Abstract
A theoretical framework for the function of the medial temporal lobe system in memory defines differential contributions of the hippocampal subregions with regard to pattern recognition retrieval processes and encoding of new information. To investigate molecular programs of relevance, we designed a spatial learning protocol to engage a pattern separation function to encode new information. After background training, two groups of animals experienced the same new training in a novel environment; however, only one group was provided spatial information and demonstrated spatial memory in a retention test. Global transcriptional analysis of the microdissected subregions of the hippocampus exposed a CA3 pattern that was sufficient to clearly segregate spatial learning animals from control. Individual gene and functional group analysis anchored these results to previous work in neural plasticity. From a multitude of expression changes, increases in camk2a, rasgrp1, and nlgn1 were confirmed by in situ hybridization. Furthermore, siRNA inhibition of nlgn1 within the CA3 subregion impaired spatial memory performance, pointing to mechanisms of synaptic remodeling as a basis for rapid encoding of new information in long-term memory.
Keywords: hippocampus, microarray, spatial learning
Episodic memory formation in humans requires intact function of the hippocampus and is well modeled in rodents using spatial memory tasks. Each of the three major hippocampal subregions, CA1, CA3, and dentate gyrus (DG), contributes to the processing of spatial information for memory encoding. Recent work examining the neuronal activity in rats has demonstrated distinctive subregional responses to spatial contexts of varying familiarity (1–4). These data have highlighted the role of the CA3 in rapidly distinguishing familiar and new contexts in spatial memory processing.
Work on the cellular and molecular mechanisms of memory has focused on the in vitro hippocampal slice preparation to isolate activity-dependent plasticity [long-term potentiation (LTP) and long-term depression]. Adjustments in synaptic strength in this model can persist over lengthy intervals, providing a candidate mechanism for long-term information storage (5, 6). A fundamental finding of the research is the molecular requirements of RNA synthesis and subsequent protein translation for both long-lasting neural plasticity at hippocampal synapses and hippocampal-dependent long-term memory (7–9).
A number of approaches have been used to identify plasticity genes with potential relevance to mechanisms of long-term memory (10, 11). Such studies have included broad molecular screening to profile gene expression by using microarray platforms in behavioral learning protocols (12–14). However, given the functional distinctions between different subregions of the hippocampal system (2, 15), a regionally specific profile could be obscured by a global assessment of the whole hippocampus. In addition, identification of molecular changes that are tied specifically to information storage can be challenging to isolate because of the difficulty in factoring out confounding effects of merely performing a task independent of the information that is acquired in learning.
Here, we used a spatial learning protocol that isolates the acquisition of spatial information from many performance-related factors. We generated profiles of learning-modulated genes in individual hippocampal subregions. An abundance of mRNA changes, with transcript increases predominating, occurred in the CA3 region, yielding a profile that includes many known targets of plasticity induction protocols and revealing numerous novel candidate plasticity genes. siRNA inhibition of one of these novel genes, neuroligin 1, impaired retention of memory for spatial information acquired at the time of gene interference, highlighting the dependence of spatial memory on intact CA3 synaptic plasticity. All gene expression data are available at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE11476. In addition, an interactive, searchable version of the CA3 data is available at http://nbc.jhu.edu/la/.
Results
Spatial Learning.
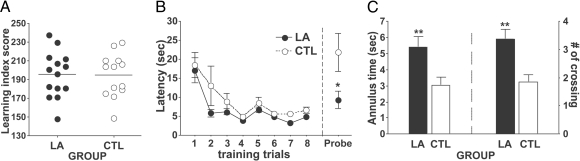
All animals were familiarized with behavioral procedures during background training in a conventional hidden platform water maze task. A learning index, based on four probe trials spaced throughout training, was computed for each rat (16) and served to equate baseline performance across groups for subsequent experiments (Fig. 1A). Rats then underwent a training protocol in a novel environment designed to isolate hippocampal spatial learning. In a new water maze environment, located at a different physical site, rats in the spatial learning-activated (LA) group were required to swim to a visible platform at a fixed location in the presence of novel spatial cues. Control (CTL) animals likewise escaped to a visible platform but without orienting spatial cues and with the platform location varying from trial to trial. Whereas rats in both the LA and CTL groups exhibited similar escape performance (Fig. 1B Left), only the rats in the LA group showed clear evidence of spatial memory in a probe trial 1 h after training (Fig. 1B Right) as evidenced by time spent in the annulus and annulus crossings during the probe trial (Fig. 1C). A separate group of rats received the identical protocol just described but were tested in a probe trial 48 h later, a time point known to require both RNA and protein synthesis (17). As before, the LA group but not the CTL group had a spatial bias for the platform location [supporting information (SI) Fig. S1] in long-term retention. In an additional test of the protocol, lesions of the dorsal hippocampus impaired the animals' ability to recall the platform location, confirming hippocampal dependency in this setting (Fig. S2).
