Abstract
The clonal expansion of mutant cells is hypothesized to be an important first step in cancer formation. To understand the earliest stages of tumorigenesis, a method to identify and analyze clonal expansion is needed. We have previously described transgenic Fluorescent Yellow Direct Repeat (FYDR) mice in which cells that have undergone sequence rearrangements (via homologous recombination events) express a fluorescent protein, enabling fluorescent labeling of phenotypically normal cells. Here, we develop an integrated one- and two-photon imaging platform that spans four orders of magnitude to permit rapid quantification of clonal expansion in the FYDR pancreas in situ. Results show that as mice age there is a significant increase in the number of cells within fluorescent cell clusters, indicating that pancreatic cells can clonally expand with age. Importantly, >90% of fluorescent cells in aged mice result from clonal expansion, rather than de novo sequence rearrangements at the FYDR locus. The spontaneous frequency of sequence rearrangements at the FYDR locus is on par with that of other classes of mutational events. Therefore, we conclude that clonal expansion is one of the most important mechanisms for increasing the burden of mutant cells in the mouse pancreas.
Keywords: aging, cancer, homologous recombination, imaging, pancreas
Cancer is caused by the accumulation of mutations within a single cell lineage. This multistep process occurs through successive rounds of clonal expansion and selection of cells that have acquired mutations that confer growth advantages (1–4). Although the clonal expansion of premalignant cells is hypothesized to be an important precursor to the development of cancer (3), no methods have been developed for studying clonal expansion within intact histologically normal tissue.
Mitotic homologous recombination (HR) events are an important class of mutations that can promote tumorigenesis (5). During the repair of DNA double strand breaks by HR, homologous DNA is used as a template for repair (for review see ref. 6). Although HR is considered to be error-free, recombination between misaligned sequences can occur, resulting in deleterious sequence rearrangements (5). Given the inherent risk of recombinational repair of DNA damage, it is not surprising that genetic and environmental factors that stimulate HR are risk factors for cancer (7–15).
To study HR in vivo, we used transgenic Fluorescent Yellow Direct Repeat (FYDR) mice, in which a HR event at an integrated transgene results in expression of a fluorescent protein (16). Our previous work shows that recombinant cells accumulate in the pancreas of FYDR mice with age as the result of de novo recombination events. In addition, results suggest that clonal expansion of previously existing recombinant cells may also contribute to the accumulation of recombinant cells (17). The extent of clonal expansion had been assessed by using standard wide-field fluorescence microscopy, but because of the inherent limitations of standard microscopy when imaging thick tissues, it was not possible to determine the number of recombinant cells per focus (i.e., per cluster of recombinant fluorescent cells). Thus, the extent of clonal expansion, an important precursor to tumor formation, could not be quantified.
Whereas wide-field fluorescence microscopy excites fluorophores based on one-photon absorption, in two-photon microscopy fluorophores are excited by the simultaneous absorption of two photons, enabling in-depth resolved imaging with micrometer-scale resolution (18). However, although two-photon microscopy offers 3D imaging of tissues, analysis of significant tissue volumes (e.g., >1 cm3) is prohibitively data intensive and slow (19), making this a suboptimal approach for the detection and quantification of rare fluorescent objects within a relatively large region of tissue (20).
Here, we have combined the speed of traditional wide-field fluorescence microscopy with the 3D resolution of two-photon microscopy to create a quantitative method for the analysis of rare fluorescent objects within intact tissue. Specifically, we applied wide-field fluorescence microscopy to rapidly map recombinant foci throughout the tissue, and subsequently applied two-photon microscopy to perform high-resolution 3D imaging and analysis. This integrated one- and two-photon imaging platform was then used to study the effect of aging on HR in the pancreas. Results reveal that as mice age recombinant pancreatic cells undergo surprisingly extensive clonal expansion and that, indeed, the vast majority of recombinant cells in aged animals arose via cell division, rather than via de novo sequence rearrangements. It is noteworthy that the spontaneous frequency of sequence rearrangements at the FYDR locus is on par with that of loss of heterozygosity events (21), making this a powerful approach for evaluating the relative contribution of de novo sequence rearrangements and cell division to the ultimate burden of mutant cells.
Results
Development of Two-Photon Imaging for Analysis of Recombinant Foci.
