Abstract
Members of the Class III homeodomain leucine zipper (Class III HD-Zip) gene family are central regulators of crucial aspects of plant development. To better understand the roles of five Class III HD-Zip genes in rice (Oryza sativa) development, we investigated their expression patterns, ectopic expression phenotypes, and auxin responsiveness. Four genes, OSHB1 to OSHB4, were expressed in a localized domain of the shoot apical meristem (SAM), the adaxial cells of leaf primordia, the leaf margins, and the xylem tissue of vascular bundles. In contrast, expression of OSHB5 was observed only in phloem tissue. Plants ectopically expressing microRNA166-resistant versions of the OSHB3 gene exhibited severe defects, including the ectopic production of leaf margins, shoots, and radialized leaves. The treatment of seedlings with auxin quickly induced ectopic OSHB3 expression in the entire region of the SAM, but not in other tissues. Furthermore, this ectopic expression of OSHB3 was correlated with leaf initiation defects. Our findings suggest that rice Class III HD-Zip genes have conserved functions with their homologs in Arabidopsis (Arabidopsis thaliana), but have also acquired specific developmental roles in grasses or monocots. In addition, some Class III HD-Zip genes may regulate the leaf initiation process in the SAM in an auxin-dependent manner.
In flowering plants, elaboration of the shoot architecture depends primarily on the activity of the shoot apical meristem (SAM). The SAM of seed plants consists of two distinct histological domains: the central zone, with a low cell division rate, and the peripheral zone, where leaf primordia initiate with a high rate of cell division (Steeves and Sussex, 1989). Once a leaf primordium initiates from the peripheral zone of the SAM, it usually displays polarities along the proximal-distal and adaxial-abaxial axes. The SAM in flowering plants first initiates at a fixed position in the embryo after the embryonic pattern is established. These patterning events are universal among these plant species, and the diversity of plant architecture largely depends on spatial and temporal modifications of subsequent events.
To build a highly complex plant form, the precise spatial and temporal expression of regulatory genes is essential. Among several other regulatory genes, members of the Class III homeodomain Leu zipper (Class III HD-Zip) family are regulators of key aspects of plant development. Class III HD-Zip genes encode transcriptional regulators containing a homeodomain for DNA binding and a Leu-zipper domain for dimer formation (Sessa et al., 1998), an N-terminal putative lipid- or steroid-binding START domain (Ponting and Aravind, 1999), and a C-terminal MEKHLA domain that potentially functions as a sensory and/or protein interaction domain (Mukherjee and Bürglin, 2006; Chandler et al., 2007). In Arabidopsis (Arabidopsis thaliana), there are five Class III HD-Zip genes: REVOLUTA (REV)/AMPHIVASAL VASCULAR BUNDLE1 (AVB1), PHABULOSA (PHB), PHAVOLUTA (PHV), CORONA (CNA)/INCURVATA4 (ICU4), and ATHB8. Phenotypic analysis of loss-of-function mutants of these genes has revealed that the genes have overlapping, antagonistic, and distinct roles in development (Emery et al., 2003; Prigge et al., 2005). REV, PHV, and PHB appear to play a role in the establishment of the SAM and the adaxial identity of the lateral organs. Although single mutations in PHB and PHV result in almost normal phenotypes, the triple mutant phb phv rev shows a single radial cotyledon and lack of the SAM in the embryo (Emery et al., 2003; Prigge et al., 2005). Dominant gain-of-function mutants of Class III HD-Zip genes in Arabidopsis show meristem and leaf polarity defects (McConnell and Barton, 1998; McConnell et al., 2001; Zhong and Ye, 2004; Ochando et al., 2006). Consistent with their role in the SAM and the adaxial identity of the lateral organs, all three of these genes are expressed in the SAM and in the adaxial domain of the lateral organs (Zhong and Ye, 1999; McConnell et al., 2001; Otsuga et al., 2001; Emery et al., 2003; Prigge et al., 2005). In contrast, CNA and ATHB8 have functions antagonistic to REV, because mutations in CNA and ATHB8 suppress meristem defects of rev mutants (Prigge et al., 2005).
Class III HD-Zip genes also regulate vascular patterning and differentiation. Loss of REV activity causes defects in the formation of interfascicular fibers that are enhanced by mutations in PHB and PHV (Talbert et al., 1995; Zhong et al., 1997; Zhong and Ye, 1999; Emery et al., 2003; Prigge et al., 2005). Dominant gain-of-function mutants of PHB, PHV, and REV show radialized vascular bundles with xylem surrounding phloem (McConnell and Barton, 1998; Emery et al., 2003; Zhong and Ye, 2004).
Polarized Class III HD-Zip expression and the roles of these genes in lateral organs and vascular patterning depend on posttranscriptional regulation by microRNA165/166 (Reinhart et al., 2002; Rhoades et al., 2002; Tang et al., 2003). Dominant gain-of-function alleles of phb-d, phv-d, rev-d, avb1, and icu4 in Arabidopsis and rld1 in maize (Zea mays) have a single nucleotide change within the miR165/166-recognition sequence, and the miR165/166-dependent cleavage of their transcripts is suppressed, resulting in the ectopic expression of their gene products (McConnell et al., 2001; Emery et al., 2003; Juarez et al., 2004; Zhong and Ye, 2004; Ochando et al., 2006).
Several genes genetically interact with Class III HD-Zip genes in Arabidopsis. KANADI (KAN) genes have an antagonistic function to Class III HD-Zip genes (Kerstetter et al., 2001; Eshed et al., 2004). KAN genes are generally expressed in domains complementary to those where Class III HD-Zip genes are expressed, and multiple loss-of-function mutants of kan genes exhibit similar polarity defects as gain-of-function mutants of Class III HD-Zip genes (Eshed et al., 2001, 2004; Kerstetter et al., 2001; Emery et al., 2003). Furthermore, in the kan1 kan2 kan4 background, PHB is expressed throughout leaf primordia (Eshed et al., 2004). Loss of the activity of the DORNRÖSCHEN (DRN) and DRN-LIKE (DRNL) genes results in embryonic defects similar to those caused by the loss of Class III HD-Zip activity and the DRN and DRNL proteins physically interact with Class III HD-Zip (Chandler et al., 2007).
The close association of KAN and DRN with auxin and/or auxin flow has been suggested. The triple mutant kan1 kan2 kan4 shows aberrant distribution of the auxin efflux carrier protein PIN1, resulting in ectopic auxin maxima and ectopic organ formation in the embryo (Izhaki and Bowman, 2007). Similarly, in the drn-1 mutant background, the abnormal expression of the auxin reporter genes DR5∷GFP and PIN1∷PIN1-GFP are observed in the embryo (Chandler et al., 2007). Although a close association between the Class III HD-Zip-mediated genetic pathway and auxin and/or auxin flow has been suggested (Baima et al., 1995; Zhong and Ye, 2001; Bowman and Floyd, 2008), it is unknown how Class III HD-Zip genes interact with auxin.
Class III HD-Zip genes are good genetic markers for understanding morphological and developmental innovations achieved during the evolutionary history of land plants, because the Class III HD-Zip gene family has been found in all land plant lineages (Floyd and Bowman, 2004; Floyd et al., 2006). In addition, expression analyses have suggested that Class III HD-Zip genes have an important developmental role in all land plants (Floyd et al., 2006; Prigge and Clark, 2006). However, the understanding of the diversity and conservation of the developmental roles of these genes is limited. In addition, there are unanswered questions concerning the roles of Class III HD-Zip genes in the leaf initiation process and their relationship to auxin.
In this study, we investigated the expression patterns, ectopic expression phenotypes, and auxin responsiveness of rice (Oryza sativa) Class III HD-Zip genes for understanding their developmental roles in rice.