Fig. 1.
Behavioral performance in the spatial LA protocol. (A) Rats assigned to LA and CTL groups were matched for background performance by learning index in standard water maze training. (B) Mean latency to the escape platform during the training trials and to the target platform location during the probe trial 1 h posttraining. Target platform location corresponded to the location of the platform for the LA group during training. (C) Time spent in the target annulus (5× of the platform size) and number of target annulus crossings made during the first 30 s of probe. **, P < 0.01 and *, P < 0.05 compared with CTL. Error bars represent SEM.
Global Profiling of mRNA Transcripts During Spatial Memory Formation.
RNA samples from CA1, CA3, and DG subregions of the hippocampus, dissected immediately after the 1-h probe trial, were interrogated by using Affymetrix RAE230A oligonucleotide microarrays, containing 15,923 probe sets. The Bioconductor package, GCRMA (18), and additional nonlinear transformations were used for quality assessment and normalization of raw Affymetrix expression intensities. Analyses of CA1 and DG data (see Fig. S3) resulted in few genes that were differentially expressed between LA and CTL subjects as compared with CA3. Only minimal overlap of such genes occurred between the three regions. Whole dataset comparisons likewise showed little correlation between CA1 and DG and the CA3 subregion. Therefore given the pronounced signal detected uniquely in CA3, a region known by other methods to be strongly engaged in rapid encoding of new spatial information, the remainder of this article will focus on those data.
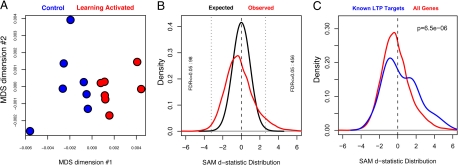
To assess the global similarity of expression profiles across individual animals, we generated a multidimensional scaling plot with each point representing a single CA3 profile in 2D space. The proximity of the points is determined by the correlation (Pearson's r) calculated for all possible comparisons between individual profiles (Fig. 2A). This unsupervised, dimension-reducing visualization shows that global expression patterns in the CA3 region are sufficient to segregate LA from CTL animals, suggesting that gene expression differences reflect learning-induced transcriptional processes.
Fig. 2.
CA3 transcriptional profile. (A) A multidimensional scaling plot depicts the global similarity between RNA expression profiles across samples. This unsupervised (blind to group labels) dimension reducing visualization reveals separation of CTL (blue) and LA (red) groups. Quantitative comparison of the distances between points reveals that the between-group difference (the distance between the two group means) is greater than the within-group variance (the mean distance between individual samples within each group). (B) SAM d-statistic distribution of the observed LA-CTL comparison (red line) and chance (black line), generated by permuting group labels and recalculating differential expression statistics. Left and right vertical dotted lines indicate statistic thresholds beyond which FDR ≤0.05. Plot labels indicate the number of probesets defined in each hippocampal subregion that fall beyond these limits. (C) Genes known to have increased expression with LTP (literature defined list of 41 genes, blue line) are differentially expressed in CA3 (red line same as in B).
Differential Expression: Individual Genes.
