Abstract
Amyloid fibers are filamentous proteinaceous structures commonly associated with mammalian neurodegenerative diseases. Nucleation is the rate-limiting step of amyloid propagation, and its nature remains poorly understood. Escherichia coli assembles functional amyloid fibers called curli on the cell surface using an evolved biogenesis machine. In vivo, amyloidogenesis of the major curli subunit protein, CsgA, is dependent on the minor curli subunit protein, CsgB. Here, we directly demonstrated that CsgB+ cells efficiently nucleated purified soluble CsgA into amyloid fibers on the cell surface. CsgA contains five imperfect repeating units that fulfill specific roles in directing amyloid formation. Deletion analysis revealed that the N- and C-terminal most repeating units were required for in vivo amyloid formation. We found that CsgA nucleation specificity is encoded by the N- and C-terminal most repeating units using a blend of genetic, biochemical, and electron microscopic analyses. In addition, we found that the C-terminal most repeat was most aggregation-prone and dramatically contributed to CsgA polymerization in vitro. This work defines the elegant molecular signatures of bacterial amyloid nucleation and polymerization, thereby revealing how nature directs amyloid formation to occur at the correct time and location.
Amyloid formation is most readily recognized as the underlying cause of neurodegenerative diseases and prion-based encephalopathies (1). At the root of these diseases is the uncontrolled conversion of soluble proteins into an insoluble fiber known as amyloid. Amyloid fibers are 4–10 nm wide, unbranched filaments possessing a characteristic cross-β sheet structure and specific tinctorial properties when stained with Congo red and thioflavin T (ThT)2 (1). The disease-associated amyloid-forming proteins have little similarity at amino acid level, although the resulting fibers are biochemically and structurally similar (1). Although the pathology of amyloid diseases is incompletely understood, there is growing evidence that soluble folding intermediates are key to cytotoxicity and disease development (2–7).
An increasing number of examples suggest that amyloid fibers, or amyloidogenic intermediates, can be utilized to facilitate a particular physiological task (8). These “functional amyloids” have been found in many organisms, including bacteria, fungi, and mammals (8). The functional amyloids provide a unique perspective on amyloidogenesis since it is assumed that the cell has evolved mechanisms to control and propagate functional amyloid formation so that the cytotoxicity normally associated with amyloid formation is minimized. A compelling example of a functional amyloid is curli, a bacterially produced extracellular fiber that is required for biofilm formation and other community behaviors (9, 10). Curli fibers exhibit the biochemical and structural properties of amyloids (11). Escherichia coli possesses at least six proteins, encoded by the csgBA and csgDEFG (csg, curli-specific genes) operons, that are dedicated to curli biogenesis. This highly regulated assembly machine ensures that the curli subunits interact at the correct time and location.
In E. coli, the polymerization of the major curli fiber subunit protein CsgA into an amyloid fiber depends on the minor curli subunit protein, CsgB (12). The mature CsgA protein has a predicted molecular mass of 13.1 kDa and at least two distinct functional domains, an N-terminal outer membrane secretion domain (13) and an amyloid core domain. The N-terminal domain is 22-amino-acids long (residues 21–42 as shown in Fig. 1A, as the first 20 amino acids are cleaved during translocation by the Sec inner membrane secretion complex) (14). The amyloid core domain (residues 43–151) contains five imperfect repeating units, each 19–23 amino acids (see Fig. 1A). The five repeating units form a protease-resistant structure (14) that is proposed to be the amyloid core of CsgA (15). These repeats are distinguished by the consensus sequence Ser-X5-Gln-X4-Asn-X5-Gln linked by 4 or 5 residues, which is found in all known CsgA homologs from Enterobacteriaceae (15). Each repeat is predicted to form a β strand-loop-strand motif, and five repeated motifs compose cross-β structure (14). In vitro, chemically synthesized oligopeptide repeating unit 1 (R1), R3, and R5 can efficiently assemble into amyloid-like fibers, suggesting that the individual repeats are amyloidogenic (15). It is plausible that these five repeating units are critical for CsgA polymerization into an amyloid fiber in vivo. However, how these repeat sequences direct CsgA polymerization remains elusive.
FIGURE 1.
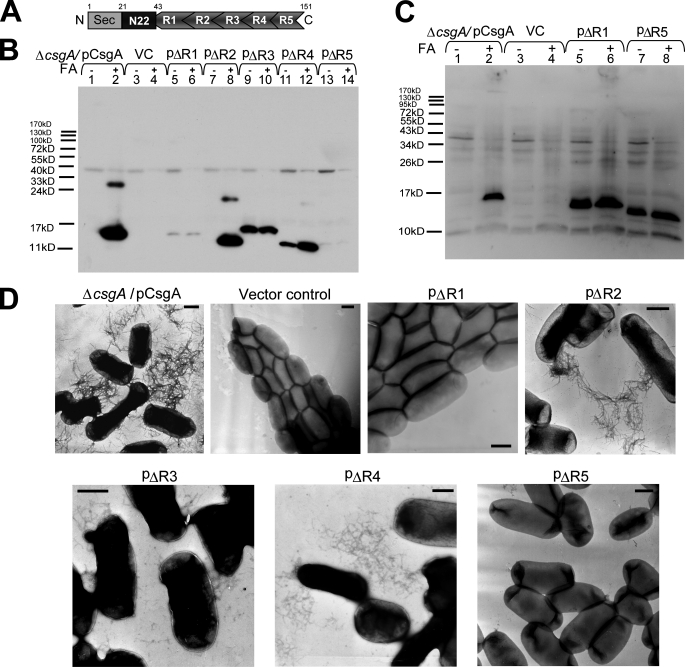
R1 and R5 are critical for CsgA in vivo polymerization into an amyloid fiber. A, schematic of CsgA including an N-terminal Sec signal peptide and the N-terminal 22 residues that precede the five repeating units. N, N terminus; C, C terminus. B, whole-cell Western blots of csgA mutant strains harboring the following plasmids: pCsgA (lanes 1 and 2), vector control (VC) (lanes 3 and 4), pΔR1 (lanes 5 and 6), pΔR2 (lanes 7 and 8), pΔR3 (lanes 9 and 10), pΔR4 (lanes 11 and 12), and pΔR5 (lanes 13 and 14). Samples were treated with (+) or without (-) FA. The blot was probed with anti-CsgA antibody. C, Western blot of whole cells and underlying agar (agar plugs) from csgA strains containing constructs pCsgA (lanes 1 and 2), vector control (lanes 3 and 4), pΔR1 (lanes 5 and 6), and pΔR5 (lanes 7 and 8) grown 48 h at 26 °C on YESCA plates. Samples were treated with (+) or without (-) FA. The blots were probed with anti-CsgA antibody. D, negative-stain EM micrographs of csgA mutant cells containing the indicated plasmids. Cells were grown on YESCA plates for 48 h at 26 °C prior to staining with uranyl acetate. Scale bars are equal to 500 nm.
