Abstract
In order to investigate the influence of temperature on the GC level of the paired sequences of ribosomal 18S RNAs in vertebrates, we have studied their base composition in cold- and warm-blooded vertebrates using a stem-by-stem comparison. We observed that a number of stems of 18S ribosomal RNAs (rRNAs) are variable among species and that the majority of such stems are GC richer in warm-blooded than in cold-blooded vertebrates. We also constructed the secondary structures of the 18S rRNAs of a polar fish, a marsupial, and a monotreme to compare them with those of temperate/tropical fishes and of eutherians, respectively. In these cases, differences similar to those already mentioned were found. We conclude that there is a correlation between stem stability and body temperature even within the relatively limited temperature range of vertebrates.
Keywords: rRNA, natural selection, warm-blooded vertebrates, cold-blooded vertebrates, nucleotide composition, thermostability
INTRODUCTION
18S ribosomal RNA (rRNA), together with 5.8S RNA and proteins, makes up the small subunit of the eukaryotic ribosome. Its secondary structure plays an important role in protein synthesis, as it interacts with ribosomal proteins, tRNA and mRNA. This role has been proposed as an explanation for the conservation of its sequence and secondary structure. In fact, because of sequence similarity among species, rRNA has been used for a long time in the study of evolutionary relationships (see, for example, Bernardi et al. 1992), although in recent years, several investigations have stressed the need of being careful when using rRNA for this purpose because of differences in evolutionary rate among sites (Abouheif et al. 1998; Telford et al. 2005; Smit et al. 2006, 2007).
The secondary structure of rRNA is characterized by stems, which are stretches of complementary base pairs (both canonical and noncanonical), and loops, junctions, and bulges, which are unpaired regions enclosed by stems. In vertebrates, the base composition of stems is unusual, since they are GC richer than any other “structural category” (Vawter and Brown 1993). GC base pairs make the stems more stable than AU, GU, or noncanonical base pairs. Moreover, GC levels are known to be correlated with the thermodynamic stability of the nucleic acids and of the encoded proteins (Bernardi and Bernardi 1986). This “thermodynamic stability hypothesis,” which was formulated in order to explain regions of elevated GC in vertebrates, was initially supported by the fact that the genomes of warm-blooded differ from those of cold-blooded vertebrates in having GC-rich isochores (for review, see Bernardi 2005) as well as increased CpG frequency and DNA methylation (Jabbari et al. 1997; Jabbari and Bernardi 1998, 2004; Varriale and Bernardi 2006a,b). Furthermore, it has also been shown that even a slight alteration in temperature can strongly influence the potential secondary structure of rRNA (Armbruster 2001). This finding highlights the role of body temperature in influencing both structure and sequence of rRNA, as well as genomic differences among species living at different temperatures.
In this study, our aim was to find whether there is thermal adaptation in vertebrate rRNAs. To do that, we reanalyzed the differences between the rRNA sequences of cold- and warm-blooded vertebrates in more detail, by carrying out stem-by-stem analyses and in some cases base-pair-by-base-pair comparisons, and by also specifically comparing those such as between polar and temperate/tropical fishes, as well as marsupials/monotremes and eutherians.
RESULTS
Analysis of the global GC level of 18S rDNA sequences
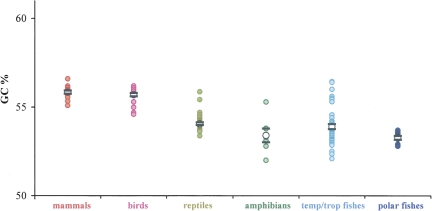
As the starting point, we calculated the global GC level of 18S rDNA sequences (both complete and partial) from species belonging to the five classes of vertebrates (Supplemental Table S1). We analyzed sequences that we also used for the stem analysis, as well as sequences whose secondary structure is absent from the European ribosomal RNA database (and that were not used, therefore, for the stem analysis). Since rRNA is a well-conserved molecule, the average GC level is similar among vertebrate classes, although with a trend toward higher GC in the vertebrates having higher body temperatures. The data are plotted in Figure 1, where one can see differences among vertebrate classes, the GC levels being generally higher for vertebrates with higher body temperatures. For example, the mean GC level of the warm-blooded vertebrates is ∼56%, whereas that of the cold-blooded vertebrates studied here is ∼53.5%. We checked the significance of the GC difference between groups having different body temperatures and that are phylogenetically more closely related, namely, birds versus reptiles, and polar versus temperate/tropical fishes. In both cases, the P-values were <0.0001. Concerning fishes, directional nucleotide substitutions, detected by aligning structures (see below), give a more evident view of the difference between species living in cold and hot environments. In fact, the results for global sequences prompted us to look at the observed contrasts in more depth and to investigate the specificity of compositional differences.
