inhibit protein synthesis in control and HFE-expressing HepG2 cells. The half-life of Zip14 decreased from 11.0 h in control cells to 7.5 h in HFE-expressing HepG2 cells (Fig. 4, A and B), indicating that HFE expression promotes Zip14 degradation in HepG2 cells.
FIGURE 4.
HFE expression promotes Zip14 degradation in HepG2 cells. A, HFE promotes Zip14 degradation in HepG2 cells. HepG2 cells and HepG2/HFE cells were treated with 100 μg/ml cycloheximide (CHX) for 0, 4, 8, and 12 h. Cell lysates were analyzed by immunoblot for Zip14 normalized to internal actin control using fluorescent secondary antibodies. The bands detected at ∼55 and ∼42 kDa represented Zip14 and actin, respectively. These results are representative of one of four experiments without significant variation between experiments. Endogenous Zip14 was degraded over time, and Zip14 degradation was promoted by HFE expression. B, quantitation of Zip14 degradation in HepG2 cells and HepG2/HFE cells. The amount of Zip14 was expressed as the ratio of the fluorescence intensity of Zip14 to actin. Zip14 expression in HepG2 cells and HepG2/HFE cells treated with cycloheximide for 0 h was set as 100%. Half-life was determined by linear regression analysis. Data are shown as average ± S.D. t½ was decreased in HFE HepG2 cells. Con, control.
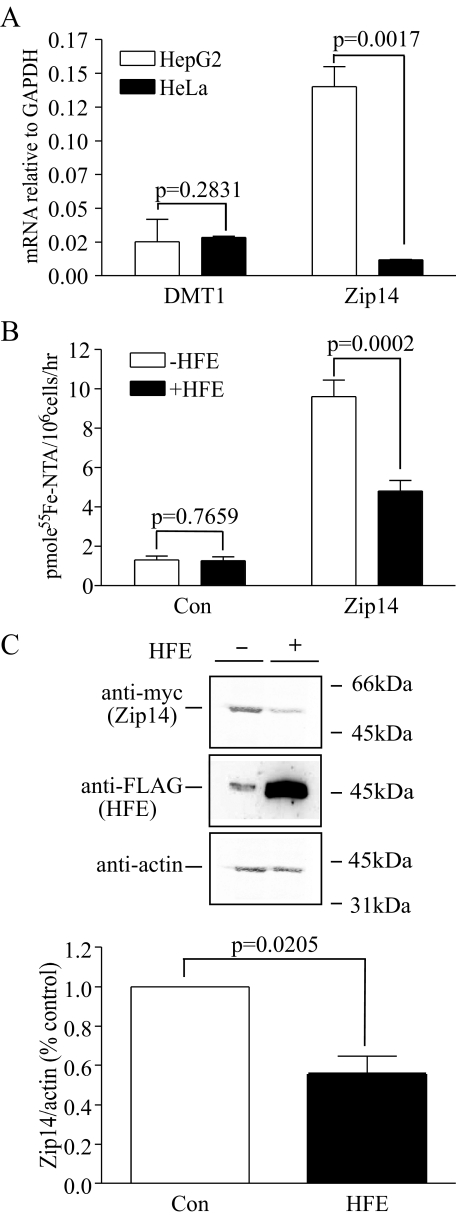
HFE Inhibits Zip14-mediated NTBI Uptake in HeLa Cells Transfected with Zip14—Since HFE has no detectable effect on NTBI uptake in HeLa cells and HT29 cells (20, 21), we wanted to test whether transfection of HeLa cells expressing HFE with Zip14 would alter NTBI uptake. First, the endogenous mRNA levels of DMT1 and Zip14 were measured in HeLa and HepG2 cells. At the mRNA level, HeLa cells expressed approximately the same amount of DMT1 as HepG2 cells but less Zip14 than HepG2 cells normalized to the GAPDH of each cell line (Fig. 5A). No Zip14 protein could be detected in HeLa cells by immunoblot (data not shown). The difference in the endogenous expression of Zip14 could be the reason why HFE inhibited NTBI uptake in HepG2 cells but had no effect in HeLa cells. Low levels of endogenous Zip14 expression makes HeLa cells a perfect tool to investigate the effect of HFE on exogenous Zip14-mediated NTBI uptake by transfection with Zip14. In HeLa/tTA-HFE-FLAG cells expressing FLAG epitope-tagged HFE under control of the tetracycline responsive promoter, induction of HFE expression had no effect on NTBI uptake (Fig. 5B), consistent with previous observations (21). Zip14 expression significantly increased NTBI uptake in HeLa cells. The increase was partially inhibited by induction of HFE expression (Fig. 5B). Induction of HFE expression also significantly decreased Zip14 protein level in HeLa/tTA-HFE-FLAG cells (Fig. 5C). Thus, in HeLa cells expressing Zip14, Zip14 increases NTBI uptake, and HFE expression inhibits Zip14-mediated NTBI uptake through the down-regulation of Zip14 at the protein level.