Most amyloids have the ability to promote their own polymerization in a process called seeding where preformed fibers provide templates for fiber elongation. This process potentially drives prion infectivity and the development of noninfectious amyloid diseases (16–18). Likewise, CsgA polymerization is seeded by preformed CsgA amyloid fibers (15). In vivo, the nucleator protein CsgB initiates the polymerization of CsgA (12). Because CsgB presents an amyloid-like template to CsgA, it was proposed that fiber-mediated self-seeding and CsgB-mediated heteronucleation are mechanistically similar processes (19). As seeding/nucleation underlies the limiting step of amyloid propagation, understanding the nature of this mechanism will shed light on how to control amyloid formation. Here, we use a powerful blend of genetics, biochemistry, and electron microscopy analysis to elucidate the sequence determinants and specificity of curli nucleation and polymerization. Although five repeats of CsgA share high sequence similarity, we found that only N- and C-terminal repeats are indispensable and responsive to CsgB-mediated heteronucleation and CsgA seeding. This work provides unique insights into the in vivo nucleated amyloid polymerization.
EXPERIMENTAL PROCEDURES
Bacterial Growth—To induce curli production, bacteria were grown on YESCA plates (1 g of yeast extract, 10 g of casamino acids, and 20 g of agar/liter) at 26 °C for 48 h (11). When needed, antibiotics were added to plates at the following concentrations: kanamycin 50 μg/ml, chloramphenicol 25 μg/ml, or ampicillin 100 μg/ml.
Strains and Plasmids—Strains and plasmids used in this study are listed in supplemental Table 1. Primer sequences used in this study are listed in supplemental Table 2. Plasmids containing repeat deletions were constructed by site-specific mutagenesis using overlapping PCR extension. The PCR products contained the relevant mutations and NcoI/BamHI sites at the 5′/3′ ends. PCR products were cloned downstream of the csgBA promoter into NcoI/BamHI sites of control vector pLR2 (13). The csgA strain LSR10 (MC4100 ΔcsgA) and expression vector pMC3 were generated previously (11). To express and purify CsgA mutant proteins, PCR-amplified mutant sequences including sequence encoding 6 histidine residues at C terminus were subcloned into pMC3 to replace sequence encoding CsgA-his. The truncated CsgA proteins and the plasmids encoding them were named to reflect the particular deletion. For example, CsgA lacking R1 was named ΔR1, and the plasmid encoding ΔR1 was named pΔR1.
Western Blot Analysis—Whole-cell and plug Western blot analysis of CsgA were described previously (11). Briefly, for whole-cell Western analysis, bacteria were grown on YESCA plates at 26 °C for 48 h. Cells were scraped off the plates and normalized by optical density at 600 nm. Cell pellets (including cell-associated protein aggregates) were resuspended in 2× SDS loading buffer either with or without prior formic acid (FA) treatment. Wild-type curli fibers require brief FA treatment prior to SDS-PAGE to depolymerize CsgA monomers from the fiber (20). For plug Western analysis, cultures were normalized by optical density at 600 nm and spotted on the YESCA plate. After growth at 26 °C for 48 h, 8-mm circular plugs that included cells and the underlying agar (plugs) were collected and resuspended in 2× SDS loading buffer either with or without prior FA treatment. Samples were electrophoresed on a 15% SDS-polyacrylamide gel and blotted onto polyvinylidene difluoride membrane using standard techniques. Western blots were probed by anti-CsgA polyclonal antibody that was raised in rabbits against purified CsgA (Proteintech, Chicago, IL) and was used at a dilution of 1:10,000. The secondary antibody was anti-rabbit antibodies conjugated to horseradish peroxidase (Sigma) and was used at a dilution of 1:7,000. The blots were developed using the Pierce SuperSignal detection system as described previously (19).
Transmission Electron Microscopy—A Philips CM10 transmission electron microscope was used to visualize the curliated cells and protein fiber aggregates. Samples (10 μl) were placed on Formvar-coated copper grids (Ernest F. Fullam, Inc., Latham, NY) for 2 min, washed with deionized water, and negatively stained with 2% uranyl acetate for 90 s.
Purification of Mutant Proteins—C-terminal hexahistidine-tagged CsgA or CsgA mutant proteins were overexpressed along with CsgG by induction with 0.25 mm isopropyl β-d-1-thiogalactopyranoside in LSR12 (C600 ΔcsgBAC and Δcsg-DEFG). In this strain, CsgA or CsgA mutant proteins were secreted to the medium and purified as described previously (15). Cell-free medium containing CsgA or CsgA mutant proteins was passed through a HIS-select™ HF nickel-nitrilotriacetic acid column (Sigma). His-tagged CsgA or CsgA mutant proteins were eluted with 100 mm imidazole in 10 mm potassium phosphate (pH 7.2). Sephadex G25 (balanced in 50 mm potassium phosphate, pH 7.2) was used to remove the imidazole in the CsgA-containing fractions. Immediately after the removal of imidazole, freshly purified proteins in 50 mm potassium phosphate (pH 7.2) were soluble and unstructured as reported previously (15). CsgA and CsgA mutant proteins were purified to homogeneity by this approach as evidenced by single monomer band on SDS-PAGE stained by Coomassie Blue. Plasmids encoding CsgA-His (11) or ΔR2-His can complement a csgA mutant in vivo (data not shown), suggesting that His tag does not disrupt amyloidogenicity of CsgA and its mutants.