RESULTS
Gene Structure and Phylogenetic Relationship of Rice Class III HD-Zip Genes
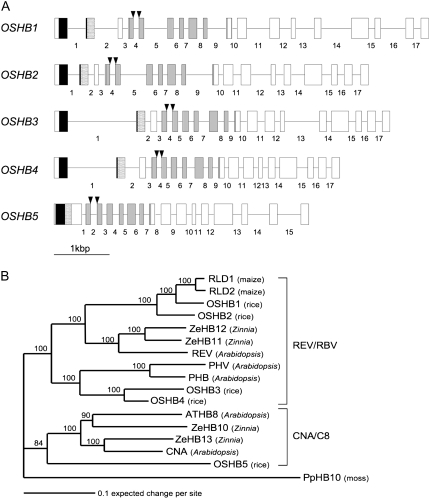
The rice genome contains five Class III HD-Zip genes: OSHB1 to OSHB5 (Zhong and Ye, 2004). Recently, a genome-wide survey of HD-Zip genes in rice also detected five Class III HD-Zip genes: Oshox10, Oshox9, Oshox33, Oshox32 and Oshox29, corresponding to OSHB1 to OSHB5, respectively (Agalou et al., 2008). We searched for full-length cDNAs of each of the five OSHB genes in a gene database, and found four full-length cDNAs corresponding to OSHB1 to OSHB4. Because no full-length cDNA for OSHB5 was found in the database, we isolated the full-length cDNA directly and determined its sequence. The coding region of the OSHB5 cDNA comprises 2,607 nucleotides, the longest of the five OSHB genes. Comparison of the cDNA and genomic sequences revealed that there are 17 internal introns in OSHB1 to OSHB4, with well-conserved splice sites, although OSHB3 and OSHB4 have longer first introns than OSHB1 and OSHB2 (Fig. 1A). In contrast to the well-conserved exon-intron organization in OSHB1 to OSHB4, OSHB5 has only 15 introns, lacking two introns corresponding to the first and second introns of the other OSHB genes.
Figure 1.
Structural and phylogenetic relationship among Class III HD-Zip genes in rice. A, Exon-intron structure of five OSHB genes. Exons are presented by boxes, and introns by bars. Introns within the cording region are numbered. Exons in black, dotted, and light gray boxes indicate the position of the homeodomain, Leu zipper, and START domains, respectively. Arrowheads indicate the miR166-binding site. B, Phylogenetic tree of HD-ZIP III proteins in several angiosperms rooted by PpHB10. Numbers above the branches indicate bootstrap values calculated from 1,000 replicates.
The miR166-binding sequence is completely conserved in the five OSHB cDNAs (Supplemental Fig. S1B), but it is interrupted by the fourth intron in OSHB1 to OSHB4 and by the second intron in OSHB5 (Fig. 1A).
All of the five OSHB proteins contain highly conserved homeodomain, Leu-zipper domains and START domains (Fig. 1A; Supplemental Fig. S1A). Phylogenetic analyses of the Class III HD-Zip genes in several angiosperm species have revealed that the genes are resolved into two monophyletic groups referred to as the REV/RBV clade and the CNA/C8 clade, although both the clades originally include gymnosperm sequences (Floyd et al., 2006; Prigge and Clark, 2006; Fig. 1B). OSHB1 to OSHB4 belong to the REV/RBV clade, and OSHB5 belongs to the CNA/C8 clade. OSHB1 and OSHB2 are orthologs of REV in Arabidopsis, together with its sister genes in Zinnia, ZeHB11, and ZeHB12. Two maize (Zea mays) genes, RLD1 and RLD2, are sisters of OSHB1. OSHB3 and OSHB4 are orthologous to both PHB and PHV in Arabidopsis. Two Arabidopsis genes, ATHB8 and CNA, are orthologous to ZeHB10 and ZeHB13 of Zinnia, respectively. However, OSHB5 is a single sister gene to both Arabidopsis and Zinnia genes in the CNA/C8 clade. This phylogenetic relationship is supported by previous reports (Zhong and Ye, 2004; Floyd et al., 2006; Prigge and Clark, 2006). These results suggest that the five OSHB genes in rice retain conserved molecular functions with other Class III HD-Zip genes of angiosperms.
Overall Expression Patterns of the OSHB Genes
We first investigated the expression levels of the OSHB genes in various organs and tissues using real-time reverse transcription (RT)-PCR. All of the five OSHB genes were expressed in all of the tissues; shoot and inflorescence apices, immature leaves, panicles, flowers, embryos, and roots, although their expression levels varied widely by gene and tissue (Supplemental Fig. S2). The transcript levels of OSHB1 and OSHB2 were low in developing embryo and root, and moderate in other tissues. The expression of OSHB3 and OSHB4 was strong in the inflorescence tissues and moderate in other tissues. OSHB5 showed a different expression profile from the other OSHB genes; its mRNA accumulated to low levels in young leaves, developing embryo and flowers, but to moderate levels in the inflorescence tissues and root. Thus, the OSHB genes may function in all the organs and tissues that we examined, but display some level of tissue-specific expression.
Expression Patterns of OSHB Genes during Embryogenesis
To better understand the spatiotemporal expression patterns of the OSHB genes, we examined their expression by in situ hybridization. First, we investigated OSHB expression in the embryo. A review of rice embryogenesis and the developmental stages of the embryo was published by Itoh et al. (2005). To summarize, for 3 d after pollination (DAP), the rice embryo remains globular, with no morphological differentiation visible. At 4 DAP, the embryonic SAM and coleoptile primordium are formed in the apical and ventral positions of the embryo, and the radicle primordium is formed in the basal region. At 5 DAP, the embryonic SAM produces the first foliage leaf, and by 8 DAP, the morphological differentiation of the rice embryo is almost complete.
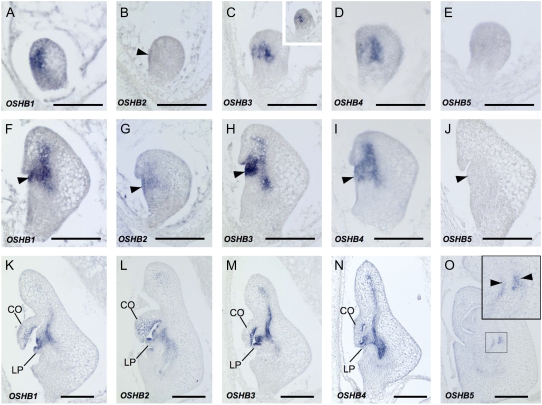
Expression of OSHB1, OSHB3, and OSHB4 was detected in the center and ventral domains of the embryo at the late globular stage (3 DAP; Fig. 2, A, C, and D). The central expression domain corresponds to the place where the vasculature will differentiate, and the ventral domain corresponds to the site where the SAM will later initiate. The central expression of OSHB3 was observed as early as the middle globular stage (2 DAP; Fig. 2C, inset). OSHB2 expression was first detected in a small region of the ventral surface corresponding to the presumptive SAM of the late globular embryo (Fig. 2B). At 4 DAP, when the coleoptile primordium and the SAM were visible, OSHB1, OSHB3, and OSHB4 were expressed in the SAM, the adaxial domain of the coleoptile, and the provascular tissue (Fig. 2, F, H, and I). OSHB2 was weakly expressed in the SAM and adaxial cells of the coleoptile (Fig. 2G). At 5 DAP, when the SAM produced the first leaf primordium, the expression of OSHB1 to OSHB4 was detected in similar domains as at 4 DAP (Fig. 2, K–N). Their expression persisted in part of the SAM, the vascular bundles, the adaxial cells of the coleoptile, and the first leaf primordium, and expression of OSHB2 was also detected over the entire surface (L1) of the SAM (Fig. 2L). In contrast, OSHB5 transcripts were first detectable at 6 to 7 DAP only in the vascular bundle of the basal portion of the radicle (Fig. 2, E, J, and O). OSHB5 expression in the radicle was observed in two separate cell files of the vascular bundle. This expression pattern in the vascular bundle was quite different from that of the other OSHB genes, whose transcripts were detected in the center (Fig. 2N). This indicates that OSHB5 is expressed in a specific cell type of the vascular bundle.