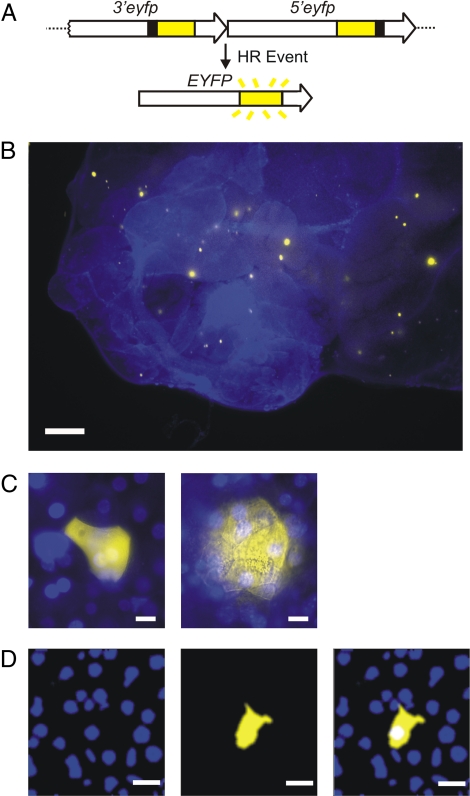
FYDR mice carry a direct repeat substrate for which HR events can restore full-length enhanced yellow fluorescent protein (EYFP) coding sequence. These sequence rearrangements, or mutations, result in the appearance of fluorescent cells (16) (Fig. 1A). To identify recombinant cells within intact pancreata of FYDR mice, we had previously developed an in situ imaging method using traditional wide-field fluorescence microscopy (17) (Fig. 1B).
Fig. 1.
FYDR system and analysis of fluorescent foci in mouse pancreata. (A) Arrangement of the FYDR transgenic recombination substrate. Large arrows indicate expression cassettes; yellow boxes show coding sequences; black boxes indicate positions of deleted sequences (deletion sizes not to scale). (B) Traditional wide-field fluorescence microscopy image of freshly excised pancreatic tissue. Overlay of EYFP (510–560 nm) and UV (420 nm) images. Brightness/contrast were optimized for each image. (C) Traditional wide-field fluorescence microscopy images of representative recombinant cell clusters in freshly excised tissue. Images show overlays of EYFP (510–560 nm) and UV (420 nm) images. (Left) Focus on the surface of the tissue sample. (Right) Focus below the surface. Brightness/contrast were optimized for each image. (D) A 2D plane imaged through a representative focus using two-photon microscopy. (Left) The blue channel images the nuclei stained with Hoechst. (Center) The yellow channel images the EYFP-expressing recombinant cell. (Right) Overlay of blue and yellow channels. Note that all images are pseudocolored. (Scale bars: B, 1 mm; C, 10 μm; D, 20 μm.)
Similar to microbial colony counting for mutation analysis where each colony is derived from a single mutation event, the number of recombination events per pancreas can be estimated by counting fluorescent foci detected within in situ images. Furthermore, the total number of recombinant cells per pancreas can be determined by evaluating the number of cells contained within all fluorescent foci. However, given the limitations of standard wide-field fluorescence microscopy, only recombinant foci located on the surface of the pancreas have distinct boundaries, enabling estimation of the number of cells within the focus (Fig. 1C Left). In contrast, for foci located below the surface, light scattering blurs the cell boundaries, making it impossible to quantify the number of cells per focus (Fig. 1C Right). Thus, to determine the number of cells contained within fluorescent recombinant foci we developed a two-photon method to create images of cross-sections of recombinant foci. By overlaying images of Hoechst stained nuclei (Fig. 1D Left) and yellow fluorescent cytoplasm (Fig. 1D Center), the number of individual cells (≈20–30 μm in diameter) contained within a single cross-section of the recombinant focus can be quantified (Fig. 1D Right). It is noteworthy that traditional microscopy often distorts the apparent size of a focus such that foci that are actually small can appear large and vice versa [see supporting information (SI) Fig. S1].
Quantification of Cell Number per Recombinant Focus Using Two-Photon Microscopy.
For our studies, the number of recombinant cells per focus is the most important parameter. To quantify the number of recombinant cells per focus, we extended an image analysis algorithm developed previously (22). Briefly, the image stack of the tissue region harboring EYFP expressing cells (yellow fluorescence) is spatially coregistered with Hoechst labeled nuclei (blue fluorescence). For each 2D plane in the 3D image stack, the segmentation of the EYFP-expressing tissue region is accomplished by an intensity thresholding method. We used a watershed-based algorithm to segment adjacent nuclei to obtain a more reliable estimate of the number of nuclei (which was verified by independent manual counting).
Counting the number of nuclei in a 3D cluster involves identification of nuclei in multiple planes. We applied our algorithm to identify and count the number of nuclei that fall within the confines of the EYFP-labeled cytosol within each plane. Subsequently, the centroids of nuclei in adjacent planes were compared. If the centroids of two nuclei on two adjacent planes fell within a critical lateral distance (as determined by the known typical nuclear size), they were marked as being from the same nucleus. This procedure ensures that the same nucleus is not counted redundantly in adjacent planes, enabling the accurate quantification of the number of recombinant cells per focus.
Integrating One- and Two-Photon Imaging Platforms.