To identify individual genes that were differentially expressed between LA and CTL, we applied Significance Analysis in Microarrays (SAM; ref. 19) across each of the probe sets on the arrays that exceeded an empirically defined low-intensity cutoff. To correct for the high number of comparisons, we used a modified false discovery rate (FDR) approach within SAM in which observed differential expression statistics (Fig. 2B, red curve) were compared with statistics calculated by using permuted group labels, resulting in an expected random distribution (Fig. 2B, black curve). A larger number of extreme statistics present in the observed data for CA3 is an indication of the magnitude and consistency of the mRNA changes induced by spatial learning in the CA3 region in this model. More than 500 probe sets are identified as differentially expressed in CA3 for which FDR ≤0.05. Table S1 presents a complete list of the 554 differentially expressed probesets that exceed the 0.05-FDR cutoff.
The majority (82%) of differentially expressed genes in CA3 were increased with learning. Among those genes are many important for synapse function such as neurotransmitter receptors (gria1, gabra4), synaptic vesicle proteins (sv2b), and other structural/adhesion molecules (nlgn1, nrcam, homer1). As expected, mRNA for inducible genes that are linked to neural plasticity mechanisms also showed increases, e.g., camk2a.
Differential Expression: Functionally Related Gene Groups.
To compare our dataset to empirically defined expression changes, we compiled a list of genes from a survey of the literature (while remaining blind to the expression data described here), whose transcript levels were found to be increased with the induction of LTP (Table S2). To assess the statistical significance of the changes in gene expression observed within this a priori defined group of genes, we compared the differential expression statistics associated with the genes in this list to the statistics for all other genes on the oligonucleotide array by using a Wilcox rank sum test. As shown in Fig. 2C, this LTP gene group is clearly increased in expression in the CA3 region of LA animals. Hence, we found that genes that are induced by LTP protocols are likewise induced during spatial memory formation in the CA3 region of behaving rats.
To extend our understanding of the functional relationship among genes, we annotated all probesets with data from Kyoto Encyclopedia of Genes and Genomes Pathways (www.genome.jp/kegg), the Gene Ontology project (www.geneontology.org), and the Pfam database (www.sanger.ac.uk/Software/Pfam), resulting in 2,680 (partially redundant and overlapping) functionally related gene groups of n ≥ 5 genes. We interrogated each of the functional groups for differential expression by conducting analyses with the same methodology as described for the LTP-induced genes. Gene groups showing increases meeting a Bonferroni correction α = 0.01 (P = 3.73e-6) highlighted primarily synaptic components for function and plasticity, including LTP, long-term depression, synaptic transmission, and Wnt signaling. Gene expression levels in these categories were elevated in the LA group as compared to the CTL with very robust P values (see Table S3).
Confirmation of Differential Expression via in Situ Hybridization.
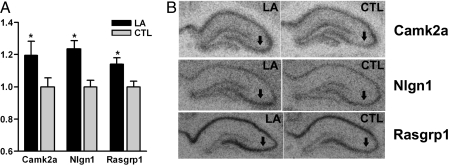
In situ hybridization histochemistry was used to confirm select genes from the microarray data in an independent set of animals given identical behavioral training. The three selected genes (camk2a, nlgn1, and rasgrp1) had exhibited significant expression changes in the CA3 region of the array analysis, with each gene up-regulated in the LA group compared with CTLs.
In situ hybridization analysis showed that all three genes were expressed in the pyramidal cell layer in the CA3 region, whereas camk2a showed expression beyond that layer, as expected (20). We restricted in situ hybridization quantification to the dorsal hippocampus (21) and included densitometry of only the pyramidal cell layers. In situ analysis for all three genes showed significant increases in the LA group as compared with the CTLs in CA3 (Fig. 3) in concordance with the microarray data. The absence of expression changes in CA1 for Nlgn1 and Rasgrp1 in in situ analyses served to control for systematic hybridization variability (data not shown). Because camk2a was expressed beyond the pyramidal cell layer, a second analysis was performed for this gene that included all layers of the CA3 region, yielding results similar to the pyramidal cell analysis (P < 0.05; data not shown).
Fig. 3.
In situ hybridization confirmation of microarray data. (A) Normalized pyramidal cell intensity after hybridization with camk2a, nlgn1, or rasgrp1 probe. *, P < 0.05 compared with CTL. (B) Representative autoradiographs of hippocampal sections used for camk2a, nlgn1, and rasgrp1 quantification. Arrows indicate CA3 pyramidal cell layer.