FIGURE 1.
Plot of GC% of the rRNA sequences listed in Supplemental Table S1. Empty points are the averages; horizontal thick bars indicate standard error.
Correspondence analysis and phylogenetic trees
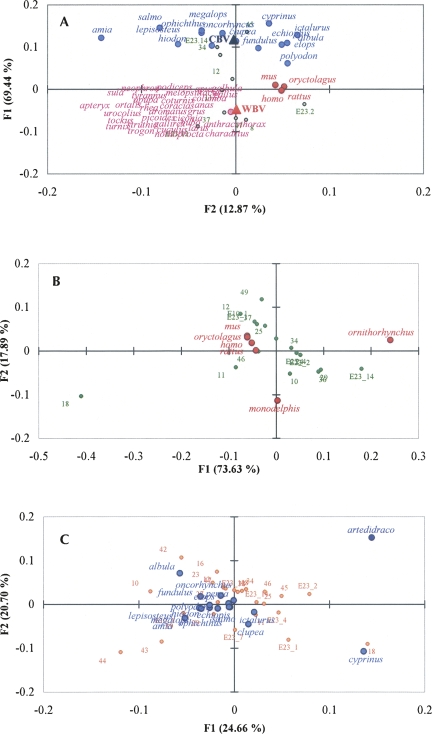
We used the correspondence analysis (CA) (see Materials and Methods) to compare the GC compositions of rRNA stems among species that experience different temperatures. By analyzing the distribution of species in terms of stem composition, we could discriminate between groups. We constructed a matrix comprising mammals, birds, fishes, and two illustrative observations (warm-blooded vertebrates [WBV]; cold-blooded vertebrates [CBV]). These last two signatures, located near the corresponding species, delimited two nonoverlapping regions (see Fig. 2A). In this representation, birds appear at a single point because their stems are essentially identical, and the four placental mammals are located very close to each other.
FIGURE 2.
Factorial plane representation of species according to the GC level of their stems. Distribution of the surveyed species as obtained in the factorial space (F1 and F2) by correspondence analysis. (A) Mammals (red points), birds (pink points), and ray-finned fishes (light-blue points) as listed in Figure 5. For species taxonomy, see Supplemental Table S1. WBV and CBV indicate warm- and cold-blooded as illustrative observations are indicated by a large red triangle and a large blue triangle, respectively. (B) Mammals (red points); (C) ray-finned fishes (light-blue points), polar fish (dark-blue point). In the figures, stems are presented in green (A,B) and orange (C).
We then focused on the more specific comparisons (marsupials/monotremes vs. placentals, and polar vs. temperate/tropical fishes). Figure 2, B and C, shows the resulting distribution of mammals and ray-finned fish species, respectively, as projected on the first factorial plane. In both cases, the species having lower body temperature are located in a different region of the plane, far from the other species with similar body temperature, and similar GC in stems, reflecting the situation that holds between the two main groups of vertebrates (cold-blooded vs. warm-blooded).
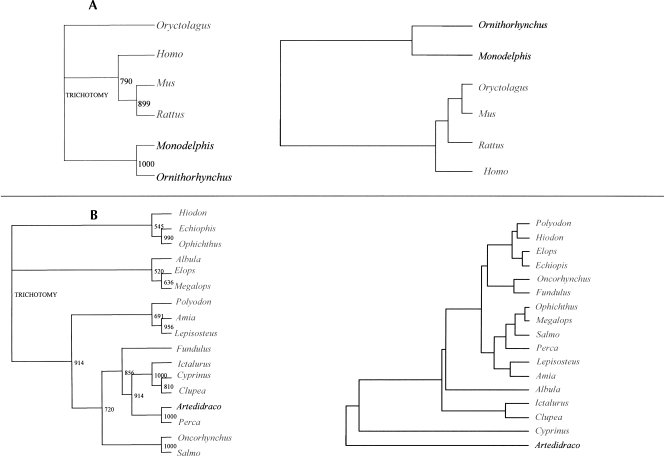
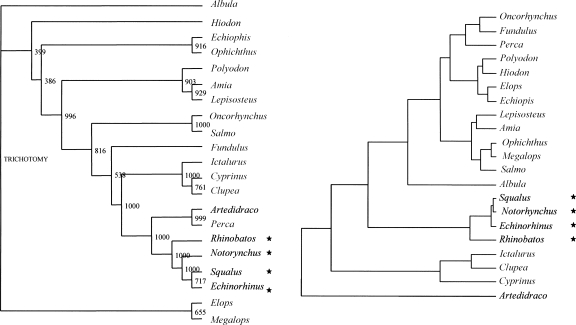
In addition, from the CA data, we constructed a hierarchical classification of species and compared it with the corresponding phylogenetic neighbor-joining (NJ) tree. The results for mammals and fishes are shown in Figure 3. Interestingly, there are differences especially in the tree of fishes, in particular in the case of the two Perciformes. Indeed, the polar Artedidraco is close to the temperate Perca in the phylogenetic tree, whereas in the GC tree, it is located far from it. In the case of mammals, the two trees appear similar, even if in this case the explanation could reside in the large similarity of the two sequences of opossum and platypus. In fact, these two species have in common an expansion of ∼30 nucleotides (nt) and some other features in the primary sequence that are absent in other mammals. As far as secondary structures are concerned, both have stems with lower GC in comparison with placentals. Such a situation leads to a “grouping” of the two species from both the phylogenetic and the GC tree. We also constructed the pair of trees for cartilaginous and ray-finned fishes (Fig. 4), and found that in the GC tree, the cartilaginous fishes appear close to other temperate/tropical fishes, in contrast to what we found for the global sequences (see Supplemental Table S1).