In Vitro Polymerization Assay—For ThT assays, freshly purified CsgA or CsgA mutant proteins were passed through a 0.02-μm Anotop 10 filter (Whatman, Maidstone, UK) before being loaded on 96-well opaque plate. ThT was added to a concentration of 20 μm. Fluorescence was measured every 10 min after shaking 5 s by a Spectramax M2 plate reader (Molecular Devices, Sunnyvale, CA) set to 438 nm excitation and 495 nm emission with a 475 nm cutoff. ThT fluorescence was normalized by (Fi - F0)/(Fmax - F0). Fi was the ThT intensity (fluorescence arbitrary unit) of samples, and F0 was the ThT background intensity. For the measurements of polymerization at different concentrations, Fmax was the maximum ThT intensity of samples at the highest concentration. For seeding reactions, Fmax was the maximum ThT intensity of the reaction in the absence of seeds. CsgA polymerization can be described as a triphasic process with a lag, growth, and stationary phase (15). We used the time intervals of lag phase (T0) and fiber growth phase (Tc) to describe the efficiency of polymerization. T0 and Tc were previously used to compare the polymerization of yeast prion Sup35p and its mutants (21). Here, the T0 was obtained by measuring the time interval between the starting point (0 h) and the intersection point of the extrapolation line from the lag phase and extrapolation line of the major portion of the growth phase. The length of the growth phase (Tc) was calculated by subtracting T0 from the time at the beginning of the stationary phase, which includes the major portion of growth phase.
Peptide Preparation—Peptides were chemically synthesized by Proteintech Group Inc., Chicago, IL. The peptides were dissolved in hexafluoroisopropanol (HFIP)/trifluoroacetic acid (1:1 v/v) as described previously (15). Alternatively, peptides were dissolved in 8.0 m guanidine hydrochloride (GdnHCl) buffered by 50 mm potassium phosphate buffer at pH 7.2 to fully denature the sample. After at least 1 h of incubation, GdnHCl was quickly removed by a Sephadex G10 column that was balanced in 50 mm potassium phosphate, pH 7.2, at room temperature. Peptides in potassium phosphate buffer (50 mm, pH 7.2) were passed through a 0.02-μm Anotop 10 filter (Whatman), and their polymerization was measured using ThT fluorescence as described for CsgA and mutant proteins. Similar results were obtained using either denaturing protocol, and the HFIP/trifluoroacetic acid denaturing protocol was used, unless indicated otherwise.
In Vitro Seeding—Mature fibers (2-week-old fibers) were sonicated using a Fisher Model 100 sonic dismembrator (Fisher) for three 15-s bursts on ice to produce seeds. Seeds were added to freshly purified or prepared samples immediately before the start of ThT florescence assay.
Overlay Assay—10 μl of freshly prepared proteins or peptides was dripped on a lawn of the CsgB+ (csgA mutant strain LSR10) and CsgB- (csgAB mutant strain LSR13) cells, which were grown on YESCA plates at 26 °C for 48 h. Samples were incubated on cells for 10 min at room temperature unless otherwise specified, stained with 0.5 mg/ml Congo red solution (50 mm potassium phosphate, pH 7.2) for 5 min, and then washed with potassium phosphate buffer. Cells incubated with protein or peptide solution were collected for transmission electron microscopy (TEM) analysis.
RESULTS
R1 and R5 Are Critical for CsgA Polymerization in Vivo—The amyloid core domain of CsgA (residues 43–151) contains five imperfect repeating units (Fig. 1A). Using a series of csgA alleles that contained precise in-frame deletions, the role of each repeating unit was determined. The ability of csgA alleles to complement a nonamyloidogenic csgA mutant strain was assessed by Western blotting and TEM. A csgA-/pCsgA strain produced curli fibers that were microscopically similar to those assembled by wild-type strain MC4100 as shown by TEM (supplemental Fig. 1). Wild-type curli are SDS-insoluble and require brief treatment with FA to liberate CsgA monomers from the fiber (20). Similarly, the CsgA produced by csgA-/pCsgA was SDS-insoluble and did not migrate into an SDS-PAGE gel without FA pretreatment (Fig. 1B, lanes 1 and 2). A csgA mutant transformed with pΔR2 produced curli fibers that were similar to those produced by csgA-/pCsgA, suggesting that R2 is dispensable for curli formation (Fig. 1B, lanes 7–8, and D, and supplemental Fig. 1). Fibers assembled by csgA-/pΔR3 and csgA-/pΔR4 appeared shorter than wild-type fibers (Fig. 1D and supplemental Fig. 1). Also, ΔR3 and ΔR4 fibers were less SDS-resistant and migrated into an SDS-PAGE gel without FA treatment (Fig. 1B, lanes 9, 10, 11, and 12). Deletion of R1 or R5 resulted in the most defective csgA alleles. Nearly no fibers were produced by csgA-/pΔR1 or csgA-/pΔR5 when analyzed by TEM (Fig. 1D). Furthermore, ΔR1 and ΔR5 proteins were nearly undetectable in whole cell lysates scraped off YESCA plates (Fig. 1B, lanes 5 and 6; lanes 13 and 14). To test the possibility that ΔR1 and ΔR5 were secreted away from the cell as soluble proteins, cells and the underlying agar were collected and analyzed by Western blotting. Both ΔR1 and ΔR5 were readily detected in samples that included the underlying agar, demonstrating that, like wild-type CsgA, ΔR1 and ΔR5 were stable and secreted to the cell surface (Fig. 1C, lanes 5–8). However, unlike CsgA, ΔR1 and ΔR5 were not assembled into an SDS-resistant fiber after secretion (Fig. 1, B and D). We also asked whether ΔR1 and ΔR5 could form fiber aggregates if cells were incubated for 100 h instead of our standard incubation time of 48 h. A small number of fibril aggregates were observed in csgA-/pΔR1 samples incubated for 100 h, although the fibers were rarely detected, and they did not associate with cells (supplemental Fig. 2). In contrast to ΔR1, ΔR5 did not form any detectable fiber aggregates, even after 100 h of incubation (supplemental Fig. 2).
We noted that ΔR1 and ΔR3 migrated more slowly than other mutant proteins on SDS-PAGE, although they are all of similar predicted molecular weights. Purified homogeneous proteins ΔR1 and ΔR3 also migrated more slowly than other repeat deletion mutant proteins, suggesting that the different migration rates resulted from the intrinsic properties of mutant proteins. The nature of these aberrant mobilities was not investigated further in this study.