Figure 2.
In situ localization of OSHB transcripts in embryo. A to O, Longitudinal sections of wild-type embryos at 3 DAP (A–E), 4 DAP (F–J), 5 DAP (K–N), and 7 DAP (O). A, F, and K, OSHB1 expression. B, G, and L, OSHB2 expression. C, H, and M, OSHB3 expression. Arrowhead in C indicates OSHB2 expression in the ventral surface of the embryo. The inset in C shows OSHB3 expression at 2 DAP. D, I, and N, OSHB4 expression. E, J, and O, OSHB5 expression. The inset in O shows a higher magnification view of the signal in the box. Signals are observed in two cell files (arrowheads) in the vascular tissue. Arrows in F to J indicate SAMs. CO, Coleoptile; LP, leaf primordium. Scale bars = 100 μm.
Expression Patterns of the OSHB Genes in Vegetative Development
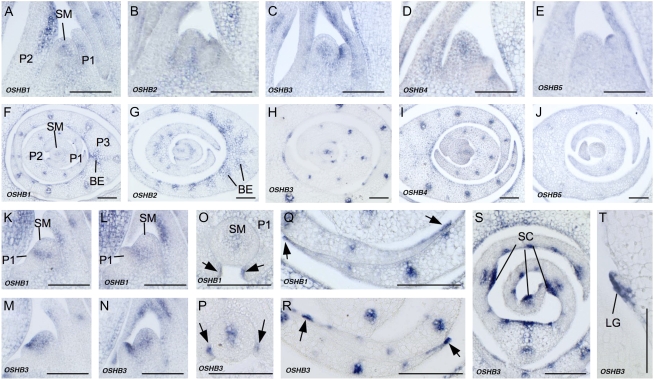
Next, we observed OSHB expression in the vegetative shoot apex. In seedlings at 2 weeks after germination, the expression of OSHB1 to OSHB4 was detected in the SAM, adaxial cells of the leaf primordia, and vascular tissue (Fig. 3, A–D and F–I), although slight differences were observed between the genes, as described below. In contrast, OSHB5 was weakly expressed in vascular bundles and in the tip of the P1 leaf primordium, but not in the SAM or adaxial cells of the leaf primordia (Fig. 3, E and J).
Figure 3.
In situ localization of OSHB transcripts in the vegetative shoot apex. A to E, K to N, and T, Longitudinal sections of 2-week-old wild-type shoot apices. F to J and O to S, Cross sections of 2-week-old wild-type shoot apices. A and F, OSHB1 expression. B and G, OSHB2 expression. C and H, OSHB3 expression. D and I, OSHB4 expression. E and J, OSHB5 expression. K, OSHB1 expression in shoot apex when the P1 primordium is just protruded. L, OSHB1 expression in the shoot apex when the P1 primordium is developing. M, OSHB3 expression in the shoot apex when the P1 primordium is just protruded. N, OSHB3 expression in the shoot apex when the P1 primordium is developing. O, OSHB1 expression in P1 leaf margins (arrows). P, OSHB3 expression in P1 leaf margins (arrows). Q, OSHB1 expression in P3 leaf margins (arrows). R, OSHB3 expression in P3 leaf margins (arrows). S, OSHB3 expression in the P3 leaf blade, shown in adaxial sclerenchymatous cells (SC). T, OSHB3 expression in the ligule primordium (LG) of the P3 leaf. SM, SAM; P1, P1 leaf primordium; P2, P2 leaf primordium; P3, P3 leaf primordium; BE, bundle sheath extension. Scale bars = 100 μm.
The expression patterns of OSHB1 to OSHB4 in the SAM appeared to change during the plastochron. In most of the shoot apices sampled, signals were limited to the central domain of the SAM and were not detected in the P0 leaf primordium (Fig. 3, A–D). Detailed examination of OSHB1 and OSHB3, however, revealed that once the P1 leaf primordium became visible as a protrusion, OSHB1 and OSHB3 expression rapidly extended from the central domain of the SAM toward the leaf initiation site (Fig. 3, K and M). This enlarged expression domain in the SAM was maintained until the middle P1 stage (Fig. 3, L and N), and then the signals became confined again to the central domain of the SAM (Fig. 3, A and C). Thus, the expression domain of OSHB1 and OSHB3 in the SAM changes drastically during the plastochron.
The expression patterns of OSHB1 to OSHB4 in leaf primordia were similar (Fig. 3, A–D and F–I). In the P1 primordium, the expression of OSHB1 and OSHB3 was restricted to the adaxial cells and a few cells in the leaf margins (Fig. 3, O and P). In the P3 leaf primordium, the expression of OSHB1 and OSHB2 extended to bundle sheath extension cells (Fig. 3, F and G), whereas OSHB3 and OSHB4 expression was limited to the adaxial surface and the vascular bundle (Fig. 3, H and I). The expression of OSHB1 and OSHB3 in the leaf margins was retained at this stage (Fig. 4, Q and R). In the P4 leaf primordium, strong OSHB3 expression was evident in the adaxial sclerenchymatous cells (Fig. 3S) and in the adaxial cells of the ligule primordium (Fig. 3T).
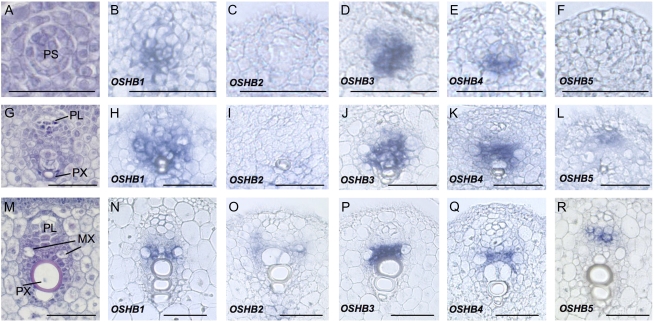
Figure 4.
In situ localization of OSHB transcripts during vascular development. A to F, Early stage of vascular development when procambial strands are evident. G to L, Middle stage of vascular development when the protophloem and protoxylem are visible. M to R, Late stage of vascular development when the metaxylem is evident. A, G, and M, Wild-type vascular bundle stained with hematoxyline, safranin, and Fast-Green FCF. B, H, and N, OSHB1 expression. C, I, and O, OSHB2 expression. D, J, and P, OSHB3 expression. E, K, and Q, OSHB4 expression. F, L, and R, OSHB5 expression. PS, Procambial strand; PL, protophloem; PX, protoxylem; MX, metaxylem. Scale bars = 100 μm.
To better understand how the rice OSHB genes are associated with vascular development, we examined the expression dynamics of these genes during vascular development (Fig. 4, A, G, and M). Signals of OSHB1, OSHB3, and OSHB4 were first observed in a group of procambium cells of late P1 to early P2 primordia (Fig. 4, B, D, and E). As cellular differentiation progressed, mRNAs of these OSHB genes accumulated in the region where the xylem would develop (Fig. 4, H, J, and K), with the expression domain of OSHB1 larger than those of OSHB3 and OSHB4 (Fig. 4, H, J, and K). At the late stage of vascular development, when cellular differentiation was being completed, expression of these genes continued in the cells between the metaxylem elements (Fig. 4, N, P, and Q). OSHB2 mRNA accumulated in a domain of the vascular tissue similar to the area where the OSHB1, OSHB3, and OSHB4 mRNAs accumulated at the late stage (Fig. 4O), but the signal was not detected at the early stage (Fig. 4, C and I). In contrast, the OSHB5 expression pattern was distinct from that of the other genes. No OSHB5 signal was observed in the early stage of vascular development (Fig. 4F), but signal was detected in the phloem tissue as cellular differentiation progressed (Fig. 4, L and R). This OSHB5 expression pattern corresponded to that in the embryo (Fig. 2O). Thus, the function of OSHB5 in vascular development may be different from that of the other OSHB genes. The expression profile of OSHB genes in leaf vasculature suggests a possibility that OSHB genes share overlapping but distinct functions in vascular development.