FIGURE 3.
(A) Phylogenetic tree and GC tree of the mammalian species studied, including one marsupial and one monotreme. (Right panel) Neighbor-joining unrooted tree obtained with the complete sequences of the rRNA. Bootstrap numbers are indicated. (Left panel) GC tree. This tree is obtained by a hierarchical classification of the organisms on the basis of their neighborhood distances. Distances between organisms are calculated in the factorial space obtained by correspondence analysis. The marsupial and the monotreme are in black, placentals in gray. (B) Phylogenetic tree and GC tree of the ray-finned fish species studied, including the two Perciformes. The polar fish is indicated in black, whereas the other ray-finned species (temperate and tropical) are in gray.
FIGURE 4.
Phylogenetic tree and GC tree of thecartilaginous (black, marked with a star) and ray-finned fish species studied (temperate/tropical, gray; polar, black). For additional explanations, see legend of Figure 3.
GC and AU level comparisons for the rRNAs of species present in the European ribosomal RNA database
The nucleotide composition was examined in the double-stranded stem regions by making use of the identification of the secondary structures written in the DCSE alignments of the European ribosomal RNA database. We chose to analyze as many species as possible in this database in order to have representative samples from the five classes of vertebrates. We avoided, however, species whose sequences contained many uncertain nucleotides (see Materials and Methods). Supplemental Table S2 presents the taxonomic list of the species analyzed for this section. Figure 5 summarizes the species analyzed, the GC values for the human stems, and the results of the comparison between the GC level of such stems and that of the other vertebrates. As far as birds, reptiles, and other mammals are concerned, we used average GC levels since these groups had similar patterns, whereas for amphibians and fishes, we presented the results for each species because they generally showed a higher heterogeneity. In Figure 5, we did not report the stems that were almost invariant in the majority of the species and of the classes. For completeness, we report GC levels for all stems of all species analyzed in Supplemental Figure S1.
FIGURE 5.
Comparison of GC levels in the rRNA stems of human and other vertebrates. Each column represents a stem, whose numbering is in the first row boxes; GC values for human are indicated in the second row boxes. The species are listed following their taxonomic order (see Supplemental Table S1) with fishes subdivided into ray-finned and Chondrichthyes. Blue, red, gray, and white boxes represent lower, higher, approximately equal (±5%), and identical GC levels compared to human.
Looking at the results, it is noticeable that there are stems that have a lower GC level in the cold-blooded compared to the warm-blooded vertebrates. It is possible to observe that there are, in particular, some stems (Fig 5, 8, 11, 12, E23_2, 37, 47) that have a lower GC level in the majority of the cold-blooded species considered, especially in fishes (Fig 5, only two stems, E23_14 and 45, show a higher GC level). One stem, 18, has a higher GC level only in a group of birds and in the monotreme (see below), but not in the other species. Other stems, instead, show in almost all the cases higher (stems 10, 29, 48) or lower GC values (stems E10_1, E23_1, 25) without an apparent trend (see Supplemental Table S2). Interestingly, reptiles seemed to be intermediate between the warm-blooded and cold-blooded vertebrates, whereas the other cold-blooded species showed the GC-poorest stems. In other words, it appears that warm-blooded vertebrates have a number of stems that have higher GC levels compared to the cold-blooded vertebrates. When calculating the variance for each of the stems listed in Figure 5 and for each species considering the two groups, warm-blooded vertebrates and ray-finned fishes, we can also observe that the former are more homogeneous (seven out of 11 stems have a lower variance) (data not shown), in spite of their phylogenetic distance. Statistical analyses of variance, then, confirmed the separation of the two main different groups.