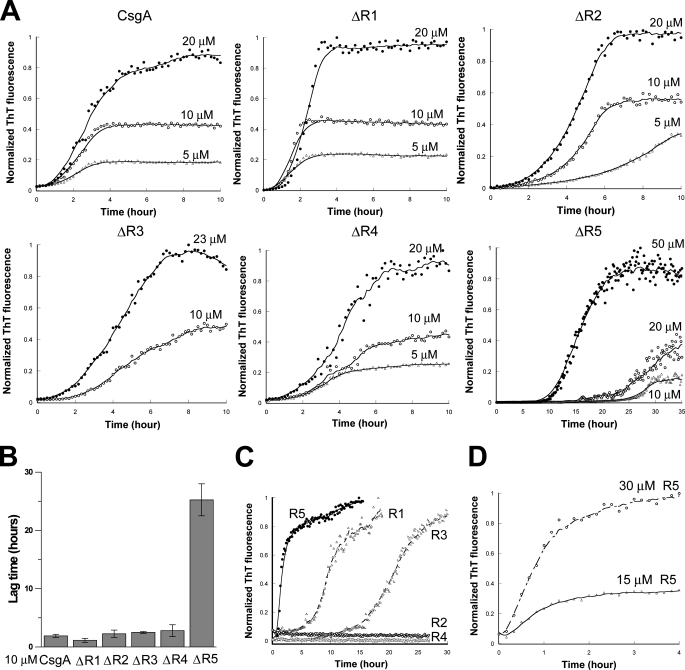
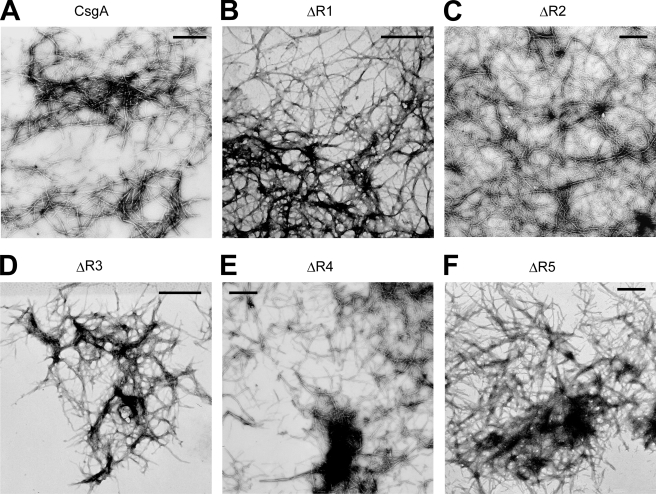
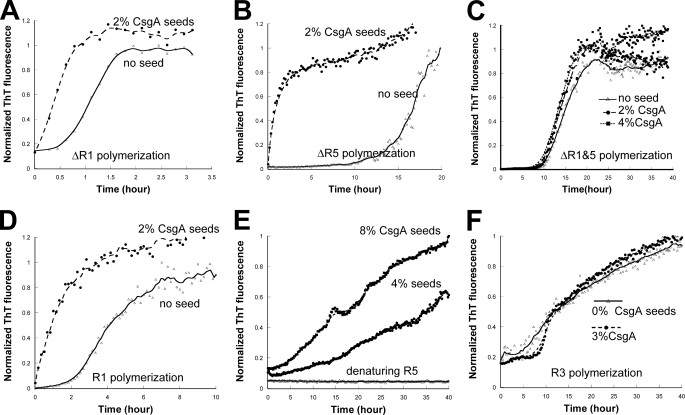
R5 Is Critical for in Vitro Self-polymerization of CsgA—At least two possibilities existed as to why ΔR1 and ΔR5 did not assemble into amyloid fibers in vivo. The first possibility is that ΔR1 and/or ΔR5 were missing CsgB-responsive domains that prevented them from participating in nucleation. Alternatively, ΔR1 and ΔR5 might inefficiently participate in intermolecular interactions with themselves. To test the ability of mutant proteins to self-assemble into an amyloid fiber, we utilized an in vitro polymerization assay that is CsgB-independent (15). ΔR1, ΔR2, ΔR3, ΔR4, and ΔR5 were purified, and their polymerization was compared with wild-type CsgA using a ThT assay described in our previous work (15). In vitro wild-type CsgA polymerization is characterized by three distinct phases: lag, growth, and stationary (15). When comparing the polymerization of CsgA mutant proteins, we utilized two simple parameters that were previously used to describe the polymerization of yeast prion Sup35p (21). The first kinetic parameter was the time period preceding rapid fiber growth, called lag phase, or T0. The second parameter was the time period encompassing fiber growth phase (Tc) (21). Interestingly, when the samples were incubated at room temperature with intermittent shaking, T0 and Tc for ΔR1, ΔR2, ΔR3, and ΔR4 were similar to those of wild-type CsgA (Fig. 2, A and B). However, both T0 and Tc for ΔR5 were dramatically increased (Fig. 2, A and B). TEM analysis confirmed that the ThT-positive aggregates formed by CsgA mutant proteins were ordered fibers (Fig. 3). Furthermore, the T0 values of CsgA, ΔR1, ΔR2, ΔR3, and ΔR4 were concentration-independent above 10 μm, whereas the T0 value for ΔR5 was inversely correlated to concentration up to 200 μm (Fig. 2A and data not shown). When the proteins were incubated without intermittent shaking, ΔR5 polymerization was even more defective relative to wild-type CsgA. At a concentration of 50 μm, T0 and Tc values of ΔR5 polymerization were 2 orders of magnitude greater than those of CsgA (supplemental Fig. 3). ΔR5 incubated at 10 μm without shaking did not assemble into fibers within 800 h (supplemental Fig. 3). These results suggest that R5 plays a critical role in CsgA self-polymerization. Peptides corresponding to each repeating unit were synthesized (15), and their polymerization kinetics was determined by ThT fluorescence (Fig. 2, C and D). The strong denaturing reagents, 8.0 m GdnHCl or HFIP/trifluoroacetic acid (1:1 v/v), were employed to remove possible seeds that would unpredictably influence polymerization kinetics. Chemically synthesized R1, R3, and R5 peptides efficiently assembled into ThT-positive fibers in vitro (15). Like CsgA, R1 and R3 peptides polymerized into ThT-positive fibers in a triphasic fashion that included a discernable lag, growth, and stationary phase (Fig. 2C). At a concentration of 0.6 mg/ml (∼240 μm), T0 values of R1 and R3 were ∼7.5 and 15 h, respectively (Fig. 2C). Unlike R1 and R3, the R5 polymerized with no apparent lag phase, becoming ThT-positive immediately after the removal of the denaturant (Fig. 2C). Even when R5 was incubated at relatively low concentrations (15 μm), there was no detectable lag phase (Fig. 2D). The Tc value of R5 was also shorter than that of R1 and R3 (Fig. 2C). Under the same conditions, R2 and R4 were never observed to polymerize as measured by ThT fluorescence and TEM (Fig. 2C and data not shown). Collectively, these results demonstrate that R5 is the most aggregation-prone of the repeating unit peptides.