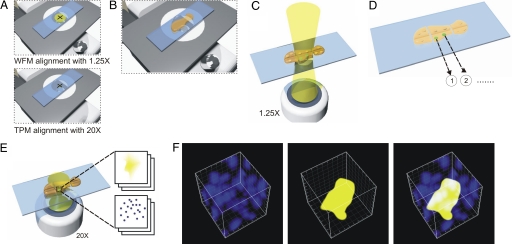
Whereas two-photon microscopy allows for effective quantification of the number of cells per recombinant focus (≈8 × 10−6 mm3), analyzing the entire mouse pancreas (≈100 mm3) would take >70 h. Therefore, to make it possible to study multiple mice under different conditions, we created an integrated wide-field fluorescence microscopy and two-photon microscopy imaging system that enables rapid data collection for both the frequency of recombinant foci and the number of cells within foci. The first step of data acquisition requires the coregistration and alignment of the fields of view for the ×1.25 wide-field fluorescence microscopy and ×20 two-photon microscopy objectives (Fig. 2A). Unfixed, freshly excised samples of pancreata are then uniformly compressed to a thickness of 0.5 mm and mounted on an automated specimen stage (Fig. 2B). The entire surface of the pancreas is then imaged under a low-powered objective using traditional wide-field fluorescence microscopy (Fig. 2C). Fluorescent recombinant foci are identified, and their x,y coordinates are mapped within the composite images (Fig. 2D). Subsequently, centering on the coordinates of the previously mapped positions, two-photon microscopy is used to create multiple sequential z-planes throughout the thickness of a given focus (Fig. 2E). To note, foci vary in thickness from ≈10 μm to >100 μm. The penetration depth of two-photon microscopy in pancreatic tissue is ≈200 μm, thus limiting our analysis to foci in positions within 200 μm of the tissue surface. In the final step, the images are deconvoluted to produce a 3D reconstruction of the focus, differentiating Hoechst-stained nuclei and EYFP (Fig. 2F). Thus, the combination of wide-field fluorescence microscopy and two-photon microscopy enables the identification and analysis of recombinant foci over the entire surface of a pancreas.
Fig. 2.
Imaging procedure for 3D site-selective tissue cytometry. (A) Alignment: Using a centering slide containing a cross-hair target, the field of view of wide-field fluorescence microscopy (WFM) and two-photon microscopy (TPM) are aligned. (B) Sample mounting: Each pancreas sample is compressed between two glass coverslips to a thickness of 0.5 mm. (C) Wide field microscopy: A series of wide-field fluorescence microscopy images are taken and are montaged to map the foci in the tissue. (D) Recording sites of interest: Positions of the foci are recorded. (E) Two-photon microscopy of sites of interest: A series of two-photon microscopy image stacks are taken at each focus location by using an automated x-y stage. (F) 3D reconstructed image cubes of a representative focus containing seven cells are shown for the blue channel (Left), the yellow channel (Center), and an overlay of blue and yellow channels (Right). The edge length of this cube is 50 μm.
The Frequency and the Sizes of Fluorescent Foci Increase with Age.
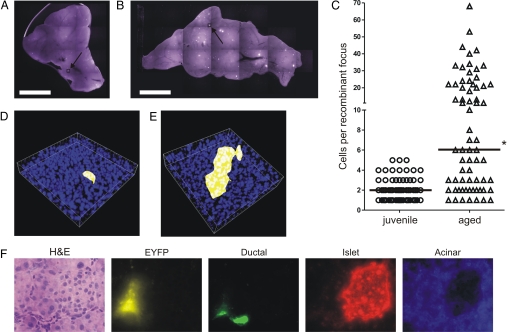
To study the effect of aging on recombinant cell frequency, we analyzed pancreata from two age groups: “juvenile” (4–5 weeks old) and “aged” (74–83 weeks old). Representative composite images collected using wide-field fluorescence microscopy show that both the frequency and the apparent sizes of the foci appear to increase with age (compare Fig. 3 A and B), which is consistent with our previous studies (see ref. 17). Two-photon analysis shows that the median number of cells per focus is statistically significantly higher in aged compared with juvenile mice, with six versus two cells per focus, respectively (Fig. 3C, note y axis scale). Of the foci analyzed in pancreata of 10 juvenile mice, the largest recombinant focus contained five cells (Fig. 3D). In contrast, ≈54% of recombinant foci analyzed in four aged mice contain more than five cells (Fig. 3C), with the largest focus containing 68 cells (Fig. 3E). Given that recombination at the FYDR substrate within pancreatic cells is a rare event [≈5 per million (17)], the probability that two neighboring recombinant cells within a large focus occurred as the result of two independent recombination events is extremely unlikely. Thus, these data clearly demonstrate that recombinant pancreatic cells clonally expand with age.
Fig. 3.