Supplemental Figure S2 is constructed like Figure 5, but in this case, the AU levels were analyzed. AU base pairs have been shown to have frequency, mutability, and “fitness” lower than GC but higher than other base pairs (Chen et al. 1999; Savill et al. 2001). AU pairs in some stems are more abundant in warm-blooded vertebrates, and in other stems in cold-blooded vertebrates. For completeness, we also reported the AU levels for all the stems of all the species analyzed in Supplemental Figure S3.
Pairwise comparisons of a bird versus fishes
In order to better understand and visualize the changes described above, we approached the problem also by comparing the number of nucleotide pair substitutions and insertions/deletions (indels) from each stem in five comparisons between fishes and a bird, the stork Ciconia nigra. In order to obtain a more precise comparison, we aligned all the stems introducing gaps or indels where required (an example is shown in Supplemental Fig. S4), and then we compared single nucleotide pairs in order to detect and count substitutions or insertions/deletions. Our results show that this comparison is in agreement with what was found before, as there are more GC pairs in the avian stems. Furthermore, such analyses show that the differences in length of some stems do not affect the global result, so we did not take such differences into consideration.
GC and AU level comparisons for the rRNA of species absent from the European ribosomal RNA database
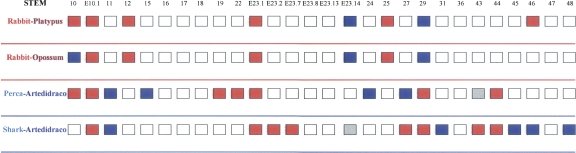
In order to get additional information on the correlation between GC level and body temperature in vertebrates, we also analyzed some species that are not represented in the European ribosomal RNA database. For mammals, we compared both the platypus and the opossum with the rabbit, which has a higher body temperature. For fishes, we compared a polar species (Artedidraco shackletoni) from the order Perciformes with a temperate species (Perca fluviatilis) from the same order and with the shark Echinorhinus cookei. This last comparison illustrates that Antarctic fishes have GC-poorer stems in comparison with Chondrichthyies even if the global sequences of the latter have a lower GC. The results of these four comparisons are shown in Figure 6. It is possible to notice that, even if only a minority of stems is variable, the stems of the species having lower body temperature are GC poorer in comparison with their counterparts. This finding, together with the result obtained by correspondence analysis, indicates that the action of temperature is more “visible” in the stems than in the overall molecule.
FIGURE 6.
Comparison of the GC levels in the stems of rRNA sequences of some vertebrate couples. The reference species are rabbit, Perca, and the shark. The color code of boxes follows that of Figure 5.
DISCUSSION
Previous studies found a positive correlation between GC levels in stems and optimal growth temperature in prokaryotes (Galtier and Lobry 1997). Since the increased GC level was found to be limited to the stems, the authors concluded that natural selection acts so as to increase the structural stability and maintain the folding of rRNA at high temperatures. Other authors (Wang et al. 2006) also showed the existence of such a correlation not only in prokaryotes, but also in Archaea and in vertebrates. Concerning the last group, they found, however, that even if rRNAs from warm-blooded vertebrates have a higher overall GC content compared to those from cold-blooded vertebrates, the correlation between GC levels in stems and temperature is less strong. Furthermore, they excluded thermal adaptation as the cause of GC differences between vertebrate lineages because they found that the difference in GC level between warm-blooded and cold-blooded vertebrates was not focused on the paired regions of rRNA, in contrast with what was found in prokaryotes. This result was explained by the smaller range of body temperatures in vertebrates compared to that of the optimal growth temperatures of bacteria. It is known, however, that loops are differently conserved in prokaryotes and eukaryotes (Smit et al. 2007) and that there are many differences in the length and expansion segments of prokaryotic and eukaryotic rRNA sequences (Doudna and Rath 2002; Dresios et al. 2006).