Connecting mRNA Changes to Memory Function.
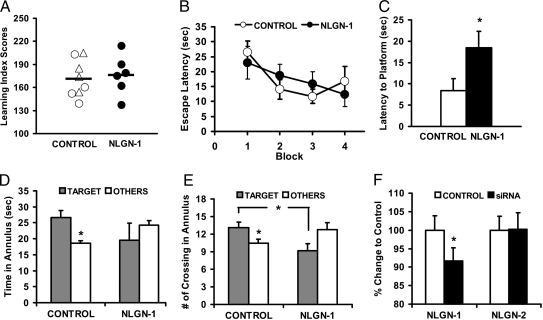
To establish the relevance of specific gene changes identified by the microarray to in vivo memory formation, we chose to pursue the functional consequences of nlgn1 inhibition through siRNA administration. siRNA specific for nlgn1 was injected bilaterally into the dorsal hippocampus to target the CA3 region of rats before training in a single session of water maze training, whereas CTL rats were administered either scrambled siRNA or saline. All rats had received standard background training without injections and were divided into groups on that basis (Fig. 4A) for subsequent training in a new environment. Infusing nlgn1 siRNA into the hippocampus 2 h before training in the new environment had no effect on performance in swimming to a hidden platform (Fig. 4B), but impaired retention when tested in a probe trial 48 h later in the absence of injections. Rats that received CTL injections (either scrambled siRNA or saline), in contrast, had good retention as indicated by significantly shorter latencies to the target location than those that received nlgn1 siRNA injection (Fig. 4C). CTL rats also spent more time (Fig. 4D) and had more crossings (Fig. 4E) in the target annulus than in other annuli, whereas nlgn1 siRNA-injected rats failed to show this search pattern. We confirmed by in situ hybridization that nlgn1 siRNA injections effectively knocked down nlgn1 mRNA expression in the CA3. Unilateral injection of the siRNA in a subset of the animals 1 week later produced a significant CA3 decrease in nlgn1 mRNA, but not nlgn2 mRNA compared with the noninjected CTL side (Fig. 4F). Nlgn1 was not significantly decreased in the CA1 region, and scrambled siRNA injections in a separate set of animals had no effect on nlgn1 mRNA level (data not shown). Thus, targeted inhibition of nlgn1 impaired the long-term retention of spatial information.
Fig. 4.
Blocking nlgn1 expression in the hippocampus impairs long-term memory. (A) Rats in CTL (○, saline; ▵, scrambled) and nlgn1 siRNA groups were matched for background performance in pretraining (F < 1). Rats in the saline and scrambled siRNA groups did not differ on any performance measure and were pooled for all subsequent analysis (CTL group). (B) There is no difference between groups in latency to locate the hidden platform during training (F < 1). Shown are blocks of two training trials each. (C) Rats that received nlgn1 siRNA took longer than those that received CTL injections (P < 0.05) to find the platform location in a probe test 48 h after training. (D and E) Rats in the CTL but not in the nlgn1 group spent more time (D; P < 0.02) and had more crossings (E; P = 0.05) in the target annulus than in other CTL annuli (averaged score shown). The CTL group also had significantly more crossings than the nlgn1 group in the target annulus (P < 0.03). There is no difference between rats that received nlgn1 and CTL injections in their latency to locate a visible platform in a cued training task given immediately after the probe test (data not shown), suggesting intact motivation and sensorimotor function. (F) In situ hybridization quantification of nlgn1 and nlgn2 after unilateral injection of nlgn1 siRNA (P < 0.05 for nlgn1).
Discussion
The results presented here demonstrate that a behavioral protocol designed to isolate spatial memory from nonspatial factors of task performance can produce a robust profile of differential gene expression in the CA3 subregion of the hippocampus. This profile is notable, not only for the number of expression changes that meet a stringent FDR, but for the parity with known or expected changes associated with models of synaptic plasticity and memory formation. Furthermore, the abundance of induced synaptic plasticity genes implies a potential functional significance for novel findings in the dataset. Although the absolute magnitude of the expression changes is small, the results with siRNA inhibition of nlgn1 in CA3 strongly support the interpretation that the detected gene induction can play a vital role in long-term memory.