FIGURE 2.
In vitro polymerization of CsgA, mutant CsgA proteins, and peptides. A, the normalized fluorescence intensity of freshly purified CsgA and repeat deletion analogues was measured at 495 nm after excitation at 438 nm in the presence of 20 μm ThT. Samples were shaken for 5 s prior to fluorescence measurements in 10-min intervals. The protein used in the assay is indicated at the top of the graph, and the protein concentration is above the relevant data points. The x axis of each graph spans from 0 h to 10 h, except for ΔR5, which spans from 0 h to 35 h. B, lag time (T0) of freshly purified CsgA and CsgA analogues incubated at 10 μm. Data were expressed as the mean ± S.E. of three independent experiments. C, the polymerization of 0.6 mg/ml (∼240 μm) chemically synthesized peptides R1, R2, R3, R4, and R5 was measured by ThT fluorescence. D, the polymerization of 15 and 30 μm R5 peptide was measured by ThT fluorescence.
FIGURE 3.
EM micrographs of in vitro polymerized fibers from CsgA and mutant proteins. A–F, negative-stain EM micrographs of in vitro polymerized fibers of CsgA (A), ΔR1(B), ΔR2 (C), ΔR3 (D), ΔR4 (E), and ΔR5 (F). Scale bars are equal to 500 nm.
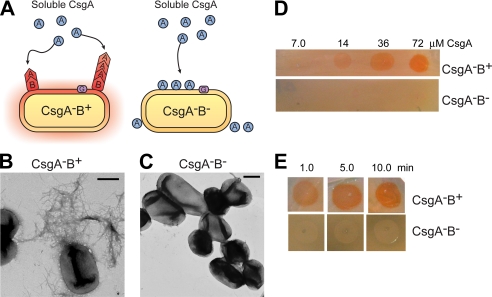
CsgA Efficiently Responds to CsgB Nucleation to Assemble into Amyloid Fibers—Although CsgA can self-assemble into amyloid fibers in vitro, CsgA polymerization in vivo is dependent on the CsgB nucleator. Direct evidence that soluble CsgA can be converted to an amyloid fiber by wild-type CsgB is missing. To test whether the cell surface-exposed CsgB can nucleate the polymerization of purified CsgA, we developed an overlay assay using CsgB+ or CsgB- cells and freshly purified soluble CsgA protein (Fig. 4A). Different concentrations of CsgA were overlaid on CsgB+ (csgA) or CsgB- (csgAB) cells, and CsgA polymerization was detected using the amyloid specific dye Congo red and TEM (Fig. 4, B–E). CsgA overlaid on CsgB+ cells efficiently assembled into amyloid-like fibers as shown by Congo red staining and TEM (Fig. 4, B and D), whereas CsgA overlaid on CsgB- cells did not induce Congo red binding and did not assemble into fibers shown by TEM (Fig. 4, C and D). CsgA fibers formed on CsgB+ cells in the overlay assay were similar to wild-type curli fibers formed by csgA-/pCsgA (compare Figs. 1D and 4B). The CsgB-mediated polymerization of purified CsgA occurred in as little as 6 min (1 min of incubation and 5 min of Congo red staining) (Fig. 4E).
FIGURE 4.
Purified CsgA is efficiently nucleated when overlaid on CsgB-expressing cells. A, a schematic presentation of the overlay assay in which freshly purified and soluble CsgA is dripped onto cells expressing the nucleator protein, CsgB. In the presence of CsgB, CsgA (shown as a circle) undergoes a conformational change and polymerizes into a fiber (shown as a chevron). In the absence of CsgB, CsgA remains soluble and does not assemble into an amyloid fiber. B and C, negative-stain EM micrographs of CsgA-B+ (B) and CsgA-B- (C) cells grown on YESCA plates for 48 h at 26 °C that were overlaid with freshly purified 40 μm CsgA. Scale bars are equal to 500 nm. D, Congo red staining of CsgA-B+ and CsgA-B- cells after being overlaid with different concentrations of soluble CsgA. E, 72 μm CsgA was overlaid on CsgA-B+ and CsgA-B- cells and incubated for the indicated time intervals before staining for 5 min with Congo red solution.
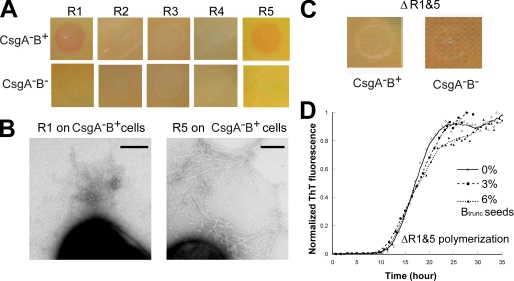
Mapping the CsgB-responsive Sequences in CsgA—The CsgB-responsive repeating units in CsgA were mapped using the overlay assay and peptides corresponding to each repeating unit. 2.0 mg/ml of freshly prepared R1, R2, R3, R4, or R5 peptides was overlaid onto CsgB+ and CsgB- cells. When either R1 or R5 was overlaid on CsgB+ cells, amyloid-like fibers were detected by Congo red binding and TEM (Fig. 5, A and B). When either R1 or R5 was overlaid on CsgB- cells for the same incubation time, no Congo red binding was observed, and no fibers were detected by TEM (Fig. 5A and data not shown). The R1 and R5 fibers formed on CsgB-presenting cells were shorter and thinner than fibers assembled by wild-type CsgA polymerization on the surface of the cell (compare Figs. 1D,4B, and 5B and supplemental Fig. 1). When 2.0 or 4.0 mg/ml R2, R3, and R4 were overlaid on CsgB+ or CsgB- cells, no fibers were formed as evidenced by the lack of Congo red binding and by TEM analysis. This suggested that R2, R3, and R4 were unable to interact with CsgB to promote fiber assembly (Fig. 5A and data not shown). Therefore, we concluded that R1 and R5 were the CsgB heteronucleation-responsive domains of CsgA.