In the reproductive phase, the expression pattern of each OSHB gene was similar to that in the vegetative phase. However, the down-regulation of OSHB1, OSHB3, and OSHB4 in the L1 and L2 layers was not observed in the inflorescence meristem (Supplemental Fig. S3, A, C, and D). In addition, OSHB2 expression was not observed in the center of the inflorescence or the floral meristems (Supplemental Fig. S3, C and G), and weak OSHB5 expression was detected in the inflorescence meristem (Supplemental Fig. S3E).
Effect of Ectopic Expression of OSHB Genes on Shoot Development
To gain further understanding of the rice Class III HD-Zip genes, we constructed ectopic-expressers of OSHB1, OSHB3, and OSHB5. Regenerated plants containing these cDNAs driven by the ACTIN promoter did not show morphological abnormalities, probably due to miR166-mediated posttranscriptional gene silencing. Therefore, we constructed miR166-resistant versions of the OSHB1, OSHB3, and OSHB5 cDNAs driven by the ACTIN promoter, each of which contained five mutations at the miR166-binding site that did not cause amino acid substitutions (Supplemental Fig. S1B), and transformed them into wild-type calli. We obtained 12, 14, and nine independent transgenic plants harboring the mutated OSHB1 (OSHB1m), OSHB3m, and OSHB5m, respectively.
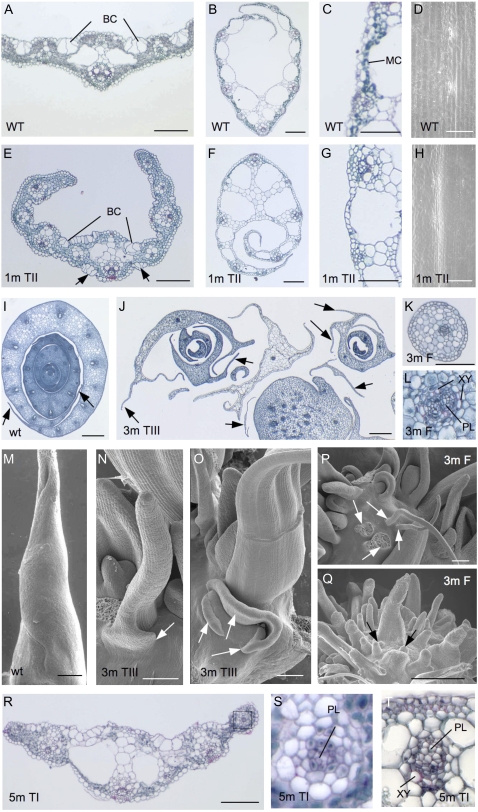
Transgenic plants containing OSHB3m showed the most severe phenotype, those containing OSHB1m showed a less severe phenotype, and those containing OSHB5m showed a weak phenotype (Fig. 5A). Two types of abnormal leaves were observed: rolled leaves and filamentous leaves (Table I). The rolled-leaf phenotype was further categorized into three types based on severity. Type I plants had adaxially rolled leaves with a normal leaf blade-sheath boundary (Fig. 5, B and F). All OSHB5m plants were categorized into this type (Table I). The intermediate phenotype, Type II, had adaxially rolled leaves with ectopic ligules. In wild-type leaves, the ligule is formed on the adaxial surface of the leaf blade-sheath boundary (Fig. 5B). In the Type II leaves, ligules formed on both the adaxial and abaxial surfaces of the leaf blade-sheath boundary (Fig. 5C). In addition, the leaf sheath of this type was twisted and whitish. Most of the OSHB1m plants showed Type II leaves (Table I). Type III plants exhibited narrow rolled leaves with ectopic ligules on the abaxial side of the blade-sheath boundary, and leaf sheath of the leaves was rough and had a complex shape (Fig. 5D). Type III plants were observed at a low frequency in OSHB1m plants and frequently in OSHB3m plants (Table I). Filamentous leaves, the most severe phenotype, were observed in half of the OSHB3m plants (Table I; Fig. 5A). Some filamentous leaves had a ring-like structure at the blade-sheath boundary (Fig. 5E), although most of these leaves did not have a ligule- and auricle-like structure and their blade-sheath boundaries were not obvious.
Figure 5.
Phenotypes of transgenic plants expressing miR166-resistant OSHB1, OSHB3, and OSHB5. A, Control and transgenic plants at 1 month after regeneration. From left to right, control plant introduced with empty vector; pACT∷OSHB1m plant showing mild phenotype; pACT∷OSHB1m plant showing severe phenotype; pACT∷OSHB3m plant showing mild phenotype; pACT∷OSHB3m plant showing severe phenotype; and pACT∷OSHB5m plant. B, Leaf blade-sheath boundary of the control leaf. LB, Leaf blade; LS, leaf sheath; LJ, lamina joint; LG, ligule. C, Blade-sheath boundary of the Type II pACT∷OSHB1m leaf. Arrows show ligules formed on the abaxial sides. D, Complex-shaped leaf sheath of the Type III pACT∷OSHB3m plant. Arrows indicate leaf margins and the arrowhead indicates an additional ridge on the abaxial side of the leaf. E, Blade-sheath boundary of the filamentous leaf in the pACT∷OSHB3m plant. Arrow indicates the ring-like swell at the leaf blade-sheath boundary. F, Blade-sheath boundary of the Type I pACT∷OSHB5m leaf. [See online article for color version of this figure.]
Table I.
Frequency of leaf phenotypes in OSHBm transgenic plants
| Transgene | Rolled Leafa
|
Filamentous Leaf | nb | ||
|---|---|---|---|---|---|
| Type I | Type II | Type III | |||
| pACT∷OSHB1m | 0 | 10 | 2 | 0 | 12 |
| pACT∷OSHB3m | 0 | 0 | 7 | 7 | 14 |
| pACT∷OSHB5m | 9 | 0 | 0 | 0 | 9 |
Rolled leaf phenotype was categorized into three types based on their severity: Type I, rolled leaf with no ectopic ligule; Type II, rolled leaf with ectopic ligules; Type III, rolled but complex-shaped leaf with ectopic ligules.
n indicates the total number of transgenic plants observed.
Next, we performed anatomical and morphological analyses on the transgenic plants. Blades of Type II OSHB1m leaves showed a reduction in abaxial sclerenchymatous cells and the ectopic formation of bulliform-like cells on the abaxial surface (Fig. 6, A and E). In the leaf sheath, most abaxial mesophyll cells were lost (Fig. 6, B, C, F, and G), which was a possible cause of the whitish appearance of the leaf sheath. Papillae were not observed on the abaxial epidermis of the OSHB1m leaf sheath, as in the adaxial epidermis of the wild-type sheath (Fig. 6, D and H). Despite these abnormalities, the vascular bundles appeared normal in the Type II leaves of OSHB1m plants. Thus, the ectopic expression of OSHB1m causes partially adaxialized leaves, but its effect is limited to the epidermis and subepidermal tissues.
Figure 6.