The distinctive engagement of the CA3 subregion with this task is consistent with our current understanding of its role in hippocampal-dependent episodic memory (1, 3, 4). Computational models of hippocampal function highlight two competing processes of pattern separation and pattern completion that are used to store and retrieve representations (15). The demands of the task in the current research, consisting of both a new environment and familiar task requirements, are designed to strongly engage a pattern separation process, which likely contributes to the prominent transcriptional profile in CA3 during memory formation. The detection of these processes may be further amplified by the unique architecture of CA3, whereby recurrent CA3 collaterals form the majority of inputs onto the CA3 neurons, potentially resulting in contributions from both presynaptic and postsynaptic structural modifications within the same dataset.
The relative paucity of differential expression in other hippocampal subregions may result, not from a lack of plasticity, but from an inability to detect it in this paradigm. For example, in studies examining plasticity of encoding as a function of changes in environmental context, CA1 responses are often more graded than those in CA3 (1, 3). Alternatively, gene induction in either CA1 or DG may lag behind that of CA3. The time point used here was selected to minimize the detection of immediate early genes [expressed within 10–30 min of a learning episode (22)] and investigate effector genes induced at later points that may give rise to the cellular and molecular alterations that occur with long-term memory formation. However, it is highly likely that we did not catch the peak of expression for all genes, irrespective of the subregion. Although the data obtained in this study do not preclude significant changes in either the dentate or CA1, the findings substantiate a vital role for CA3 in pattern separation.
The extensive differential expression in CA3, as analyzed by functional gene groups and compared with known targets of LTP induction, reveal a program of transcript changes related to plasticity. Our follow-up analysis confirmed expression changes for one such well known plasticity gene, camk2a. The reproducibility of this and two other novel gene changes by in situ hybridization provide biological replication of expression differences, as an independent set of animals was tested. These data, supporting the quality and relevance of the microarrays, can also be more broadly tied to other existing findings in the literature. For example, neurogranin (23) and calcineurin (24) have been widely implicated as proteins of major importance in learning and memory via LTP and behavioral studies targeting expression. Both were induced with learning in our dataset; the calcineurin increase in particular was detected by three separate probesets that met the FDR. Thus, expression increases in the current study encompass transcripts with well established roles in memory mechanisms.
In addition to camk2a, two other genes were confirmed by in situ hybridization, nlgn1 and rasgrp1, both uniquely induced within the CA3 region. Neither rasgrp1 nor nlgn1 have been experimentally implicated in memory formation previously; however, their brain distribution and cellular functions are biologically consistent with such a finding. Indeed rasgrp1, a diacylglycerol and calcium-responsive activator of the oncogene Ras (25) may be a novel link between molecules activated in behavioral paradigms such as phospholipase C and the well known Ras–MAPK pathway (26).
Changes in transcript levels in response to learning may occur through mechanisms that alter mRNA stability but more likely require modulation of gene transcription, particularly because learning is known to activate a number transcription factors (e.g., CREB). Transcription of new mRNA may occur, in part, to provide a substrate for proteins that are used in signaling and pathways for induction of plasticity; however, our follow-up work reported here focused on gene transcripts potentially needed to stabilize and maintain a neural basis for long-lasting storage of information. Neuroligins are developmentally important, transsynaptic cell adhesion molecules. Recent work strongly supports a hypothesis in which neuroligins direct the stabilization and specification of newly formed synapses, but not synapse formation per se (27, 28). Induction of nlgn1 with learning, as we have described here, could contribute to the stabilization of functional connections, newly formed or modified, which are needed to consolidate new information in memory. We tested a potential requirement for nlgn1 expression in memory processes by inhibiting the mRNA by using a published nlgn1 siRNA sequence (29). The protocol, designed as a single administration of the siRNA just before training, was intended to block newly synthesized mRNA but not to chronically deplete existing protein. This acute treatment had no effect on acquisition performance during training, but clearly impaired the ability to remember the platform location 48 h later. These data demonstrate experimentally that induction of nlgn1 with a behavioral protocol is relevant to the biological mechanisms that support memory at a long retention interval.