Effects of aging on the size of recombinant cell clusters in the pancreas. (A and B) Wide-field fluorescence microscopy images of pancreata from a juvenile (A) and an aged (B) mouse. Examples of pancreata with the highest number of recombinant foci for each cohort are shown. Arrows indicate cell clusters containing the most recombinant cells for each cohort. Brightness/contrast were optimized for each image. (C) Number of cells per recombinant focus as determined by two-photon microscopy for foci from 10 juvenile (n = 79) and 4 aged (n = 68) mice. *, Aged cohort is statistically significantly higher than juvenile cohort (P < 0.0001). Medians are indicated by black bars. (D) 3D reconstruction of the largest fluorescent recombinant focus (five cells) found in pancreata of 10 juvenile FYDR mice imaged by two-photon microscopy. The dimensions of this 3D volume are 240 × 240 × 41 μm along x, y, and z directions. (E) 3D reconstruction of the largest fluorescent recombinant focus (68 cells) found in pancreata of four aged FYDR mice imaged by two-photon microscopy. The dimension of this 3D reconstructed volume is 240 × 240 × 53 μm along x, y, and z directions. (F) Analysis of cell type of fluorescent foci in mouse pancreata. Images show one representative focus for (left to right): H&E staining; recombinant focus taken using an EYFP (510–560 nm) filter; immunohistochemical staining for ductal cells (DBA lectin); immunohistochemical staining for islet cells (α-insulin antibody); and immunohistochemical staining for acinar cells (α-amylase antibody). Brightness/contrast were optimized for each image. Note that images are pseudocolored. (Scale bars: A and B, 5 mm.) (Magnification: F, ×40.)
The pancreas contains multiple cell types, including ductal, acinar, and islet cells, which are able to express the FYDR substrate (17). Previously, we had analyzed cells within 43 recombinant foci from FYDR mice, and all were found to be acinar cells (17). Given that ≈80% of the pancreas is composed of acinar cells (23), we wondered whether additional analysis would reveal recombinant islet and/or ductal cells. An additional 78 foci were analyzed in frozen sections from aged FYDR mice to amass data from >100 independent foci. The cell types within these sections were determined by using antibodies specific for ductal, islet, and acinar cells (Fig. 3F). All of the recombinant foci analyzed contained acinar cells (Fig. 3F). Occasionally, we observed foci that appeared to contain both acinar and ductal cells (6 of 78 foci), but we never observed recombinant ductal cells in isolation. Thus, the vast majority of fluorescent cells seen within pancreata of FYDR mice are acinar cells that have undergone a recombination event at the FYDR substrate.
Discussion
For some tumors, it may take 20–40 years from the initial formation of a cancer progenitor cell until the appearance of a detectable tumor (24–27). During this time, the population of precancerous cells can increase as the result of clonal expansion (3). However, these clonal cell populations can appear to be phenotypically normal, even though they have acquired mutations in tumor suppressors or oncogenes that may provide a survival and proliferative advantage. The ability to detect clonal expansion in situ within histologically normal tissue enables the identification of these clonal cell populations, and studying these cells may provide insights into of the earliest stages of cellular transformation, before the clinical appearance of a tumor.
Here, we have combined the ability of 2D wide-field fluorescence microscopy to rapidly identify rare fluorescent objects with the high resolution of 3D two-photon microscopy to develop an imaging platform that rapidly identifies rare fluorescent cells within a tissue (centimeter scale) and subsequently analyzes these cells at high resolution (micrometer scale). Using the FYDR mice, it is now possible to detect the accumulation of cells harboring rare sequence rearrangements while simultaneously studying the extent to which conditions foster clonal expansion of such mutant cells. Although a few studies have analyzed clonal expansion of mutant cells within tissue sections (28–30), most other mutation assays require tissue disaggregation followed by analysis of either cultured primary cells or genomic DNA [e.g., RMC, Aprt, Tk, Big Blue, Muta Mouse, GptΔ (31–34)], thus limiting information about the clonal relationship among mutant cells. In addition, being able to detect a single fluorescent cell within an entire pancreas [≈3 × 107 cells (35)], makes this imaging technique one of the most sensitive methods for detecting rare DNA sequence changes. Taken together, integrated one- and two-photon imaging provides a highly sensitive method for detecting and analyzing cells that contain small sequence rearrangements within intact tissues.
Although the formation of new mutations can promote tumorigenesis, it has been suggested that susceptibility to clonal expansion of preexisting mutant cells is also important to carcinogenesis (3). Here, results show that, if anything, clonal expansion plays a more important role in contributing to the burden of cells harboring a mutation at a particular locus than do de novo sequence rearrangements. More specifically, in this study we observed that within the 68 recombinant foci analyzed in aged mice, there were a total of 898 recombinant cells. Given that each independent focus originated from a single recombination event, then 68 of these 898 recombinant cells were caused by de novo recombination events and 830 of the 898 cells arose as the result of clonal expansion of previously existing recombinant cells. Therefore, >90% of recombinant cells in aged animals are the result of clonal expansion. Together, these data show that clonal expansion is by far the most significant contributor to the overall increase in the number of pancreatic cells harboring DNA sequence rearrangements with age.