Leaf and shoot apex morphology of pACT∷OSHBm transgenic plants. A, Cross section of the wild-type leaf blade. Bulliform cells (BC) are observed at the adaxial surface. B, Cross section of the wild-type leaf sheath. C, Close-up view of the abaxial side of the wild-type leaf sheath with dark-stained mesophyll cells (MC). D, Adaxial surface view of the wild-type leaf sheath lacking papillae. E, Cross section of the pACT∷OSHB1m leaf blade, showing ectopic formation of bulliform cells (BC) on the abaxial side. F, Cross section of the pACT∷OSHB1m leaf sheath. G, Close-up view of the abaxial side of the pACT∷OSHB1m leaf sheath. H, Abaxial surface view of the pACT∷OSHB1m leaf sheath lacking papilla. I, Cross section of the wild-type shoot apex. Arrows show membranous leaf margins. J, Cross section of the pACT∷OSHB3m shoot apex. Arrows show ectopic leaf margins. K, Cross section of the pACT∷OSHB3m filamentous leaf. L, Close-up view of the vascular bundle of the pACT∷OSHB3m filamentous leaf. Phloem (PL) is surrounded by xylem (XY) tissue. M, SEM image of wild-type leaf primordium. N, SEM image of the pACT∷OSHB3m shoot apex, in which the ectopic leaf margin (arrow) is formed. O, SEM image of the pACT∷OSHB3m shoot apex showing ectopic formation of leaf margin-like structures (arrows) that are formed independently from the abaxial base of the leaf. P, SEM image of the pACT∷OSHB3m plant showing multiple shoots and filamentous leaves (arrows) without shoot meristem at their axils. Q, SEM image of the pACT∷OSHB3m plant with enormous spike-like leaves. Leaf primordia-like structures (arrows) are observed between the preexisting leaves. R, Cross section of the pACT∷OSHB5m leaf blade. S, Close-up view of the vascular bundle of the pACT∷OSHB5m leaf in R with phloem (PL) surrounded by bundle sheath cells and underdeveloped xylem tissue. T, Close-up view of the vascular bundle of the pACT∷OSHB5m leaf with fully differentiated phloem (PL) and reduced xylem (XY) tissue. Scale bar = 100 μm in A, B, E, F, J, M to R; 50 μm in C, D, G, H, and K; 500 μm in I. Phenotypic categories and genotypes are labeled in the images; WT, Wild type; 1 m, OSHB1m; 3 m, OSHB3m, 5 m, OSHB5m; TI, Type I; TII, Type II; TIII, Type III; F, filamentous leaf (see Table I). [See online article for color version of this figure.]
Cross sections and scanning electron microscope (SEM) views of complex-shaped leaves in Type III OSHB3m plants revealed that the cause of the complexity was the formation of ectopic leaf margin-like structures from the leaves (Fig. 6, I, J, and M–O). Most of the ectopic structures resembled the membranous margin of wild-type leaves (Fig. 6, I and J). In an extreme case, leaf margin-like membranous structures formed directly from the base of the preexisting leaf primordia (Fig. 6, J and O). Because membranous structure of leaf margins in the wild type are established at the margins of SAM-derived leaf primordia, the ectopic margins are likely recruited independent of the main part of the leaf.
The filamentous leaves observed in OSHB3m plants were radialized (Fig. 6K). The vascular bundles were also radialized, with xylem surrounding phloem (Fig. 6L). The radialized leaves are likely produced ectopically from the stem, because no SAM-like structures were apparent at the axils of these filamentous leaves (Fig. 6P). Two OSHB3m plants formed spike-like leaves. Most were positioned randomly and appeared to be radialized (Fig. 6Q).
Type I leaves of OSHB5m plants did not show obvious abnormalities (Fig. 6R). However, abnormal vascular bundles were observed in some leaves (Fig. 6, S and T). The vascular bundles are sometimes radialized, and xylem tissues are poorly differentiated. This indicates that OSHB5 has less of a function in leaf polarity, but is involved in vascular bundle patterning and differentiation. This is in contrast with OSHB1m plants, in which leaf abnormalities were observed mainly in the epidermis and subepidermal tissues but not in the vascular tissues.
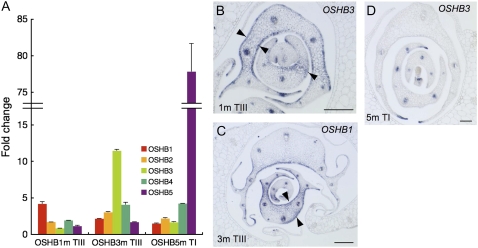
To clarify how the introduction of OSHBm transgenes affects leaf polarity, we first examined the expression levels of the OSHB genes using real-time RT-PCR. In OSHB5m plants, the expression levels dramatically increased (Fig. 7A). However, in OSHB1m and OSHB3m plants, a relatively low level of increase in expression was observed. Next, we examined the expression of OSHB3 in severe OSHB1m shoots, by in situ hybridization. OSHB3 transcripts were ectopically detected in the adaxial and abaxial epidermis of P2 to P3 leaf primordia (Fig. 7B). Considering that the OSHB3 transcript did not increase quantitatively in OSHB1m shoots (Fig. 7A), leaf adaxialization in OSHB1m is a direct cause of the ectopic expression and/or overexpression of OSHB1m, and the ectopic OSHB3 expression in OSHB1m leaves is a secondary effect of the leaf adaxialization. Reciprocally, OSHB1 transcripts in OSHB3m shoots were distributed in both the adaxial and abaxial epidermis of P2 to P3 leaf primordia, indicating that the ectopic expression and/or overexpression of OSHB3m caused leaf adaxialization (Fig. 7C). However, the adaxialization did not occur in all leaf tissues, and the adaxial/abaxial polarity was maintained in the leaf margins. No ectopic expression of OSHB3 was detected on the abaxial sides of the OSHB5m leaves. However, the level of OSHB3 expression in the xylem tissue of the several vascular bundles was lowered (Fig. 7D). Thus, the introduction of the OSHB1m and OSHB3m transgenes driven by the ACTIN promoter affects the adaxial-abaxial patterning of leaves. However, ectopically expressed OSHB5m does not affect leaf polarity, but influences vascular development.
Figure 7.
Effect of pACT∷OSHBm expression on leaf polarity. A, Real-time RT-PCR analysis of five OSHB genes in pACT∷OSHB1m plants showing Type III phenotype (see Table I; left), in Type III of pACT∷OSHB3m (middle), and in Type I of pACT∷OSH5m (right). Fold-change relative to the vector control plant is shown. Expression level was normalized to that of eEF-1α. Bars indicate sd. B, In situ localization of OSHB3 transcripts in Type III pACT∷OSHB1m shoot apex. Arrowheads indicate OSHB3 expression on both the adaxial and abaxial sides of the leaf primordia. C, In situ localization of OSHB1 transcripts in Type III pACT∷OSHB3m shoot apex. Arrowheads indicate OSHB1 expression on both the adaxial and abaxial sides of the leaf primordia. D, In situ localization of OSHB3 transcripts in Type I pACT∷OSHB5m shoot apex. Scale bars in B to D =100 μm.
Effect of Auxin Treatment on OSHB Expression
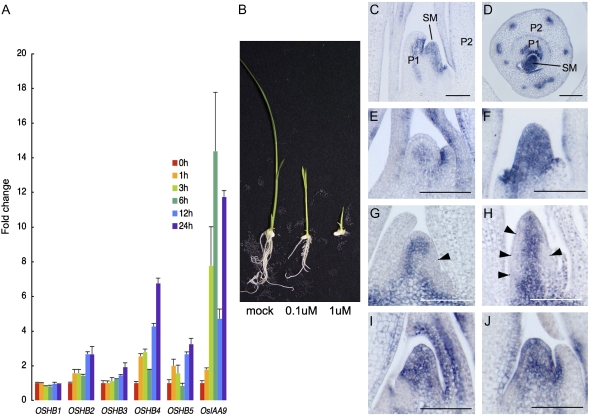
The expression of one of the Arabidopsis Class III HD-Zip genes, ATHB8, is induced by auxin (Baima et al., 1995), and a possible interaction between auxin and/or auxin flow and Class III HD-Zip genes in Arabidopsis has been proposed (Bowman and Floyd, 2008). To examine the effect of auxin on the expression of the OSHB genes, we first treated rice seedlings with 1 μm 2,4-dichlorophenoxyacetic acid (2,4-D) for 3 to 24 h, and examined the expression levels of the five OSHB genes using real-time RT-PCR. The expression of the OSHB1 and OSHB3 genes was not significantly affected by 2,4-D treatment, whereas the expression of OSHB2, OSHB4, and OSHB5 had increased at 12 and 24 h after 2,4-D treatment. In contrast, the expression of OsIAA9, one of the early auxin response AUX/IAA-family genes (Jain et al., 2006), increased dramatically at 3 and 6 h after 2,4-D treatment (Fig. 8A).