FIGURE 5.
R1 and R5 are responsive to CsgB heteronucleation. A, 2.0 mg/ml of freshly prepared peptide solution was overlaid on CsgA-B+ and CsgA-B- cells. After a 5-min incubation, cells were stained with 0.5 mg/ml Congo red solution for 5 min and washed once by 50 mm potassium phosphate buffer. B, negative stain EM micrographs of CsgA-B+ cells overlaid with freshly prepared R1 (left) and R5 (right). Conditions are the same as in A. Scale bars are equal to 200 nm. C, freshly purified 105 μm ΔR1&5 was overlaid onto CsgA-B+ and CsgA-B- cells. ΔR1&5 was incubated on cells for 10 h before staining with Congo red. D, 3 and 6% CsgBtrunc by weight was added to freshly purified 50 μm ΔR1&5. The polymerization was measured by ThT fluorescence.
To further test the notion that R1 and R5 were the only CsgB-responsive domains of CsgA, a csgA allele without both R1 and R5 (ΔR1&5) was engineered, and its CsgB responsiveness was measured. ΔR1&5 was unable to complement a csgA mutant for curli biogenesis (data not shown). Unlike CsgA, ΔR1&5 did not respond to CsgB nucleation on CsgB+ cells measured with the overlay assay even after extended incubation (Fig. 5C). In vitro, ΔR1&5 polymerized into ThT-positive fibers, with T0 and Tc values comparable with those of ΔR5 (compare Figs. 2A and 5D). Fiber aggregates formed by ΔR1&5 in vitro were short and protofibril-like, similar to fibers assembled by peptide R3 by TEM (data not shown). A nucleation-competent CsgB truncation mutant (CsgBtrunc) was purified as described previously (19). CsgBtrunc has been demonstrated to seed wild-type CsgA (19). Unlike CsgA, the polymerization of ΔR1&5 was not stimulated by CsgBtrunc in vitro (Fig. 5D). Collectively, these results demonstrate that a csgA allele missing R1 and R5 is not responsive to CsgB-mediated heteronucleation in vivo and in vitro.
R1 and R5 Are CsgA Seeding-responsive Domains—CsgA polymerization is a self-propagating process in which CsgA seeds can completely eliminate the lag phase (15). However, the determinants of CsgA seeding specificity are poorly understood. Our results demonstrate that R1 and R5 are CsgB-responsive domains. To test whether R1 and R5 also play an important role in the growth of CsgA fibers, we tested CsgA seeding on ΔR1, ΔR5, and ΔR1&5 in vitro. CsgA seeds eliminated the lag phase of ΔR1 and ΔR5 polymerization (Fig. 6, A and B), whereas ΔR1&5 polymerization was not promoted by CsgA seeding (Fig. 6C). These findings demonstrated that both R1 and R5 are involved in the response to CsgA seeding and that specificity is encoded in R1 and R5 sequences.
FIGURE 6.
R1 and R5 are responsive to CsgA seeding. A–C, indicated percentages of sonicated CsgA fibers were added to freshly prepared 17 μm ΔR1 (A), 50 μm ΔR5 (B), and 50 μm ΔR1&5 (C). The polymerization was measured by ThT fluorescence. D, 2% sonicated CsgA fibers were added to freshly prepared 1.0 mg/ml R1 peptide solution. The polymerization was measured by ThT fluorescence. E, R5 was solubilized in HFIP/trifluoroacetic acid. Upon the removal of HFIP/trifluoroacetic acid, R5 was resuspended in 1.0 m GdnHCl buffered by 50 mm potassium phosphate buffer at pH 7.2. 4 and 8% sonicated CsgA fibers were added to 0.2 mg/ml R5 solution in the presence of 1.0 m GdnHCl and mixed with ThT at 20 μm, and fluorescence was measured. F, the indicated percentages of sonicated CsgA fibers were added to freshly prepared 1.0 mg/ml R3 peptide solution. The polymerization was measured by ThT fluorescence.
In support of the hypothesis that only R1 and R5 are CsgA seeding-responsive domains, each repeating unit peptide was incubated with wild-type CsgA fiber seeds. The lag phase of R1 polymerization was completely eliminated when 2% preformed CsgA fibers were added to the reaction (Fig. 6D). To test the responsiveness of R5 to CsgA seeding, we needed to slow the aggregation of R5 so that a discernable lag phase could be observed. In the presence of 1.0 m GdnHCl, 0.2 mg/ml R5 remains unpolymerized for over 40 h (Fig. 6E). CsgA seeds eliminated the lag phase of R5 even under these mildly denaturing conditions (Fig. 6E). However, CsgA seeds did not accelerate R3 polymerization (Fig. 6F). CsgA seeds were also unable to stimulate the polymerization of R2 and R4 peptides, as both R2 and R4 remained unpolymerized for 48 h detected by ThT assay and TEM (data not shown).
DISCUSSION
Amyloid formation has been associated with neurodegenerative diseases for over a century, yet very little is known about the mechanism of nucleated polymerization in vivo. Curli provide a sophisticated genetic and molecular tool set with which to explore amyloid nucleation and polymerization. We have described the molecular determinants of a highly evolved and elegant amyloid assembly pathway. CsgB-mediated heteronucleation is requisite for in vivo curli fiber assembly. We extensively investigated the CsgA sequence determinants for nucleation and polymerization. We found that the N- and C-terminal repeats (R1 and R5) are indispensable for curli biogenesis in vivo. R5 is the most aggregation-prone repeating unit, and it contributes dramatically to CsgA polymerization in vitro. R1 and R5 play critical roles in CsgA seeding and CsgB heteronucleation response.