Measuring HR events provides information on both the formation of double-strand breaks and the ability of cells to use HR as a repair pathway. Although HR is generally error-free, recombination between misaligned sequences can cause insertions, deletions, and translocations that promote cancer. In fact, the FYDR mice specifically detect sequence rearrangements that result from misalignments at a repeat sequence (for mechanisms, see ref. 36), which occur not only at the FYDR locus, but throughout the genome [of which ≈40% is composed of repeat elements (37)]. In addition, the results of these studies can be generalized to other classes of mutations. For example, the recombination events captured by the FYDR substrate occur at a frequency that is very similar to the frequency of loss of heterozygosity (21) and the frequency of spontaneous mutations within a gene (31). Thus, one would expect to see similar accumulations of other known tumor-promoting sequence changes, such as loss of heterozygosity events and points mutations, in pancreatic cells with age.
Within the FYDR pancreas, recombinant cells are most likely acinar cells. Acinar cells are part of the exocrine pancreas and secrete hydrolytic enzymes into the duodenum that aid in digestion. The secretory unit within the exocrine pancreas is a grape-like structure called an acinus, which contains 10–40 acinar cells that secrete enzymes into a system of epithelial ducts (38). Interestingly, acinar cells appear to play an important role in tumor formation. Evidence suggests that acinar to ductal transdifferentation can occur in vivo and in vitro, leading to the formation of pancreatic intraepithelial neoplasias and pancreatic ductal adenocarcinoma (39–45). In the adult pancreas, acinar cells exhibit extremely low levels of proliferation [≈1% of cells are in S phase (46)] and persist for long periods of time [turnover time of mouse acinar cells is estimated to be ≈500 days (47)]. Thus, the dramatic accumulation of recombinant acinar cells with age is most likely caused by a combination of de novo recombinant events, clonal expansion, and a relatively long persistence of acinar cells within the tissue. Here, we observed one recombinant focus from an aged mouse that contained 68 cells, indicating that significant clonal expansion (at least seven population doublings) can occur within the acinar cell compartment of the adult pancreas. These data provide additional information regarding the proliferative capacity of adult acinar cells in vivo, and it will be interesting to further apply these techniques to learn more about how injury affects clonal expansion of pancreatic cells.
It is well established that mutations cause cancer. Thus, it is of great interest to know what causes mutations in mammals. In the absence of technologies that reveal clonal expansion, our attention has generally been directed toward de novo mutagenesis, and relatively few studies have asked the relative importance of de novo mutagenesis compared with mutations that arise as a consequence of clonal expansion from preexisting mutant cells. With the development of these approaches that combine 2D wide-field fluorescence microscopy and 3D two-photon microscopy imaging, mutant cells can be visualized within intact tissue. Here, we show that in at least one mammalian tissue the major underlying cause of a high burden of mutant cells is not de novo mutagenesis but rather clonal expansion. It will be interesting to learn about the underlying causes of such clonal expansion; of interest is the possibility that injury and/or infection could promote cell division. Taken together, the techniques described here thus open the doors to studies of how our genes and environment impact one of the most significant mechanisms of mutation accumulation in the pancreas: clonal expansion.
Materials and Methods
One- and Two- Photon Microscopy Platform.
Wide-field fluorescence microscopy and two-photon microscopy are combined into a single system as shown in Fig. 2. The instrument is implemented on a Zeiss Axiovert 110 microscope. An automated x-y stage (H101; Prior Scientific) allows stage scanning. A control computer coordinates both the image acquisition electronics of both wide-field fluorescence microscopy and two-photon microscopy and automated specimen stage.
Wide-field fluorescence microscopy uses a green diode laser (BTG-2S; Beam of Light Technologies) as its excitation source. The excitation beam is magnified by a beam expander and is focused at the back focal plane of the ×1.25, 0.04-NA objective (Plan Apo; Olympus), producing uniform illumination of the specimen. A Pixera Pro 150ESM Pander CCD camera acquires wide-field images with a field of view of 4.7 × 6 mm. The typical image integration time is 10 s per frame.
The two-photon microscope uses a femtosecond Ti:Sapphire laser, (Tsunami; Spectra Physics) as its light source. The typical power at the specimen is <25 mW. The laser beam is raster scanned across the sample plane with an x-y galvanometric scanner (Cambridge Technology) producing 256 × 256-pixel images. The expanded laser beam reflected by a dichroic mirror overfills the back aperture of the microscope objective. A Zeiss ×20, 0.75-NA objective was used with a field of view of 240 × 240 μm. The fluorescence signal is collected by the objective and is transmitted through the dichoric mirror and the IR blocking filter. The signal is separated into blue and green channels via an appropriate filter combination to detect Hoechst and YFP fluorescence. These signals are measured by photomultiplier detectors (R3896; Hamamatsu) using single photon counting electronics and are recorded by the control computer. A piezoelectric objective translator (Jena) translates the objective axially over 300-μm range for axial scanning. The control computer reconstructs 3D image stacks by correlating the measured signal intensity with the known x-y-z raster scan pattern. The typical image acquisition time is ≈4 s for a 2D image slice.