This study has shown that microarray profiling can be used to detect global changes in gene expression in a relevant hippocampal subregion in response to spatial learning. Confirming differential expression of specific genes in biologically independent samples demonstrates the reproducibility of novel changes while the siRNA inhibition of nlgn1 extends such changes to an experimental approach for testing the functional significance of such genes in memory formation. Although much of the data implicating individual genes or pathways is preliminary, the findings reported here offer many avenues to pursue in determining important molecular players in memory acquisition and consolidation within this model system.
Methods
Detailed methods can be found in SI Text.
Animal Subjects.
Young (3–4 months old) male Long-Evans rats (Charles River Laboratories) were individually housed on a 12:12-hr light/dark cycle with ad libitum access to food and water. Animal procedures were conducted in accordance with approved institutional animals care procedures and National Institutes of Health guidelines, and all efforts were made to reduce the number of animals used.
Behavioral Procedure.
All rats received the same standard water maze spatial training as detailed by Gallagher et al. (16). After 2–3 weeks, the rats were given a single training session [eight trials with 8-min intertrial interval (ITI)] in a new water maze environment located at a different site. The LA group received training in the presence of orienting spatial cues and in which a visible escape platform remained at the same location. The CTL group received training in which the location of the visible platform varied across the trials and the environment lacked any informative orienting cues. One hour after the last training trial, all rats were given 90-s probe trial without the escape platform. Data were analyzed with a video tracking system (HVS Image Analyzing VP-116) and an IBM computer with software developed by HVS Imaging.
Microarray Hybridization and Analysis.
Rats were killed immediately after the probe trial. The CA1, CA3, and DG were microdissected from the hippocampus, and total RNA was extracted by homogenization in TRIzol reagent (Invitrogen) followed by application to Qiagen RNeasy columns. RNA samples were sent to the Johns Hopkins Microarray core facility for cRNA labeling, and hybridization to Affymetrix RAE230A microarrays used standard Affymetrix-recommended procedures.
All quality-control, normalization, differential expression, and exploratory analysis of microarray data were performed by using the open-source R statistical language (www.r-project.org). The quality of microarray data were assessed on many levels, resulting in the omission of two hybridizations from the analysis, leaving n = 6 in the CTL and LA groups in the CA1 data and n = 7 in each of the two groups in the CA3 and DG data. The gcrma package in Bioconductor (www.bioconductor.org) (18) was used to normalize microarray data, and mild biases in mean ratios across intensity were balanced by using a loess function in R. For global analysis each sample was visualized as a single point in 2D space. Using all 15,923 gene expression measures generated from each microarray, the pairwise correlation (r) between all possible sample pairs was calculated, and an MDS algorithm was used to represent all pairwise distances (defined to be 1 − r). SAM d-statistics (19) coupled with a low-intensity cut-off were used to assess differential expression across the two groups of animals. An FDR was calculated by comparing the observed differential expression statistics to those expected by chance (estimated by permuting the group labels of the data many times and recalculating differential expression statistics).
Differential Expression of Functional Gene Groups.
The assessment of differential expression in defined groups of functionally related genes (Fig. 2C and Table S3) was carried out by using the implementation of the Wilcox rank sum test in the geneSetTest function in the limma package in Bioconductor (30). As shown in Fig. 2C, we interrogated a single group of 41 a priori defined genes (represented by 71 probesets on the array) known to be trancriptional targets after hippocampal LTP as defined by published literature (Table S2). For additional functional analysis, probesets measuring genes within groups defined by The Gene Ontology, The Kyoto Encyclopedia of Genes and Genomes, and the Pfam databases were organized into 2,680 partially overlapping functional groups and assessed as before by using geneSetTest. Although P values listed in Table S3 (5 ≤ n < 500) are uncorrected for multiple comparisons, all exceed a Bonferroni correction α = 0.01.
In Situ Hybridization.