In this study, we introduced the parameter temperature to discriminate between the classes of vertebrates and provided evidence that the rule of thermal adaptation found in prokaryotes also applies to vertebrates. It should be stressed here that our methodological approach was different from that used in previous studies since we also evaluated the difference in GC (or AU) in a stem-by-stem comparison between the rRNA of cold-blooded and warm-blooded vertebrates. In contrast, both Galtier and Lobry (1997) for prokaryotes and Wang et al. (2006) for prokaryotes and vertebrates calculated the GC levels of both stems and loops as cumulative averages for each class. Unfortunately, this approach did lack an appropriate filtering of sequences that were identical or contained a high number of uncertain nucleotides (indicated as N or S).
We expected warm-blooded vertebrates to show contrasting rRNA patterns compared to cold-blooded fish, amphibians, and reptiles, given that the GC level is known to be regionally higher in the nuclear genomes of warm-blooded as compared with cold-blooded vertebrates (for review, see Bernardi 2005). In agreement with this expectation, we found that the average GC level of both rRNA sequences and of a number of their individual stems was higher in warm-blooded vertebrates. In fact, we observed a trend of increasing GC with increasing temperature already from the rough data of the global sequences, warm-blooded vertebrates having the highest GC level and Antarctic fishes the lowest one (together with Chondrichthyes, which, however, have more GC-rich stems compared with the ray-finned fishes). A comparison between the GC levels of global rRNA sequences of cold-blooded and warm-blooded vertebrates was already done with a similar result (Xia et al. 2003), but at that time, the available sequences were few, and differences between fishes living in cold or hot environments were not taken into consideration.
By comparing the differences at the level of individual stems and base pairs, we found that the majority of stems showing the highest GC levels are present in the avian sequences (birds have the highest body temperature among vertebrates), whereas, on the contrary, the majority of stems showing the lowest GC levels were found in the Antarctic fish studied. It is also interesting to point out that we found a small difference in GC level at both the global sequence and at the stem level between polar and tropical/temperate fishes, and that this difference is accompanied by differences in the DNA methylation level (Varriale and Bernardi 2006a). Reptiles seem to be in an intermediate position, in agreement with previous results on their genomes that concerned nucleotide composition, band asymmetry in cesium chloride, compositional heterogeneity (for review, see Bernardi 2005), karyotypic evolutionary rates (Olmo et al. 2002; Olmo 2005), and DNA methylation (Varriale and Bernardi 2006b).
Along the same line, the cases of marsupials and monotremes are outstanding. In particular, marsupials and monotremes have very different genomic GC composition (A. Varriale and G. Bernardi, in prep.; data not shown), but very similar rRNA sequence and structure, a point in agreement with the marsupionta hypothesis (Janke et al. 1997, 2002). Such similarity concerned both nucleotide composition and the presence of an insertion (Janke et al. 2002) and followed what we found here concerning the correlation between temperature and GC. Both marsupials and monotremes, in fact, have a lower body temperature (33°C–35°C) compared with eutherians (∼37°C), and have a GC-poorer rRNA for the majority of the stems that are different from those of eutherians. All these findings concerning stem differences related to temperature, together with the fact that the GC level of both global rRNA sequences and of other “structural categories” (junctions, ends, and bulges) are positively correlated with temperature (Smit et al. 2006; Wang et al. 2006), could suggest that temperature acts on the underlying global sequence and eventually on secondary structural categories.
Another important point concerns phylogenesis. rRNA has been extensively used for phylogenetic studies, but recently the need to be aware of different evolutionary rates and GC levels in the structural categories has been highlighted. We have shown that the environment has a role in the molding of nucleotide composition of secondary structure in rRNA, and that environmental parameters can group together species that are not necessarily related phylogenetically. CA is an efficient method to extract significant properties and trends from large-scale genomic data and to integrate them with qualitative features. Together with hierarchical classification, it represents a methodological approach that is useful to classify genomes. In our case, CA based on the GC level of stems revealed a clear-cut segregation of species following the life style–phylogenetic classes (warm-blooded and cold-blooded vertebrates), and, within them, placentals and marsupials/monotremes, on the one hand, and temperate/tropical and polar fishes, on the other. Using CA, in fact, we obtained a tree based on the GC levels of stems, and this tree was different from the phylogenetic tree.
The case of birds is interesting. Although a phylogenetic tree was presented (van Tuinen et al. 2002) that indicated a certain distance among species, if we consider a potential GC tree made with CA, all the avian species are clustered together as if we were looking at only one species. Along the same line, Xia et al. (2003) already reported the case of a phylogenetic tree in which birds appeared closer to mammals than to reptiles, contrary to other molecular and morphological results, and to their tree in which the rRNAs were aligned with the aide of the secondary structure. From these results and by referring to literature (Bernardi 1993), they suggested that such discrepancy could have been due to the common high body temperature and GC level in birds and mammals.