Mice.
Ten Juvenile (4–5 weeks old) and 4 aged (74–83 weeks old) female C57BL/6 FYDR mice (28) were retroorbitally injected with 0.04 mg/g body weight of 5 mg/ml Hoechst 33342 (Sigma–Aldrich) in PBS. Twenty minutes after injection, mice pancreata were isolated and placed in ice-cold PBS containing 0.01% soybean trypsin inhibitor (Sigma–Aldrich). Whole pancreata were pressed between glass slides separated by 0.5-mm spacers and imaged as described above.
Immunohistochemistry.
Immunohistochemistry was performed on frozen sections (5 μm thick) of pancreas taken from an aged (73 weeks old) female FYDR mouse. Slides were dried at room temperature for 30 min, fixed in 10% formalin for 10 min, and permeabilized for 10 min in 0.2% Triton X-100, and 0.05% Tween 20 in TBS, and blocking was performed for 30 min in 1% BSA in TBS. Sections were incubated with primary antibodies against amylase (rabbit, 1:200; Sigma–Aldrich) for acinar cells (48) and against insulin (guinea pig, prediluted 30.5 mg/liter; Invitrogen) for islet cells (49) at 4°C in a humidity chamber overnight. After washes with TBS, sections were incubated with secondary antibodies (Alexa Fluor 350 goat anti-rabbit, Alexa Fluor 594 goat anti-guinea pig; Invitrogen) diluted 1:500 for 30 min at room temperature, washed extensively with TBS, and imaged by using a fluorescent microscope. Sections were then stained with DBA lectin (Fluorescein or Biotin conjugated, 1:100; Vector Laboratories), a marker specific for epithelial and ductal cells (50), at room temperature for 30 min. For sections stained with biotinylated DBA, sections were incubated with secondary antibody (streptavidin Alexa Fluor 594; Invitrogen). Sections were washed extensively with TBS and imaged with a fluorescent microscope. After imaging, sections were H&E-stained.
Supplementary Material
Acknowledgments.
We thank the Massachusetts Institute of Technology Division of Comparative Medicine, Dr. Arlin Rogers and Kathleen Cormier for assistance, Dr. Werner Olipitz and Evelyn Park for contributions, Dr. Seok Chung for assistance in 3D drawing, and the Massachusetts Institute of Technology Center for Environmental Health Sciences (National Institute on Environmental Health Sciences Grant P30 ES001209-26A1). This work was primarily supported by National Institutes of Health Grant R33 CA091354 with additional support from Department of Energy Grant FG01-04ER04--21. This work was partly supported by the Singapore–Massachusetts Institute of Technology Alliance. D.M.W.-B. was supported by National Institutes of Health/ National Institute of General Medical Sciences Interdepartmental Biotechnology Training Program Grant GM008334 and National Institutes of Health Grant T32 ES007020.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804346105/DCSupplemental.
References
- 1.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 2.Prehn RT. A clonal selection theory of chemical carcinogenesis. J Natl Cancer Inst. 1964;32:1–17. [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 4.Kim U, Depowski MJ. Progression from hormone dependence to autonomy in mammary tumors as an in vivo manifestation of sequential clonal selection. Cancer Res. 1975;35:2068–2077. [PubMed] [Google Scholar]
- 5.Bishop AJ, Schiestl RH. Role of homologous recombination in carcinogenesis. Exp Mol Pathol. 2003;74:94–105. doi: 10.1016/s0014-4800(03)00010-8. [DOI] [PubMed] [Google Scholar]
- 6.Helleday T, Lo J, van Gent DC, Engelward BP. DNA double-strand break repair: From mechanistic understanding to cancer treatment. DNA Repair (Amst) 2007;6:923–935. doi: 10.1016/j.dnarep.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Chaganti RS, Schonberg S, German J. A manyfold increase in sister chromatid exchanges in Bloom's syndrome lymphocytes. Proc Natl Acad Sci USA. 1974;71:4508–4512. doi: 10.1073/pnas.71.11.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prince PR, Emond MJ, Monnat RJ., Jr Loss of Werner syndrome protein function promotes aberrant mitotic recombination. Genes Dev. 2001;15:933–938. doi: 10.1101/gad.877001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bishop AJ, Kosaras B, Sidman RL, Schiestl RH. Benzo(a) pyrene and x-rays induce reversions of the pink-eyed unstable mutation in the retinal pigment epithelium of mice. Mutat Res. 2000;457:31–40. doi: 10.1016/s0027-5107(00)00118-4. [DOI] [PubMed] [Google Scholar]