Figure 8.
Effect of auxin treatment on OSHB gene expression. A, Real-time RT-PCR analysis of five OSHB genes and OsIAA9. Fold-change relative to the control plant at 0 h after 2,4-D treatment for each gene is shown. Expression level was normalized to that of eEF-1α. Bars indicate sd. B, Phenotypes of 2,4-D-treated plants 1 week after germination grown in culture media containing 0 μm (left), 0.1 μm (middle), and 1 μm (left) 2,4-D. C and D, OSHB3 expression in longitudinal and cross sections of the shoot apex at 1 week after 2,4-D treatment. SM, SAM; P1, P1 leaf primordium; P2, P2 leaf primordium. E, OSHB3 expression in the shoot apex without 2,4-D treatment. F, OSHB3 expression in the SAM at 1 week after 2,4-D treatment. G, OSH1 expression in the shoot apex without 2,4-D-treatment. Arrow shows down-regulation of OSH1 on one side of the flank of the SAM. H, OSH1 expression in the SAM at 1 week after 2,4-D treatment. Arrows indicate down-regulation of OSH1 (arrow) along the peripheral domain of the SAM. I, OSHB3 expression in the SAM at 6 h after 2,4-D treatment. J, OSHB3 expression in the SAM at 24 h after 2,4-D treatment. Scale bars in C to J =100 μm.
We next investigated the long-term effect of auxin treatment on rice seedling development and OSHB expression. Seedlings grown for 2 weeks in culture medium containing 1 μm 2,4-D showed reduced leaf length, the formation of thick and tube-like leaves, and the inhibition of leaf initiation and root elongation (Fig. 8B). In these plants, the expression of OSHB3 in the SAM was dramatically enhanced (Fig. 8, C and D; compare with Fig. 3, C and H). Unlike the localized expression of OSHB3 in the central domain of the untreated SAM, the expression was extended to the whole region of the 2,4-D-treated SAM (Fig. 8, E and F). Concurrently, the SAM was vertically elongated. However, OSHB3 expression in the adaxial domain and the vascular bundles of young leaf primordia was not affected (Fig. 8, C and D). In addition, polarity defects were not observed in either the leaf primordia or the vascular bundles, although the shape of the leaf primordia was abnormal and the vascular bundles were unequally distributed along the lateral direction of the leaf (Fig. 8D). A similar expression pattern was observed when the OSHB1 and OSHB4 genes were used as a probe (data not shown).
To better understand how leaf primordia initiate in the 2,4-D-treated SAM, we examined the expression pattern of the Class I KNOX gene OSH1, whose down-regulation is a marker of P0 leaf primordium (Sentoku et al., 1999). Down-regulation of OSH1 was observed in a small group of cells (P0 primordium) in the normal SAM (Fig. 8G). However, in 2,4-D-treated plants, OSH1 down-regulation occurred in the vertically extended domain of the flank of the SAM (Fig. 8H). This expression pattern indicates that leaf founder cells required for the leaf primordium are distributed in a large cylinder-like domain of the entire SAM. This cylinder-like distribution of leaf founder cells is consistent with the thick and tube-like leaf primordia produced by 2,4-D treatment (Fig. 8, C and D).
To demonstrate that the aberrant leaf initiation was caused by ectopic OSHB3 expression and not the result of geometrical alternation of the SAM or the long-term effect of auxin treatment, we observed OSHB3 expression soon after the onset of auxin treatment. Ectopic expression of OSHB3 in the SAM was observed as early as 6 h after 2,4-D treatment, when morphological changes in the SAM were not yet observed; the expression was maintained at least 24 h after the treatment (Fig. 8, I and J). This result indicates that ectopic OSHB3 expression is quickly induced by auxin in the SAM, which may affect the normal leaf initiation processes.
DISCUSSION
Functional Conservation, Redundancy, and Differences among Rice Class III HD-Zip Genes
Five Class III HD-Zip genes of rice encode possible transcriptional regulators with well-conserved motifs that are present in all plant Class III HD-Zip genes. OSHB1 to OSHB4 are similar in both gene structure and expression patterns, suggesting their functional redundancy. In fact, a loss-of-function mutant of OSHB4 having a Tos17 insertion (Miyao et al., 2003) showed no obvious phenotype (data not shown). However, a detailed examination of the expression profiles of these genes revealed that they differ from one another. Phylogenetic analysis showed that OSHB5 is distantly related to other OSHB genes. This finding is supported by an analysis of the gene structure and the expression profile.
Transgenic analysis also revealed diverse functions for the OSHB genes. Adaxially rolled leaves were commonly observed in the OSHB1m, OSHB3m, and OSHB5m plants, but OSHB5m did not exhibit leaf polarity defects. Ectopic ligules were observed in both the OSHB1m and OSHB3m plants, but radialized leaves were observed only in the OSHB3m plants. Thus, OSHB3 may contribute more to leaf polarity than OSHB1. The radially symmetric vascular bundles of the OSHB3m and OSHB5m plants indicate that OSHB3 and OSHB5 are involved in vascular patterning and differentiation. In contrast, the OSHB1m plants exhibited no vascular defects. The expression level of OSHB1 in OSHB1m transgenic plants and OSHB3 in OSHB3m was not dramatically elevated; one possible explanation for this is that strong OSHB1m and OSHB3m expression may be lethal to regenerating plants.
The conservation of miR166-binding sequences among the five OSHB genes indicated that all of the genes are potentially regulated by miR166. Indeed, transgenic plants containing the OSHB1, OSHB3, or OSHB5 genes driven by the ACTIN promoter showed no obvious phenotypes, whereas when OSHB1, OSHB3, and OSHB5 with mutations in the miR166-binding site were introduced, the resulting transgenic plants exhibited abnormal phenotypes. Thus, miR166-mediated gene silencing contributes to the regulation of OSHB expression. However, our transgenic analysis using a constitutive ACTIN promoter, makes it difficult to know how OSHB genes are spatially and quantitatively regulated by miR166. In fact, phenotypes of gain-of-function allele of the maize RLD1 gene, which is an ortholog of OSHB1, are considerably weaker than those of OSHB1m (Juarez et al., 2004). Thus, analyses of similar transgenic experiments using OSHB endogenous promoters and of their gain-of-function mutants are needed for further understanding of the miR166-mediated regulation.
Role of OSHB Genes in Embryogenesis
The OSHB3 gene was first expressed in a small group of cells in the center of the early globular embryo, and additional expression of OSHB1, OSHB3, and OSHB4 appeared in the ventral domain of the embryo, where the SAM and coleoptile primordium will initiate. This expression pattern in the early embryo is different from that of the Arabidopsis orthologs of the gene. In Arabidopsis, the REV, PHB, and PHV genes are expressed at the apical and central poles of early globular embryos, which is important for the radial patterning of the apical region and SAM formation (McConnell et al., 2001; Emery et al., 2003; Prigge et al., 2005). The early expression in the embryo indicates that the OSHB genes may also participate in radial pattern formation in the rice embryo. If so, the central expression of OSHB is involved in radial patterning in the early globular embryo, and subsequent additional ventral expression is necessary to establish the SAM or shoot axis. This expression pattern suggests that Class III HD-Zip proteins function in radial patterning and SAM establishment could be separable. This is consistent with the analysis of shootless mutants of rice, which lack the ventral expression of the OSHB genes in the globular embryo (Nagasaki et al., 2007). In the shootless mutants, despite the complete loss of the embryonic shoot, embryo polarity and the other embryonic organs are normal (Satoh et al., 1999; Nagasaki et al., 2007). Accordingly, one of the functions of OSHB in embryogenesis is SAM establishment in the ventral domain of the embryo.