In conclusion, we found a correlation between temperature and GC level in the double-stranded stretches of rRNA, showing that temperature has an influence not only on the nucleotide composition (Bernardi 2005), the open or closed chromatin structure in the nucleus (Saccone et al. 2002), and DNA methylation (Varriale and Bernardi 2006a,b), but also on the structure of rRNA, through the molding of nucleotide composition.
MATERIALS AND METHODS
Calculation of the global GC level from rDNA sequences
Both partial and (whenever available) complete rDNA sequences from species belonging to the five classes of vertebrates were downloaded from GenBank and by making use of the BLASTN tool (http://www.ncbi.nlm.nih.gov/BLAST/). The GC level of genes was calculated with a Perl script (implemented in our laboratory).
Correspondence analysis
This classical method for the multivariate exploration of large-scale data (Benzecri 1973; Greenacre 1984; Tekaia et al. 1999, 2002; Tekaia and Yeramian 2006) allows visual inspection of significant trends in genome analysis by projecting simultaneously the properties of the observations and the variables on the factorial planes. In brief, the dimensions of the considered planes are relevant to the number of variables and observations involved, which have to be listed on a contingency matrix. In our case, the species are observations, whereas the variables are represented by the stems that change within the species under analysis. We took into consideration only warm-blooded vertebrates and ray-finned fishes, which are the groups containing more sequences in the database. For both the construction of the factorial plane representation and the hierarchical classification, we used a trial version of the program XLSTAT 2008 (Addinsoft SARL). We constructed phylogenetic trees through the program ClustalX starting from the sequences of the rRNA and chose unrooted NJ with default parameters and 1000 bootstrap replicates.
Secondary structure data
The rRNA sequence data were downloaded from the European ribosomal RNA Database (http://www.psb.ugent.be/rRNA/index.html) (Wuyts et al. 2004). The sequence files from this database are text files with a special distribution format (DCSE) that specifies secondary structure information. The format uses square brackets to define a helix, braces to enclose an internal loop, and parentheses for the nonstandard base pairs. Furthermore, in the DCSE, the secondary structures are given together with the helix numbering and the start/end points of stems so that it is possible to get length and AU and GC level of all the stems for each species. For example, in order to construct stem number 8, one can take sequence 8 and align it with the complementary inverted sequence 8′. Thus, one can get all the stems and make pairwise comparisons with those of a reference species. We chose the human rRNA as a reference because the human genome is a typical mammalian genome and because its sequence is complete.
In the comparisons, we detected all the stems showing differences in composition and/or length by counting all nucleotide pairs and by calculating the percent of both GC and AU. We did not take into account the first seven stems and the last one because these sequences were absent in a vast proportion of cold-blooded vertebrates. However, in the cases where we found them, we noticed that they were invariant. The data were filtered by retaining only the first entry for each species in the case of species having multiple entries, and by excluding sequences containing more than two uncertain nucleotides (N, S). This filtering left rRNA data for a total of 64 vertebrate species, 36 of which are warm-blooded vertebrates including 32 birds and four mammals, as well as 24 species from cold-blooded vertebrates including two reptiles, three amphibians, 15 ray-finned fishes, and four chondrichthyes. The taxonomic list of the species studied is presented in Supplemental Table S1. The manual count and a Perl script (implemented in our laboratory) were used together for cross-checking the results.
Construction of new secondary structures
In order to compare some vertebrate species whose rRNA structures were absent from the European ribosomal RNA database, we constructed such structures through a comparative method. Briefly, we put our starting sequence into BLASTN (http://www.ncbi.nlm.nih.gov/BLAST/) and then explored the distance tree of the results in order to obtain the phylogenetically closest species whose structure was available in the European ribosomal RNA database. We then manually aligned it with the starting sequence (using the application SeAl version 2.0a11 Carbon) (http://evolve.zoo.ox.ac.uk), so that we could construct the new structure and make pairwise comparisons. This approach, in fact, limits the possibility of ambiguous alignments when used between closely related species.
For the platypus (a monotreme, Ornithorhynchus anatinus), we first aligned its sequence with that of the rabbit, Oryctolagus cuniculus, then detected the indels and subsequently superimposed such sequence onto the rabbit structure. The newly constructed structure of the platypus was then used to align the sequence of the marsupial Monodelphis domestica (gray short-tailed opossum), and finally the latter was compared to the sequence of another marsupial, Vombatus ursinus. As the latter two sequences differed in a few nucleotides only, the structure of Monodelphis was used. The same procedure was used for two fishes belonging to the order Perciformes that we wanted to compare, the Antarctic Artedidraco shackletoni and the temperate Perca fluviatilis. We aligned both sequences to the closely related Sebastes brevispinis (order Scorpaeniformes) whose structure, in turn, had been superimposed to that of Ophichthus rex (order Anguilliformes, present in the European ribosomal RNA database). All the newly constructed structures are available from the authors upon request.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We are grateful to Oliver Clay, Giacomo Bernardi, and one anonymous referee for the critical reading of the manuscript and for suggestions. We also thank Fabio Auletta for the implementation of a Perl script.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.957108.