FIGURE 5.
HFE decreases Zip14 mediated-NTBI uptake in HeLa/tTA-HFE-FLAG cells by down-regulation of Zip14. A, HeLa cells express less Zip14 than HepG2 cells. Total RNA was isolated from HepG2 and HeLa cells. DMT1 and Zip14 expression was measured by qRT-PCR and normalized to internal GAPDH control. All samples were run in triplicate in three independent experiments. Data are shown as average ± S.D. HeLa cells expressed 11 times less Zip14 than HepG2 cells, whereas HeLa and HepG2 cells expressed similar amounts of DMT1. B, HFE decreases Zip14-mediated NTBI uptake in HeLa/tTA-HFE-FLAG cells. The HeLa/tTA-HFE-FLAG cells express FLAG epitope-tagged HFE under control of the tetracycline-repressible promoter. Withdrawal of dox induced HFE expression. NTBI uptake was measured in pCMV/Zip14-Myc-transfected or untransfected HeLa/tTA-HFE-FLAG cells with or without dox. Cells were treated with 100 nm 55Fe-NTA in DMEM, supplemented with 20 mm Hepes (pH 7.4) and 2 mg/ml ovalbumin for 1 h. Cells were washed with 2 ml of 5 mm EDTA plus phosphate-buffered saline, and the internal 55Fe was counted. Results are expressed as average ± S.D. in pmol of 55Fe/106 cells/h for six samples per cell line. The data shown represent one of three independent experiments with minimal variation between experiments. HFE expression through withdrawal of dox had no effect on NTBI uptake in HeLa/tTA-HFE-FLAG cells. Zip14 expression significantly increased NTBI uptake in HeLa cells, and this increase was inhibited by induction of HFE expression. C, induction of HFE expression decreases the level of Zip14 in HeLa/tTA-HFE-FLAG cells. Cell lysates (50 μg) from pCMV/Zip14-Myc-transfected HeLa/tTA-HFE-FLAG cells in the presence or absence of dox were analyzed by immunoblot for Zip14 and HFE. Fluorescent secondary antibodies detected ∼55 and ∼45 kDa bands that represent Zip14 and HFE-FLAG, respectively. Actin was used as a loading control (Con). These results are representative of one of four experiments without significant variation between experiments. Induction of HFE expression through withdrawal of dox significantly decreased Zip14 protein. Zip14 levels decreased by an average of 45%.
DISCUSSION
HFE appears to function at several different levels in the liver. Genetic evidence shows that it plays a role in the regulation of hepcidin in hepatocytes (29, 30). In the latter study, the authors speculated that HFE might increase the transcription of hepcidin through its interaction with TfR2. Earlier studies on the function of HFE demonstrated that HFE also decreases iron uptake in some cell types and lowers iron efflux in other cell types (reviewed in Ref. 31). HepG2 cells possess many of the key features of hepatocytes, including the ability to polarize and secrete hepatocyte-specific proteins, such as albumin, transferrin, and hepcidin (32). Because of their similarity to hepatocytes, we used HepG2 cells to observe the effect of HFE on cellular iron homeostasis. Stable transfection of HFE in HepG2 cells decreased ferritin levels and lowered levels of iron uptake. Notably, HFE expression not only reduced TBI uptake but also NTBI uptake, which is different from the observation in HeLa cells, where HFE reduced only TBI uptake, leaving NTBI uptake unaffected (21). DMT1 and Zip14 are iron transporters for NTBI (3, 7, 33). Expression of HFE in HepG2 cells resulted in a lower amount of Zip14 protein. This was partially due to a decreased stability of Zip14 rather than changes in the mRNA levels. The insensitivity of iron uptake to HFE expression after Zip14 knockdown by siRNA implies that HFE has a direct effect on Zip14-mediated iron transport. These results suggest that in HepG2 cells, Zip14 is involved in NTBI uptake, and the reduction in NTBI uptake by HFE expression may be mediated through Zip14.