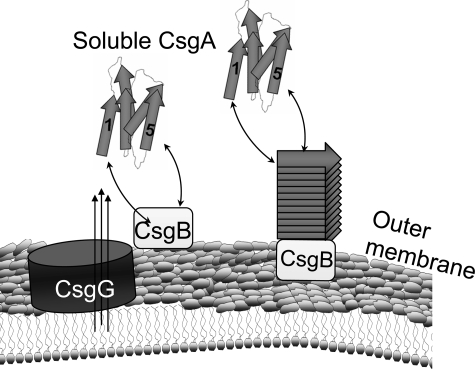
Molecular Details of Curli Assembly—Curli biogenesis occurs via a nucleation-precipitation mechanism, where surface-exposed CsgB nucleates secreted CsgA into an insoluble fiber (12, 19). We found that R1 and R5 are indispensable for in vivo curli assembly (Fig. 1). We have provided direct evidence that surface-exposed CsgB promotes the transition of CsgA from a soluble protein to an insoluble amyloid-like fiber (Fig. 4). Both R1 and R5 of CsgA were demonstrated to respond to CsgB heteronucleation (Fig. 5). Similarly, preformed CsgA fibers (in the absence of CsgB) were able to recruit soluble CsgA into the fiber in a process called seeding (15). Seeding was also dependent on R1 and R5 (Fig. 6). Based on these results, we propose a promiscuous nucleation model to describe the molecular details of curli assembly (Fig. 7). CsgA is not amyloidogenic within or outside the cell until it encounters outer membrane-localized CsgB. This temporal and spatial control might prevent the proteotoxicity of amyloid formation. After CsgA is secreted from cells, either R1 or R5 of CsgA interacts with CsgB or the growing fiber tip, facilitating CsgA polymerization (Fig. 7). Collectively, the curli fiber assembly machinery on the cell surface is composed of multiple nucleation templates (CsgB and CsgA fiber tip) and multiple responsive domains of fiber subunit CsgA (R1 and R5), which makes nucleation polymerization an efficient process in vivo.
FIGURE 7.
The promiscuous nucleation model of CsgA in vivo polymerization. Curli assembly is governed by surface-localized CsgB (heteronucleation) and by the growing fiber tip (homonucleation). After secretion from the periplasm to extracellular space, R1 and R5 of CsgA can interact with CsgB or fiber tips, initiating curli formation.
Microbes assemble many proteinaceous fibers on their cell surface. One challenge to assembling extracellular fibers is prevention of subunit diffusion to the extracellular milieu. Therefore, the assembly of curli, pili, and flagella must be regulated so that efficient subunit incorporation into the fiber occurs. P and type-I pilus assembly is dependent on the chaperone/usher pathway, where periplasmic chaperone proteins deliver the pilus subunits to the outer membrane-localized usher protein. On the periplasmic side of the usher, pilus subunits are added to the base of the growing pilus. Because pilus subunits never leave the periplasmic space before incorporation into the pilus, there is no chance for diffusion of unassembled subunits to the extracellular space (22). Flagella are another common extracellular fiber assembled by Gram-negative bacteria. Soluble flagellin is secreted through the growing fiber until it is bound by the cap protein and polymerized onto the distal end (23). In contrast to flagella and pili assembly, the major curli subunit CsgA is secreted from the cell as a soluble protein, which subsequently polymerizes into a fiber. To ensure that secreted subunits are efficiently incorporated into an amyloid fiber, curli assembly is dependent on the CsgB nucleator and multiple CsgA-responsive domains. Nucleation, the stalwart of curli formation, is also important for disease-associated amyloid formation. Understanding the molecular signatures of nucleation will lead to new insights into disease-associated protein aggregation.
Distinction between in Vivo and in Vitro Bacterial Amyloid Formation—Although CsgA is amyloidogenic in vitro, it remains soluble in the extracellular milieu in the absence of nucleator CsgB in vivo (12). Like a csgB mutant, csgA alleles missing one or both CsgB-responsive domains (R1 and R5) are unable to efficiently polymerize in vivo. For example, ΔR1 is completely defective in curli assembly in vivo, although ΔR1 polymerizes with similar kinetics as wild-type CsgA in vitro (Figs. 1 and 2). ΔR1 was mostly incapable of forming fiber aggregates in vivo, even if cells were incubated for over 100 h instead of the standard 48 h. A small amount of ΔR1 short fibril aggregates not associated with cells were observed by TEM, whereas ΔR5 was never observed to form fiber aggregates by TEM (supplemental Fig. 2). These observations were consistent with the in vitro polymerization results; ΔR1 polymerization was much more efficient than ΔR5 polymerization (Fig. 2, A and B). This suggests that within natural cellular environments, the amyloidogenicity of CsgA is tightly controlled by the CsgB heteronucleator. Therefore, these intermolecular interactions between CsgA and CsgB initiate bacterial amyloid formation at the correct place and time.
Nucleation Specificity of Amyloid Formation—Nucleation/seeding is the rate-limiting step of amyloid formation (24). The mechanism and specificity of nucleation is poorly understood. Nucleation specificity is arguably the best understood in the yeast prion Sup35p. One small region (amino acids 9–39) located in the N-terminal prion domain of Sup35p governs self-recognition and species-specific seeding activity (25). The residues located in this region are critical for prion propagation (21). Here, we demonstrated that bacterial amyloid protein CsgA also contains two small regions (R1 and R5) that control nucleation response. It may be a general rule that only small portions (nucleation sites) of amyloid proteins govern the nucleation response and specificity. The residues contained in the nucleation site determine the nucleation response and specificity. Mutagenesis analyses on Sup35p (21) and CsgA support this notion. The nucleation sites of Sup35p and CsgA are located within the protease-resistant amyloid core of the proteins, and peptides containing the nucleation sites of Sup35p and CsgA are sufficient to form amyloid fibers in vitro (15, 26).