REFERENCES
- Abouheif E., Zardoya R., Meyer A. Limitations of metazoan 18S rRNA sequence data: Implications for reconstructing a phylogeny of the animal kingdom and inferring the reality of the Cambrian explosion. J. Mol. Evol. 1998;47:394–405. doi: 10.1007/pl00006397. [DOI] [PubMed] [Google Scholar]
- Armbruster G.F.J. Temperature-based variation of rRNA secondary structure models: A case study in the insect Drosophila simulans, the land snail Isabellaria adriani, and the crustacean Daphnia pulex . Can. J. Zool. 2001;79:334–345. [Google Scholar]
- Benzecri J.P. L'analyse des données, L'Analyse des Correspondances, 2. Dunod; Paris, France: 1973. [Google Scholar]
- Bernardi G. The vertebrate genome: Isochores and evolution. Mol. Biol. Evol. 1993;10:186–204. doi: 10.1093/oxfordjournals.molbev.a039994. [DOI] [PubMed] [Google Scholar]
- Bernardi G. Structural and evolutionary genomics. Natural selection in genome evolution. Elsevier; Amsterdam, The Netherlands: 2005. [Google Scholar]
- Bernardi G., Bernardi G. Compositional constrains and genome evolution. J. Mol. Evol. 1986;24:1–11. doi: 10.1007/BF02099946. [DOI] [PubMed] [Google Scholar]
- Bernardi G., Sordino P., Powers D.A. Nucleotide sequence of the 18S ribosomal ribonucleic acid gene from two teleosts and two sharks and their molecular phylogeny. Mol. Mar. Biol. Biotechnol. 1992;1:187–194. [PubMed] [Google Scholar]
- Chen Y., Carlini D.B., Baines J.F., Parsch J., Braverman J.M., Tanda S., Stephan W. RNA secondary structure and compensatory evolution. Genes Genet. Syst. 1999;74:271–286. doi: 10.1266/ggs.74.271. [DOI] [PubMed] [Google Scholar]
- Doudna J.A., Rath V.L. Structure and function of the eukaryotic ribosome. The next frontier. Cell. 2002;109:153–156. doi: 10.1016/s0092-8674(02)00725-0. [DOI] [PubMed] [Google Scholar]
- Dresios J., Panopoulos P., Synetos D. Eukaryotic ribosomal proteins lacking a eubacterial counterpart: Important players in ribosomal function. Mol. Microbiol. 2006;59:1651–1663. doi: 10.1111/j.1365-2958.2006.05054.x. [DOI] [PubMed] [Google Scholar]
- Galtier N., Lobry J.R. Relationships between genomic G+C content, RNA secondary structures, and optimal growth temperature in prokaryotes. J. Mol. Evol. 1997;44:632–636. doi: 10.1007/pl00006186. [DOI] [PubMed] [Google Scholar]
- Greenacre M.J. Theory and applications of Correspondence Analysis. 1st ed. Academic Press; London, UK: 1984. [Google Scholar]
- Jabbari K., Bernardi G. CpG doublets, CpG islands and ALU repeats in long human DNA sequences from different isochore families. Gene. 1998;224:123–127. doi: 10.1016/s0378-1119(98)00474-0. [DOI] [PubMed] [Google Scholar]
- Jabbari K., Bernardi G. Cytosine methylation and CpG, TpG (CpA) and TpA frequencies. Gene. 2004;333:143–149. doi: 10.1016/j.gene.2004.02.043. [DOI] [PubMed] [Google Scholar]
- Jabbari K., Cacciò S., Pais de Barros J.P., Desgres J., Bernardi G. Evolutionary changes in CpG and methylation levels in the genome of vertebrates. Gene. 1997;205:109–118. doi: 10.1016/s0378-1119(97)00475-7. [DOI] [PubMed] [Google Scholar]
- Janke A., Xu X., Arnason U. The complete mitochondrial genome of the wallaroo (Macropus robustus) and the phylogenetic relationship among Monotremata, Marsupialia, and Eutheria. Proc. Natl. Acad. Sci. 1997;94:1276–1281. doi: 10.1073/pnas.94.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke A., Magnell O., Wieczorek G., Westerman M., Arnason U. Phylogenetic analysis of 18S rRNA and the mitochondrial genomes of the wombat, Vombatus ursinus, and the spiny anteater, Tachyglossus aculeatus: Increased support for the Marsupionta hypothesis. J. Mol. Evol. 2002;54:71–80. doi: 10.1007/s00239-001-0019-8. [DOI] [PubMed] [Google Scholar]