Zip14 (SLA39A14) is a member of the SLC39A metal ion transporter family, which was initially characterized as a zinc transporter. A recent study indicated that NTBI uptake increases in HEK 293 cells and Sf9 insect cells transfected with Zip14 and decreases in AML12 mouse hepatocytes when Zip14 is knocked down by siRNA (7). Moreover, Zip14 is expressed at the plasma membrane of hepatocytes (34) and can efficiently transport iron at pH 7.4 (7), the normal pH at the plasma membrane surface of hepatocytes. In contrast, DMT1, the first iron transporter to be identified, transports iron optimally at pH 5.5 (35) and is readily detected in endosomes but not on the plasma membrane (36). These observations suggest that Zip14, rather than DMT1, plays the predominant role in NTBI uptake by hepatocytes.
The variation in phenotypes of patients with the HFE mutation has led to the hypothesis that there are HFE modifiers, including iron importers (37–39). Interestingly, the survival of Slc11a2-/- mice that lack DMT1 is significantly improved when HFE alleles were inactivated. This finding suggests that in Slc11a2-/- mice, lacking HFE might lead to the up-regulation of another iron importer (40). Zip14 expression in duodenum tissue is higher in Hfe-/- mice than that of control mice by microarray analysis (41). In that case, the mRNA levels for Zip14 increase. No such change was noted in the mRNA levels of Zip14 in the liver sample, consistent with our results. Given the findings in the present study, Zip14 is a potential candidate for an HFE modifier involved in TBI and NTBI uptake in HepG2 cells.
To investigate the mechanism by which HFE lowers NTBI in HepG2 cells, we found that knockdown of Zip14 abolished the inhibitory effect of HFE on NTBI uptake. This suggested that the reduction of NTBI uptake by HFE expression was mediated through Zip14 in HepG2 cells. We further observed the effect of HFE on Zip14 expression in HepG2 cells by immunoblot and qRT-PCR. HFE significantly reduces Zip14 protein with no change in the level of Zip14 mRNA. We also found that HFE reduced Zip14 half-life from 11.0 h to 7.5 h in HepG2 cells. These results imply that HFE lowers Zip14-mediated NTBI uptake by decreasing Zip14 stability. In HeLa/tTA-HFE-FLAG cells, inducing HFE expression by withdrawal of dox does not affect NTBI uptake (21). Transfection of this cell line with Zip14 increased NTBI uptake 7-fold over untransfected cells. This increase in NTBI uptake was inhibited by HFE expression. Analysis of Zip14 expression by immunoblot demonstrated that HFE expression reduces Zip14 protein level in HeLa/tTA-HFE-FLAG cells transfected with Zip14. These observations were consistent with the results obtained in HepG2 cells, which endogenously express Zip14.
The evidence that HFE reduced both TBI uptake and NTBI in HepG2 cells raises the question of whether both TBI uptake and NTBI share a common pathway in HepG2 cells (9). HFE reduces NTBI uptake in Chinese hamster ovary cells lacking endogenous TfR1 and TBI uptake in Chinese hamster ovary cells transfected with TfR1 (18). A single divalent iron transporter could explain the shared pathway for TBI and NTBI uptake, which functions in the endosome for TBI uptake and on cell membrane for NTBI uptake. DMT1 overexpression in hepatoma (HLA) cells does not change TfR1-dependent iron uptake (36). These results suggest that Zip14, which can be down-regulated by HFE, mediates TBI and NTBI uptake in HepG2 cells. Future studies on how HFE increases the turnover of Zip14, the localization and the pH dependence of Zip14, and a direct association of HFE and Zip14 will be necessary to test these possibilities.