Probe templates were synthesized de novo by PCR from whole hippocampal RNA and modified to contain SP6 and T7 RNA polymerase binding sites. Thirty-micrometer sections from perfused brains were taken through the hippocampus (LA: n = 7; CTL: n = 6) and stored free-floating in 4% paraformaldehyde until hybridization with 35S-UTP-labeled probe generated from the templates. Hybridized sections were exposed to film and quantified by using NIH Image. Detailed methods can be found in SI Text.
siRNA Behavioral Experiment.
Rats received pretraining as detailed above before bilateral cannulae implantation into the hippocampus. Coordinates for the guide cannulae were 3.8 mm posterior to bregma, 3.0 mm lateral to midline, and 3.3 mm ventral to the skull surface. Internal cannula used for injection protruded 0.5 mm beyond the tip of the guide cannula. Rats were allowed 2 weeks to recover before being trained in a new water maze environment. On the day of training, each rat received bilateral injections of either nlgn1 siRNA (29), scrambled siRNA, or saline. Lyophilized siRNA was resuspended in sterile saline at a concentration of 1 mM for the injections. The injection volume was 1 μl per side (0.25 μl·min−1). Two hours after injection, rats received eight training trials (60 s each; ITI 8 min) to locate a hidden escape platform in the water maze. The retention interval was 48 h; no injection was given before the probe test (120 s). At the end of the experiment, all cannula tracks were verified by histology (Fig. S4).
Supplementary Material
Acknowledgments.
We thank Alexandra Lunt, Jubeen Moaven, Joseph McQuail, and Weidong Hu for technical assistance, Giovanni Parmigiani for bioinformatics consultation, and Peter Rapp for helpful comments on the manuscript. This work was supported by National Institute on Aging/National Institutes of Health Grant P01AG09973.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804292105/DCSupplemental.
References
- 1.Lee I, Yoganarasimha D, Rao G, Knierim JJ. Comparison of population coherence of place cells in hippocampal subfields CA1 and CA3. Nature. 2004;430:456–459. doi: 10.1038/nature02739. [DOI] [PubMed] [Google Scholar]
- 2.Leutgeb JK, Leutgeb S, Moser M-B, Moser EI. Pattern separation in the dentate gyrus and CA3 of the hippocampus. Science. 2007;315:961–966. doi: 10.1126/science.1135801. [DOI] [PubMed] [Google Scholar]
- 3.Leutgeb S, Leutgeb JK, Treves A, Moser M-B, Moser EI. Distinct ensemble codes in hippocampal areas CA3 and CA1. Science. 2004;305:1295–1298. doi: 10.1126/science.1100265. [DOI] [PubMed] [Google Scholar]
- 4.Vazdarjanova A, Guzowski JF. Differences in hippocampal neuronal population responses to modifications of an environmental context: Evidence for distinct, yet complementary, functions of CA3 and CA1 ensembles. J Neurosci. 2004;24:6489–6496. doi: 10.1523/JNEUROSCI.0350-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lynch MA. Long-term potentiation and memory. Physiol Rev. 2004;84:87–136. doi: 10.1152/physrev.00014.2003. [DOI] [PubMed] [Google Scholar]
- 6.Raymond CR. LTP forms 1, 2 and 3: Different mechanisms for the “long” in long-term potentiation. Trends Neurosci. 2007;30:167–175. doi: 10.1016/j.tins.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 7.Bourtchouladze R, et al. Different training procedures recruit either one or two critical periods for contextual memory consolidation, each of which requires protein synthesis and PKA. Learn Mem. 1998;5:365–374. [PMC free article] [PubMed] [Google Scholar]
- 8.Igaz LM, Vianna MR, Medina JH, Izquierdo I. Two time periods of hippocampal mRNA synthesis are required for memory consolidation of fear-motivated learning. J Neurosci. 2002;22:6781–6789. doi: 10.1523/JNEUROSCI.22-15-06781.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bailey CH, Montarolo P, Chen M, Kandel ER, Schacher S. Inhibitors of protein and RNA synthesis block structural changes that accompany long-term heterosynaptic plasticity in Aplysia. Neuron. 1992;9:749–758. doi: 10.1016/0896-6273(92)90037-e. [DOI] [PubMed] [Google Scholar]