- Olmo E. Rate of chromosome changes and speciation in reptiles. Genetica. 2005;125:185–203. doi: 10.1007/s10709-005-8008-2. [DOI] [PubMed] [Google Scholar]
- Olmo E., Capriglione T., Odierna G. Different genomic evolutionary rates in the various reptile lineages. Gene. 2002;295:317–321. doi: 10.1016/s0378-1119(02)00685-6. [DOI] [PubMed] [Google Scholar]
- Saccone S., Federico C., Bernardi G. Localization of the gene-richest and the gene-poorest isochores in the interphase nuclei of mammals and birds. Gene. 2002;300:169–178. doi: 10.1016/s0378-1119(02)01038-7. [DOI] [PubMed] [Google Scholar]
- Savill N.J., Hoyle D.C., Higgs P.G. RNA sequence evolution with secondary structure constraints: Comparison of substitution rate models using maximum likelihood methods. Genetics. 2001;157:399–411. doi: 10.1093/genetics/157.1.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit S., Yarus M., Knight R. Natural selection is not required to explain universal compositional patterns in rRNA secondary structure categories. RNA. 2006;12:1–14. doi: 10.1261/rna.2183806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit S., Widmann J., Knight R. Evolutionary rates vary among rRNA structural elements. Nucleic Acids Res. 2007;35:3339–3354. doi: 10.1093/nar/gkm101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekaia F., Yeramian E. Evolution of proteomes: fundamental signatures and global trends in amino acid compositions. BMC Genomics. 2006;7:307. doi: 10.1186/1471-2164-7-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekaia F., Lazcano A., Dujon B. The genomic tree as revealed from whole proteome comparisons. Genome Res. 1999;9:550–557. [PMC free article] [PubMed] [Google Scholar]
- Tekaia F., Yeramian E., Dujon B. Amino acid composition of genomes, lifestyles of organisms, and evolutionary trends: A global picture with correspondence analysis. Gene. 2002;297:51–60. doi: 10.1016/s0378-1119(02)00871-5. [DOI] [PubMed] [Google Scholar]
- Telford M.J., Wise M.J., Gowri-Shankar V. Consideration of RNA secondary structure significantly improves likelihood-based estimates of phylogeny: Examples from the bilateria. Mol. Biol. Evol. 2005;22:1129–1136. doi: 10.1093/molbev/msi099. [DOI] [PubMed] [Google Scholar]
- van Tuinen M., Sibley C.G., Hedges S.B. The early history of modern birds inferred from DNA sequences of nuclear and mitochondrial ribosomal genes. Mol. Biol. Evol. 2000;17:451–457. doi: 10.1093/oxfordjournals.molbev.a026324. [DOI] [PubMed] [Google Scholar]
- Varriale A., Bernardi G. DNA methylation and body temperature in fishes. Gene. 2006a;385:111–121. doi: 10.1016/j.gene.2006.05.031. [DOI] [PubMed] [Google Scholar]
- Varriale A., Bernardi G. DNA methylation in reptiles. Gene. 2006b;385:122–127. doi: 10.1016/j.gene.2006.05.034. [DOI] [PubMed] [Google Scholar]
- Vawter L., Brown W.M. Rates and patterns of base change in the small subunit ribosomal RNA gene. Genetics. 1993;134:597–608. doi: 10.1093/genetics/134.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.C., Xia X., Hickey D. Thermal adaptation of the small subunit ribosomal RNA gene: A comparative study. J. Mol. Evol. 2006;63:120–126. doi: 10.1007/s00239-005-0255-4. [DOI] [PubMed] [Google Scholar]
- Wuyts J., Perrière G., Van de Peer Y. The European ribosomal RNA database. Nucleic Acids Res. 2004;32:D101–D103. doi: 10.1093/nar/gkh065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X., Xie Z., Kjer K.M. 18S ribosomal RNA and tetrapod phylogeny. Syst. Biol. 2003;52:283–295. doi: 10.1080/10635150390196948. [DOI] [PubMed] [Google Scholar]