Acknowledgments
We thank Juxing Chen, Maria Chloupkova, Julia Maxson, and An-Sheng Zhang for helpful suggestions on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants DK072166 (to C. A. E.) and DK065064 (to M. D. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TBI, transferrin-bound iron; NTBI, non-transferrin-bound iron; DMEM, Dulbecco's modified Eagle's medium; dox, doxycycline; HeLa/tTA-, HeLa cells in which expression of the transfected plasmid is controlled by the tetracycline-repressible promoter; Tf, transferrin; siRNA, small interfering RNA; NTA, nitrilotriacetic acid; qRT, quantitative reverse transcription; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
References
- 1.Ohgami, R. S., Campagna, D. R., Greer, E. L., Antiochos, B., McDonald, A., Chen, J., Sharp, J. J., Fujiwara, Y., Barker, J. E., and Fleming, M. D. (2005) Nat. Genet. 37 1264-1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knutson, M. D. (2007) Nutr. Rev. 65 335-340 [DOI] [PubMed] [Google Scholar]
- 3.Gunshin, H., Mackenzie, B., Berger, U. V., Gunshin, Y., Romero, M. F., Boron, W. F., Nussberger, S., Gollan, J. L., and Hediger, M. A. (1997) Nature 388 482-488 [DOI] [PubMed] [Google Scholar]
- 4.Graham, R. M., Chua, A. C., Herbison, C. E., Olynyk, J. K., and Trinder, D. (2007) World J. Gastroenterol. 13 4725-4736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor, K. M., Morgan, H. E., Johnson, A., and Nicholson, R. I. (2005) FEBS Lett. 579 427-432 [DOI] [PubMed] [Google Scholar]
- 6.Eide, D. J. (2004) Pflugers Arch. 447 796-800 [DOI] [PubMed] [Google Scholar]
- 7.Liuzzi, J. P., Aydemir, F., Nam, H., Knutson, M. D., and Cousins, R. J. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 13612-13617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarkar, B. (1970) Can. J. Biochem. 48 1339-1350 [DOI] [PubMed] [Google Scholar]
- 9.Chua, A. C., Olynyk, J. K., Leedman, P. J., and Trinder, D. (2004) Blood 104 1519-1525 [DOI] [PubMed] [Google Scholar]
- 10.Fletcher, L. M., and Halliday, J. W. (2002) J. Intern. Med. 251 181-192 [DOI] [PubMed] [Google Scholar]
- 11.Hentze, M. W., Muckenthaler, M. U., and Andrews, N. C. (2004) Cell 117 285-297 [DOI] [PubMed] [Google Scholar]
- 12.Feder, J. N., Tsuchihashi, Z., Irrinki, A., Lee, V. K., Mapa, F. A., Morikang, E., Prass, C. E., Starnes, S. M., Wolff, R. K., Parkkila, S., Sly, W. S., and Schatzman, R. C. (1997) J. Biol. Chem. 272 14025-14028 [DOI] [PubMed] [Google Scholar]
- 13.Zhou, X. Y., Tomatsu, S., Fleming, R. E., Parkkila, S., Waheed, A., Jiang, J., Fei, Y., Brunt, E. M., Ruddy, D. A., Prass, C. E., Schatzman, R. C., O'Neill, R., Britton, R. S., Bacon, B. R., and Sly, W. S. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 2492-2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lebron, J. A., Bennett, M. J., Vaughn, D. E., Chirino, A. J., Snow, P. M., Mintier, G. A., Feder, J. N., and Bjorkman, P. J. (1998) Cell 93 111-123 [DOI] [PubMed] [Google Scholar]
- 15.Feder, J. N., Penny, D. M., Irrinki, A., Lee, V. K., Lebron, J. A., Watson, N., Tsuchihashi, Z., Sigal, E., Bjorkman, P. J., and Schatzman, R. C. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 1472-1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giannetti, A. M., and Bjorkman, P. J. (2004) J. Biol. Chem. 279 25866-25875 [DOI] [PubMed] [Google Scholar]
- 17.Andrews, N. C., and Schmidt, P. J. (2007) Annu. Rev. Physiol. 69 69-85 [DOI] [PubMed] [Google Scholar]
- 18.Carlson, H., Zhang, A. S., Fleming, W. H., and Enns, C. A. (2005) Blood 105 2564-2570 [DOI] [PubMed] [Google Scholar]