CsgA homologues from different bacteria share high sequence similarity, suggesting cross-species nucleation might occur. Salmonella typhimurium CsgA and CsgB homologues are 74.8 and 82.1% identical to E. coli CsgA and CsgB, respectively. S. typhimurium csgA and csgB genes can complement E. coli csgA and csgB mutations in terms of fiber assembly (27). Because curli subunits are secreted into the extracellular milieu prior to incorporation into the fiber, complex bacterial communities may have members that secrete CsgA-like molecules and others that produce CsgB-like nucleators. It is plausible that nucleation promiscuity facilitates interspecies fiber formation in complex communities, allowing curli to act as a structural signal that links different species together.
Supplementary Material
Acknowledgments
We thank the Hultgren laboratory for supplying us with the plasmid pLR5. We also thank members of the Chapman laboratory and Ryan Frisch for helpful discussions and review of this manuscript. Special thanks to Bryan McGuffie for the technical help preparing the figures.
This work was supported, in whole or in part, by National Institutes of Health Grant AI073847-01. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains three supplemental figures and two supplemental tables.
Footnotes
The abbreviations used are: ThT, thioflavin T; GdnHCl, guanidine hydrochloride; HFIP, hexafluoroisopropanol; FA, formic acid; TEM, transmission electron microscopy.
References
- 1.Chiti, F., and Dobson, C. M. (2006) Annu. Rev. Biochem. 75 333-366 [DOI] [PubMed] [Google Scholar]
- 2.Bucciantini, M., Giannoni, E., Chiti, F., Baroni, F., Formigli, L., Zurdo, J., Taddei, N., Ramponi, G., Dobson, C. M., and Stefani, M. (2002) Nature 416 507-511 [DOI] [PubMed] [Google Scholar]
- 3.Hartley, D. M., Walsh, D. M., Ye, C. P., Diehl, T., Vasquez, S., Vassilev, P. M., Teplow, D. B., and Selkoe, D. J. (1999) J. Neurosci. 19 8876-8884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lambert, M. P., Barlow, A. K., Chromy, B. A., Edwards, C., Freed, R., Liosatos, M., Morgan, T. E., Rozovsky, I., Trommer, B., Viola, K. L., Wals, P., Zhang, C., Finch, C. E., Krafft, G. A., and Klein, W. L. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 6448-6453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lesne, S., Koh, M. T., Kotilinek, L., Kayed, R., Glabe, C. G., Yang, A., Gallagher, M., and Ashe, K. H. (2006) Nature 440 352-357 [DOI] [PubMed] [Google Scholar]
- 6.Roher, A. E., Chaney, M. O., Kuo, Y. M., Webster, S. D., Stine, W. B., Haverkamp, L. J., Woods, A. S., Cotter, R. J., Tuohy, J. M., Krafft, G. A., Bonnell, B. S., and Emmerling, M. R. (1996) J. Biol. Chem. 271 20631-20635 [DOI] [PubMed] [Google Scholar]
- 7.Walsh, D. M., Klyubin, I., Fadeeva, J. V., Cullen, W. K., Anwyl, R., Wolfe, M. S., Rowan, M. J., and Selkoe, D. J. (2002) Nature 416 535-539 [DOI] [PubMed] [Google Scholar]
- 8.Fowler, D. M., Koulov, A. V., Balch, W. E., and Kelly, J. W. (2007) Trends Biochem. Sci. 32 217-224 [DOI] [PubMed] [Google Scholar]
- 9.Barnhart, M. M., and Chapman, M. R. (2006) Annu. Rev. Microbiol. 60 131-147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerstel, U., and Romling, U. (2003) Res. Microbiol. 154 659-667 [DOI] [PubMed] [Google Scholar]
- 11.Chapman, M. R., Robinson, L. S., Pinkner, J. S., Roth, R., Heuser, J., Hammar, M., Normark, S., and Hultgren, S. J. (2002) Science 295 851-855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammar, M., Bian, Z., and Normark, S. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 6562-6566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson, L. S., Ashman, E. M., Hultgren, S. J., and Chapman, M. R. (2006) Mol. Microbiol. 59 870-881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collinson, S. K., Parker, J. M., Hodges, R. S., and Kay, W. W. (1999) J. Mol. Biol. 290 741-756 [DOI] [PubMed] [Google Scholar]
- 15.Wang, X., Smith, D. R., Jones, J. W., and Chapman, M. R. (2007) J. Biol. Chem. 282 3713-3719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harper, J. D., and Lansbury, P. T., Jr. (1997) Annu. Rev. Biochem. 66 385-407 [DOI] [PubMed] [Google Scholar]
- 17.Shorter, J., and Lindquist, S. (2005) Nat. Rev. Genet. 6 435-450 [DOI] [PubMed] [Google Scholar]
- 18.Prusiner, S. B., Scott, M. R., DeArmond, S. J., and Cohen, F. E. (1998) Cell 93 337-348 [DOI] [PubMed] [Google Scholar]
- 19.Hammer, N. D., Schmidt, J. C., and Chapman, M. R. (2007) Proc. Natl. Acad. Sci. U. S. A.
- 20.Collinson, S. K., Emody, L., Muller, K. H., Trust, T. J., and Kay, W. W. (1991) J. Bacteriol. 173 4773-4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DePace, A. H., Santoso, A., Hillner, P., and Weissman, J. S. (1998) Cell 93 1241-1252 [DOI] [PubMed] [Google Scholar]
- 22.Sauer, F. G., Remaut, H., Hultgren, S. J., and Waksman, G. (2004) Biochim. Biophys. Acta 1694 259-267 [DOI] [PubMed] [Google Scholar]
- 23.Yonekura, K., Maki-Yonekura, S., and Namba, K. (2002) Res. Microbiol. 153 191-197 [DOI] [PubMed] [Google Scholar]
- 24.Jarrett, J. T., and Lansbury, P. T., Jr. (1993) Cell 73 1055-1058 [DOI] [PubMed] [Google Scholar]
- 25.Tessier, P. M., and Lindquist, S. (2007) Nature 447 556-561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glover, J. R., Kowal, A. S., Schirmer, E. C., Patino, M. M., Liu, J. J., and Lindquist, S. (1997) Cell 89 811-819 [DOI] [PubMed] [Google Scholar]
- 27.Romling, U., Bian, Z., Hammar, M., Sierralta, W. D., and Normark, S. (1998) J. Bacteriol. 180 722-731 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.