- 10.Bill CA, Nickoloff JA. Spontaneous and ultraviolet light-induced direct repeat recombination in mammalian cells frequently results in repeat deletion. Mutat Res. 2001;487:41–50. doi: 10.1016/s0921-8777(01)00101-x. [DOI] [PubMed] [Google Scholar]
- 11.Thompson LH, Schild D. Recombinational DNA repair and human disease. Mutat Res. 2002;509:49–78. doi: 10.1016/s0027-5107(02)00224-5. [DOI] [PubMed] [Google Scholar]
- 12.Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 13.Little MP. Risks associated with ionizing radiation. Br Med Bull. 2003;68:259–275. doi: 10.1093/bmb/ldg031. [DOI] [PubMed] [Google Scholar]
- 14.Brash DE, et al. A role for sunlight in skin cancer: UV-induced p53 mutations in squamous cell carcinoma. Proc Natl Acad Sci USA. 1991;88:10124–10128. doi: 10.1073/pnas.88.22.10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phillips DH. Polycyclic aromatic hydrocarbons in the diet. Mutat Res. 1999;443:139–147. doi: 10.1016/s1383-5742(99)00016-2. [DOI] [PubMed] [Google Scholar]
- 16.Hendricks CA, et al. Spontaneous mitotic homologous recombination at an enhanced yellow fluorescent protein (EYFP) cDNA direct repeat in transgenic mice. Proc Natl Acad Sci USA. 2003;100:6325–6330. doi: 10.1073/pnas.1232231100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiktor-Brown DM, Hendricks CA, Olipitz W, Engelward BP. Age-dependent accumulation of recombinant cells in the mouse pancreas revealed by in situ fluorescence imaging. Proc Natl Acad Sci USA. 2006;103:11862–11867. doi: 10.1073/pnas.0604943103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Göppert-Mayer M. About elementary events with two quantal jumps (Translated from German) Ann Phys. 1931;5:273–294. [Google Scholar]
- 19.So PT, Dong CY, Masters BR, Berland KM. Two-photon excitation fluorescence microscopy. Annu Rev Biomed Eng. 2000;2:399–429. doi: 10.1146/annurev.bioeng.2.1.399. [DOI] [PubMed] [Google Scholar]
- 20.Ragan T, et al. High-resolution whole organ imaging using two-photon tissue cytometry. J Biomed Opt. 2007;12 doi: 10.1117/1.2435626. 014015. [DOI] [PubMed] [Google Scholar]
- 21.Van Sloun PP, et al. Determination of spontaneous loss of heterozygosity mutations in Aprt heterozygous mice. Nucleic Acids Res. 1998;26:4888–4894. doi: 10.1093/nar/26.21.4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim KH, et al. Three-dimensional tissue cytometer based on high-speed multiphoton microscopy. Cytometry A. 2007;71:991–1002. doi: 10.1002/cyto.a.20470. [DOI] [PubMed] [Google Scholar]
- 23.Pour PM, Pandey KK, Batra SK. What is the origin of pancreatic adenocarcinoma? Mol Cancer. 2003;2:13–22. doi: 10.1186/1476-4598-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Voelter-Mahlknecht S, et al. Skin tumors among employees of a tar refinery: The current data and their implications. Int Arch Occup Environ Health. 2007;80:485–495. doi: 10.1007/s00420-006-0155-y. [DOI] [PubMed] [Google Scholar]
- 25.Marinaccio A, Branchi C, Massari S, Scarselli A. National epidemiologic surveillance systems of asbestos-related disease and the exposed workers register. Med Lav. 2006;97:482–487. [PubMed] [Google Scholar]
- 26.Agalliu I, et al. Prostate cancer incidence in relation to time windows of exposure to metalworking fluids in the auto industry. Epidemiology. 2005;16:664–671. doi: 10.1097/01.ede.0000173266.49104.bb. [DOI] [PubMed] [Google Scholar]
- 27.Dropkin G. Low-dose radiation and cancer in A-bomb survivors: Latency and nonlinear dose–response in the 1950–1990 mortality cohort. Environ Health. 2007;6:1–25. doi: 10.1186/1476-069X-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rebel H, et al. Early p53-positive foci as indicators of tumor risk in ultraviolet-exposed hairless mice: Kinetics of induction, effects of DNA repair deficiency, and p53 heterozygosity. Cancer Res. 2001;61:977–983. [PubMed] [Google Scholar]
- 29.Schlake G, et al. Single-cell immunohistochemical mutation load assay (SCIMLA) using human paraffin-embedded tissues. Environ Mol Mutagen. 2003;42:206–215. doi: 10.1002/em.10193. [DOI] [PubMed] [Google Scholar]