- 10.Pfenning A, Schwartz R, Barth A. A comparative genomics approach to identifying the plasticity transcriptome. BMC Neurosci. 2007;8:20. doi: 10.1186/1471-2202-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hevroni D, et al. Hippocampal plasticity involves extensive gene induction and multiple cellular mechanisms. J Mol Neurosci. 1998;10:75–98. doi: 10.1007/BF02737120. [DOI] [PubMed] [Google Scholar]
- 12.Cavallaro S, D'Agata V, Manickam P, Dufour F, Alkon DL. Memory-specific temporal profiles of gene expression in the hippocampus. Proc Natl Acad Sci USA. 2002;99:16279–16284. doi: 10.1073/pnas.242597199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robles Y, et al. Hippocampal gene expression profiling in spatial discrimination learning. Neurobiol Learn Mem. 2003;80:80–95. doi: 10.1016/s1074-7427(03)00025-x. [DOI] [PubMed] [Google Scholar]
- 14.Dubnau J, et al. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr Biol. 2003;13:286–296. doi: 10.1016/s0960-9822(03)00064-2. [DOI] [PubMed] [Google Scholar]
- 15.Guzowski JF, Knierim JJ, Moser EI. Ensemble dynamics of hippocampal regions CA3 and CA1. Neuron. 2004;44:581–584. doi: 10.1016/j.neuron.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 16.Gallagher M, Burwell R, Burchinal M. Severity of spatial learning impairment in aging: Development of a learning index for performance in the Morris water maze. Behav Neurosci. 1993;107:618–626. doi: 10.1037//0735-7044.107.4.618. [DOI] [PubMed] [Google Scholar]
- 17.Kandel ER. The molecular biology of memory storage: A dialogue between genes and synapses. Science. 2001;294:1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- 18.Irizarry RA, et al. Exploration, normalization, and summaries of high-density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 19.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts LA, et al. Increased expression of dendritic mRNA following the induction of long-term potentiation. Brain Res Mol Brain Res. 1998;56:38–44. doi: 10.1016/s0169-328x(98)00026-6. [DOI] [PubMed] [Google Scholar]
- 21.Bannerman DM, et al. Double dissociation of function within the hippocampus: A comparison of dorsal, ventral, and complete hippocampal cytotoxic lesions. Behav Neurosci. 1999;113:1170–1188. doi: 10.1037//0735-7044.113.6.1170. [DOI] [PubMed] [Google Scholar]
- 22.Lanahan A, Worley P. Immediate-early genes and synaptic function. Neurobiol Learn Mem. 1998;70:37–43. doi: 10.1006/nlme.1998.3836. [DOI] [PubMed] [Google Scholar]
- 23.Huang KP, et al. Neurogranin/RC3 enhances long-term potentiation and learning by promoting calcium-mediated signaling. J Neurosci. 2004;24:10660–10669. doi: 10.1523/JNEUROSCI.2213-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Havekes R, Nijholt IM, Luiten PGM, Van der Zee EA. Differential involvement of hippocampal calcineurin during learning and reversal learning in a Y-maze task. Learn Mem. 2006;13:753–759. doi: 10.1101/lm.323606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stone JC. Regulation of Ras in lymphocytes: Get a GRP. Biochem Soc Trans. 2006;34:858–861. doi: 10.1042/BST0340858. [DOI] [PubMed] [Google Scholar]
- 26.Buckley CT, Caldwell KK. Fear conditioning is associated with altered integration of PLC and ERK signaling in the hippocampus. Pharmacol Biochem Behav. 2004;79:633. doi: 10.1016/j.pbb.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 27.Varoqueaux F, et al. Neuroligins determine synapse maturation and function. Neuron. 2006;51:741–754. doi: 10.1016/j.neuron.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 28.Chubykin AA, et al. Activity-dependent validation of excitatory versus inhibitory synapses by neuroligin-1 versus neuroligin-2. Neuron. 2007;54:919–931. doi: 10.1016/j.neuron.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chih B, Engelman H, Scheiffele P. Control of excitatory and inhibitory synapse formation by neuroligins. Science. 2005;307:1324–1328. doi: 10.1126/science.1107470. [DOI] [PubMed] [Google Scholar]
- 30.Smyth G. K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.