The involvement of OSHB in SAM initiation is also suggested by the OSHB3m transgenic plants, in which multiple shoots are produced not only at the axils of leaves but also at irregular positions. This means that the overexpression or ectopic expression of OSHB3m can induce the de novo formation of the SAM. In accordance with this, overexpressed OSHB1m can induce the formation of adventitious shoots from the callus of rice shoot organization mutants, which are defective in shoot regeneration (Nagasaki et al., 2007). Thus, OSHB genes should be associated with SAM formation not only in embryogenesis but also in the regeneration process.
Role of the OSHB Genes in Leaf Development
In Arabidopsis, three of the five Class III HD-Zip genes (REV, PHB, and PHV) regulate leaf polarity (Prigge et al., 2005). OSHB1 to OSHB4 are also involved in leaf polarity, as evidenced by the adaxialized leaves of the OSHB1m and OSHB3m plants. Radially symmetric leaves in severe OSHB3m plants did not seem to differentiate from the SAM. The spatial arrangement of filamentous leaves did not follow any obvious phyllotaxis, and no SAM-like structures were ever observed at the axils of filamentous leaves. Accordingly, OSHB3 has the ability to induce leaf initiation independent of the SAM. A similar situation has been observed in triple mutants in KAN genes in Arabidopsis (Izhaki and Bowman, 2007); kan1 kan2 kan4 embryos allow ectopic auxin maxima on the hypocotyls, resulting in leaf-like outgrowths without the expression of meristem markers. In Arabidopsis, activity of Class III HD-Zip genes and KAN genes is complementary and antagonistic (Eshed et al., 2001, 2004; Kerstetter et al., 2001; Emery et al., 2003). Although the relationship between the rice KAN orthologs and OSHB genes is unknown, KAN orthologs of maize are also expressed in domains complementary to those of the Class III HD-Zip genes (Henderson et al., 2006). Thus, it is possible that the genetic pathway is conserved between Arabidopsis and rice, and the ectopic expression of OSHBm affects the expression of the KAN orthologs, allowing the initiation of radialized and ectopic leaves.
The expression of all rice Class III HD-Zip genes in vascular tissues suggests a possibility that these genes have some role in vascular development. The expression of OSHB3 in xylem tissue and radialized and the formation of amphivasal vascular bundles in OSHB3m leaves suggest that OSHB3 has a function in vascular polarity and patterning. In contrast to the other OSHB genes, OSHB5 expression was observed in phloem tissues. However, OSHB5m plants did not show defects in phloem tissue, but rather showed aberrant xylem tissue differentiation and vascular polarity. OSHB5 may be necessary for the proper patterning of vascular bundles through its expression in phloem tissue. Although Arabidopsis and Zinnia genes belonging to the same clade as OSHB5 are expressed in the xylem and procambium tissue and promote xylem differentiation (Baima et al., 2001; Ohashi-Ito and Fukuda, 2003; Ohashi-Ito et al., 2005), CNA and ATHB8 have functions antagonistic to REV in Arabidopsis in some cases (Prigge et al., 2005), and one of the lycophytes Class III HD-Zip genes, SkC3HDZ1 of Selaginella kraussiana, is expressed in tissues complementary to those of SkC3HDZ2 (Floyd and Bowman, 2006; Floyd et al., 2006). Thus, Class III HD-Zip genes could have achieved intraspecific divergence of their functions, and the expression of OSHB5 may reflect antagonistic roles to the rest of the Class III HD-Zip genes in vascular patterning, although the evolutionary meaning is unclear.
Induction of OSHB Expression by Auxin and Its Effect on the Leaf Initiation Process
The expression of the Arabidopsis Class III HD-Zip genes PHB and REV is localized in the center of the SAM and in the peripheral region where leaf initiation will occur (McConnell et al., 2001). However, the regulatory mechanism of the expression pattern is unknown. Our analysis revealed that OSHB expression is up-regulated rapidly in the SAM flank at the early stage of leaf initiation, and then disappears again from the flank in the middle P1 stage. This dynamic change of expression is reminiscent of the leaf determination process regulated by local auxin accumulation in the SAM. The leaf initiation site in the SAM is determined by the local auxin maximum established by polar auxin transport through the L1 layer (Reinhardt et al., 2003). Recent studies on the PIN1 protein in the SAM have also shown a possible relationship between the auxin concentration and REV expression (Heisler et al., 2005; Bowman and Floyd, 2008).
The induction of OSHB3 expression by auxin in the SAM suggests that excess auxin disturbs the local auxin distribution in the SAM, resulting in the ectopic expression of OSHB3, which may specify all peripheral SAM cells as leaf founder cells. In addition, OSHB3m promotes leaf initiation. Therefore, we propose a simple model with regard to the relationship of auxin, Class III HD-Zip genes, and the leaf initiation process in the SAM. In the wild type, the local concentration of auxin established by polar auxin transport induces Class III HD-Zip gene expression, and then Class III HD-Zip genes promote the initiation of leaf founder cells. A role of Class III HD-Zip genes in organ initiation has been proposed by expression analysis in angiosperms and lycophytes (Prigge and Clark, 2006), and supported by the ectopic induction of radialized leaves in the OSHB3m plants. However, the promotion of leaf initiation by Class III HD-Zip genes may not be a direct regulation, but a result of the elimination of other genetic factors such as the KAN and Class I KNOX genes.
In situ experiments revealed that auxin treatment affected OSHB expression in the SAM and thus leaf initiation, but not in leaves. Altered auxin distribution affects the leaf initiation process, but has less effect on organ polarity (Reinhardt et al., 2000, 2003). The difference in the auxin sensitivity of the processes of leaf initiation and leaf development may be associated with the differential actions of Class III HD-Zip genes in the SAM and the leaf. Although it is not known how auxin induces OSHB3 expression only in the SAM, one possibility is that the auxin responsiveness depends on posttranscriptional mRNA silencing of Class III HD-Zip genes by miR166. miR166 accumulation has been observed in the abaxial domains of leaves, but not in the SAM of maize or rice (Juarez et al., 2004; Nagasaki et al., 2007). Consequently, the auxin responsiveness of Class III HD-Zip genes may be displayed in the SAM but masked in the leaf due to miR166-dependent mRNA degradation.
Conservation and Divergence of Class III HD-Zip Gene Functions in Rice and Arabidopsis
In Arabidopsis, Class III HD-Zip genes play roles in embryo patterning, SAM formation, organ polarity, and vascular development (Prigge et al., 2005). The present analysis revealed that OSHB genes are expressed in domains similar to those in which Arabidopsis Class III HD-Zip genes are expressed: in part of the SAM, in the adaxial cells of leaf primordia, and in vascular tissues. It is difficult to make a direct examination of the functional conservation between Arabidopsis and rice orthologs due to the lack of loss-of-function mutants in rice. However, the transgenic analysis may suggest that the functions of Class III HD-Zip genes are largely conserved between rice and Arabidopsis, because the order of phenotypic severity on leaf polarity between OSHB3m, PHB/PHV ortholog, and OSHB1m, REV ortholog, is in parallel with that between gain-of-function PHB/PHV and REV in Arabidopsis (Kerstetter et al., 2001; Zhong and Ye, 2004).
However, remarkable differences between the genes in these two organisms are also evident. OSHB genes are involved in leaf margin development, as evidenced by the OSHB1 and OSHB3 expression in leaf margins and the independent formation of leaf margin-like structures from the preexisting leaves in OSHB3m plants. In addition, shoot organization mutants of rice exhibit reduced levels of Class III HD-Zip expression and the loss of specific leaf domains, including leaf margins (Itoh et al., 2000). Unlike typical eudicots, the leaves of grasses and other monocots have a membranous leaf margin that is clonally distinct from the main part of the leaf (Scanlon and Freeling, 1997). Thus, Class III HD-Zip genes were probably utilized in the morphological innovation of the leaf during evolution, in which the membranous leaf margin characteristic of grasses and other monocots was established.