- 30.Jonason AS, et al. Frequent clones of p53-mutated keratinocytes in normal human skin. Proc Natl Acad Sci USA. 1996;93:14025–14029. doi: 10.1073/pnas.93.24.14025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bielas JH, et al. Human cancers express a mutator phenotype. Proc Natl Acad Sci USA. 2006;103:18238–18242. doi: 10.1073/pnas.0607057103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hendricks CA, Engelward BP. “Recombomice”: The past, present, and future of recombination-detection in mice. DNA Repair (Amst) 2004;3:1255–1261. doi: 10.1016/j.dnarep.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 33.Nohmi T, Suzuki T, Masumura K. Recent advances in the protocols of transgenic mouse mutation assays. Mutat Res. 2000;455:191–215. doi: 10.1016/s0027-5107(00)00077-4. [DOI] [PubMed] [Google Scholar]
- 34.Lambert IB, Singer TM, Boucher SE, Douglas GR. Detailed review of transgenic rodent mutation assays. Mutat Res. 2005;590:1–280. doi: 10.1016/j.mrrev.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 35.Taga R, Bispo LB, Bordin RA, Hassunuma RM. Morphometric and allometric study of the mouse exocrine pancreas growth during the postnatal life. Okajimas Folia Anat Jpn. 1998;74:271–278. doi: 10.2535/ofaj1936.74.6_271. [DOI] [PubMed] [Google Scholar]
- 36.Jonnalagadda VS, Matsuguchi T, Engelward BP. Interstrand cross-link-induced homologous recombination carries an increased risk of deletions and insertions. DNA Repair (Amst) 2005;4:594–605. doi: 10.1016/j.dnarep.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 37.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 38.Motta PM, Macchiarelli G, Nottola SA, Correr S. Histology of the exocrine pancreas. Microsc Res Tech. 1997;37:384–398. doi: 10.1002/(SICI)1097-0029(19970601)37:5/6<384::AID-JEMT3>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 39.Guerra C, et al. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11:291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 40.Grippo PJ, et al. Preinvasive pancreatic neoplasia of ductal phenotype induced by acinar cell targeting of mutant Kras in transgenic mice. Cancer Res. 2003;63:2016–2019. [PubMed] [Google Scholar]
- 41.Wagner M, et al. Malignant transformation of duct-like cells originating from acini in transforming growth factor transgenic mice. Gastroenterology. 1998;115:1254–1262. doi: 10.1016/s0016-5085(98)70098-8. [DOI] [PubMed] [Google Scholar]
- 42.Wagner M, et al. A murine tumor progression model for pancreatic cancer recapitulating the genetic alterations of the human disease. Genes Dev. 2001;15:286–293. doi: 10.1101/gad.184701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Means AL, et al. Pancreatic epithelial plasticity mediated by acinar cell transdifferentiation and generation of nestin-positive intermediates. Development. 2005;132:3767–3776. doi: 10.1242/dev.01925. [DOI] [PubMed] [Google Scholar]
- 44.Bockman DE, et al. Origin and development of the precursor lesions in experimental pancreatic cancer in rats. Lab Invest. 2003;83:853–859. doi: 10.1097/01.lab.0000074918.31303.5a. [DOI] [PubMed] [Google Scholar]
- 45.Song SY, et al. Expansion of Pdx1-expressing pancreatic epithelium and islet neogenesis in transgenic mice overexpressing transforming growth factor α. Gastroenterology. 1999;117:1416–1426. doi: 10.1016/s0016-5085(99)70292-1. [DOI] [PubMed] [Google Scholar]
- 46.de Dios I, et al. Cell-cycle distribution of pancreatic cells from rats with acute pancreatitis induced by bile-pancreatic obstruction. Cell Tissue Res. 2000;300:307–314. doi: 10.1007/s004410000194. [DOI] [PubMed] [Google Scholar]
- 47.Cameron IL. Cell renewal in the organs and tissues of the nongrowing adult mouse. Tex Rep Biol Med. 1970;28:203–248. [PubMed] [Google Scholar]
- 48.Dor Y, Brown J, Martinez OI, Melton DA. Adult pancreatic β-cells are formed by self-duplication rather than stem-cell differentiation. Nature. 2004;429:41–46. doi: 10.1038/nature02520. [DOI] [PubMed] [Google Scholar]
- 49.Falkmer S. Immunocytochemical studies of the evolution of islet hormones. J Histochem Cytochem. 1979;27:1281–1282. doi: 10.1177/27.9.383830. [DOI] [PubMed] [Google Scholar]
- 50.Kobayashi H, et al. Lectin as a marker for staining and purification of embryonic pancreatic epithelium. Biochem Biophys Res Commun. 2002;293:691–697. doi: 10.1016/S0006-291X(02)00278-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.