- 19.Gross, C. N., Irrinki, A., Feder, J. N., and Enns, C. A. (1998) J. Biol. Chem. 273 22068-22074 [DOI] [PubMed] [Google Scholar]
- 20.Davies, P. S., and Enns, C. A. (2004) J. Biol. Chem. 279 25085-25092 [DOI] [PubMed] [Google Scholar]
- 21.Roy, C. N., Penny, D. M., Feder, J. N., and Enns, C. A. (1999) J. Biol. Chem. 274 9022-9028 [DOI] [PubMed] [Google Scholar]
- 22.Zhang, A. S., Xiong, S., Tsukamoto, H., and Enns, C. A. (2004) Blood 103 1509-1514 [DOI] [PubMed] [Google Scholar]
- 23.Zhang, A. S., Anderson, S. A., Meyers, K. R., Hernandez, C., Eisenstein, R. S., and Enns, C. A. (2007) J. Biol. Chem. 282 12547-12556 [DOI] [PubMed] [Google Scholar]
- 24.Laemmli, U. K. (1970) Nature 227 680-685 [DOI] [PubMed] [Google Scholar]
- 25.Hentze, M. W., Caughman, S. W., Rouault, T. A., Barriocanal, J. G., Dancis, A., Harford, J. B., and Klausner, R. D. (1987) Science 238 1570-1573 [DOI] [PubMed] [Google Scholar]
- 26.Leibold, E. A., and Munro, H. N. (1988) Proc. Natl. Acad. Sci. U. S. A. 85 2171-2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haile, D. J., Rouault, T. A., Harford, J. B., Kennedy, M. C., Blondin, G. A., Beinert, H., and Klausner, R. D. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 11735-11739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Owen, D., and Kuhn, L. C. (1987) EMBO J. 6 1287-1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt, P. J., Toran, P. T., Giannetti, A. M., Bjorkman, P. J., and Andrews, N. C. (2008) Cell Metab. 7 205-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vujic Spasic, M., Kiss, J., Herrmann, T., Galy, B., Martinache, S., Stolte, J., Grone, H. J., Stremmel, W., Hentze, M. W., and Muckenthaler, M. U. (2008) Cell Metab. 7 173-178 [DOI] [PubMed] [Google Scholar]
- 31.Enns, C. A. (2006) Biol. Res. 39 105-111 [DOI] [PubMed] [Google Scholar]
- 32.Bokhari, M., Carnachan, R. J., Cameron, N. R., and Przyborski, S. A. (2007) J. Anat. 211 567-576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gruenheid, S., Cellier, M., Vidal, S., and Gros, P. (1995) Genomics 25 514-525 [DOI] [PubMed] [Google Scholar]
- 34.Liuzzi, J. P., Lichten, L. A., Rivera, S., Blanchard, R. K., Aydemir, T. B., Knutson, M. D., Ganz, T., and Cousins, R. J. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 6843-6848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garrick, M. D., Kuo, H. C., Vargas, F., Singleton, S., Zhao, L., Smith, J. J., Paradkar, P., Roth, J. A., and Garrick, L. M. (2006) Biochem. J. 398 539-546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shindo, M., Torimoto, Y., Saito, H., Motomura, W., Ikuta, K., Sato, K., Fujimoto, Y., and Kohgo, Y. (2006) Hepatol. Res. 35 152-162 [DOI] [PubMed] [Google Scholar]
- 37.Gouya, L., Muzeau, F., Robreau, A. M., Letteron, P., Couchi, E., Lyoumi, S., Deybach, J. C., Puy, H., Fleming, R., Demant, P., Beaumont, C., and Grandchamp, B. (2007) Gastroenterology 132 679-686 [DOI] [PubMed] [Google Scholar]
- 38.Cruz, E., Melo, G., Lacerda, R., Almeida, S., and Porto, G. (2006) Blood Cells Mol. Dis. 37 33-39 [DOI] [PubMed] [Google Scholar]
- 39.Jacolot, S., Le Gac, G., Scotet, V., Quere, I., Mura, C., and Ferec, C. (2004) Blood 103 2835-2840 [DOI] [PubMed] [Google Scholar]
- 40.Gunshin, H., Fujiwara, Y., Custodio, A. O., Direnzo, C., Robine, S., and Andrews, N. C. (2005) J. Clin. Invest. 115 1258-1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coppin, H., Darnaud, V., Kautz, L., Meynard, D., Aubry, M., Mosser, J., Martinez, M., and Roth, M. P. (2007) Genome Biol. 8 R221.1-R221.16 [DOI] [PMC free article] [PubMed] [Google Scholar]