Alternatively, the functions of Class III HD-Zip genes associated with leaf margin development in rice may be related to stipule formation in some eudicots. In Arabidopsis, at least one Class III HD-Zip gene, PHB, seemed to be expressed in stipules (McConnell et al., 2001). In addition, ectopic stipules surrounding the entire leaf base have been observed in phb-d and kan1 kan2, both of which show excess amounts of Class III HD-Zip transcripts (Eshed et al., 2001, 2004). If ectopic stipule formation in Arabidopsis and ectopic leaf margin formation in rice are equivalent events, the leaf margin in grasses is homologous to the stipule in eudicots. This interpretation is consistent with the characteristics of the narrow sheath mutant in maize (Nardmann et al., 2004). Although this issue must still be resolved, it is clear that Class III HD-Zip genes are involved in the diversification of leaves between rice and Arabidopsis.
CONCLUSION
Our results suggest that rice Class III HD-Zip genes are involved in key developmental processes that are conserved between rice and Arabidopsis, including embryo patterning, SAM initiation, leaf polarity, and vascular development. However, our findings indicate that rice Class III HD-Zip genes also regulate the leaf initiation process in an auxin-dependent manner. Furthermore, we have shown that Class III HD-Zip genes contribute to the morphological innovation and diversification of leaves in flowering plants.
MATERIALS AND METHODS
Plant Material
Rice plants (Oryza sativa ‘Taichung 65’) were used for this study. The plants were grown in a field or in a greenhouse at 30°C (day) and 25°C (night).
Isolation of the OSHB5 Gene
Because only a partial cDNA corresponding to the OSHB5 gene was found in the full-length cDNA database (KOME; http://cdna01.dna.affrc.go.jp/cDNA), a full-length OSHB5 cDNA was amplified using one primer specific to the 3′ untranslated region of the partial OSHB5 cDNA and another primer corresponding to a putative 5′ untranslated region sequence. The amplified fragment was inserted into pCR4 (Invitrogen) and sequenced.
Sequence and Phylogenetic Analysis
Full-length cDNAs of the OSHB1 to OSHB4 genes were identified in the rice full-length cDNA database, KOME. The splice sites of the OSHB genes were predicted by comparing the genomic sequences and their full-length cDNAs. The miR166-binding sequences of the five OSHB cDNAs were determined using the ClustalW program at the DNA Data Bank of Japan (http://www.ddbj.nig.ac.jp/). The Class III HD-Zip amino acid sequences of several species were identified by searching the public databases at the DNA Data Bank of Japan and the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/). Class III HD-Zip amino acid sequences were aligned using the ClustalW program at the DNA Data Bank of Japan, and then manually adjusted to optimize alignments using GENETYX software (Genetyx). The phylogenetic tree was constructed based on a full-length protein alignment generated with the neighbor-joining method using ClustalW and GENETYX, and then viewed and rooted using TREEVIEW (Page, 1996). The bootstrap values at the branching points were calculated from 1,000 replicates.
In Situ Hybridization
Samples were fixed in 4% paraformaldehyde in 0.1 m sodium phosphate buffer for 24 h at 48°C, and then dehydrated in a graded ethanol series. The dehydrated samples in 100% ethanol were replaced with xylene and embedded in Paraplast Plus (McCormick Scientific). Paraffin sections (8-μm thick) were applied to microscope slides coated with 3-aminopropyl triethoxysilane (Matsunami Glass). For the OSH1 probe, the full-length cDNA was used as a template. The OSHB1 and OSHB2 probes were prepared as described elsewhere (Nagasaki et al., 2007). For the OSHB3, OSHB4, and OSHB5 probes, a cDNA fragment was amplified by PCR and subcloned into pCR2.1 (Invitrogen) using primers specific to each gene (Supplemental Table S1). The probe lengths for OSHB1 to OSHB5 are 949 bp, 571 bp, 740 bp, 895 bp, and 750 bp, respectively. Among the five probes, the OSHB1 probe shows the highest similarity with OSHB2 mRNA (84.1%). Because no perfect overlap of the expression domains were observed between OSHB1 and OSHB2, a possibility of cross-hybridization among the genes is weak in our experimental condition. Digoxigenin-labeled antisense and sense riboprobes were generated by digestion with SpeI and transcription with T7 RNA polymerase and DIG-RNA labeling mix (Roche). In situ hybridization and immunological detection of the hybridization signals were performed as described by Kouchi and Hata (1993).
Generation of Transgenic Plants
The mutant constructs of OSHB1, OSHB3, and OSHB5 were generated from plasmids containing each OSHB cDNA by introducing five mutations into the predicted miR166-binding site (Supplemental Fig. S1) using a PCR-based site-directed mutagenesis method (Hemsley et al., 1989). The mutated cDNA fragments were inserted into the binary vector pBIAct1nos downstream of the Act1 promoter in the sense orientation between the XbaI and SmaI sites (Kamiya et al., 2003). Expression vectors and control vectors that lacked the cDNA fragments were introduced into calli of the cultivar Taichung 65 via Agrobacterium-mediated genetic transformation (Hiei et al., 1994). Transgenic plants that regenerated from calli were placed and grown in Murashige and Skoog medium aseptically for 1 month, and then observed and sampled.
Histological Analysis
For paraffin sectioning, samples were fixed in formaldehyde-acetic acid solution (formaldehyde:glacial acetic acid:ethanol [1:1:18]) for 24 h at 4°C, dehydrated in graded ethanol series, and embedded as described above. Microtome sections (8-μm thick) were stained with Delafield's hematoxylin, safranin, or Fast-Green FCF and then observed with a light microscope. Dehydrated samples in 100% ethanol were infiltrated with 3-methyl-butylacetate, critical-point dried, sputter-coated with platinum, and observed under an SEM (S-4000; Hitachi) at an accelerating voltage of 10 kV.
Real-Time RT-PCR
Total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. One microgram of RNA after DNase-I digestion was used for first-strand cDNA synthesis, and an RT reaction was performed using the High Capacity RNA-to-cDNA Master Mix (Applied Biosystems). The cDNA products were brought to a final volume of 200 μL, and 1.5 μL of the cDNA solution was subjected to amplification by real-time PCR. For quantification of the genes, Taq-Man assay was performed, using the TaqMan Fast Universal PCR Master Mix and FAM-labeled TaqMan MGB probes for each gene (Applied Biosystems), by the StepOnePlus real-time PCR system (Applied Biosystems). The expression level of each sample was normalized to that of an internal control, eEF-1α. The primers and Taq-Man MGB probes used to specifically detect OSHB1 to OSHB5, OsIAA9, and eEF-1α are listed in Supplemental Table S1.
Auxin Treatment
For short-term treatment, 7-d-old seedlings were placed in sterilized water containing 1 μm 2,4-D whose stock had been dissolved in ethanol. The seedlings were sampled at 0, 3, 6, 12, and 24 h after treatment. As a negative control, we used 7-d-old seedlings treated for 24 h with sterilized water containing the same amount of ethanol as the 2,4-D treatment. For long-term treatment, seeds were sterilized in 2% sodium hypochlorite and inoculated and grown aseptically for 7 d on Murashige and Skoog medium containing 3% Suc; 1% agar (pH 5.8); and 0, 0.1, or 1 μm 2,4-D in a plant box at 28°C.
The GenBank accession numbers for the sequences described in the text and in Figure 1 and Supplemental Figure S1 are OSHB1, AK102378; OSHB2, AK102603; OSHB3, AK102183; OSHB4, AK103284; OSHB5, AB374207; RLD1, AY501430; RLD2, ABB89930; ZeHB-10, AB084380; ZeHB-11, AB084381; ZeHB-12, AB084382; ZeHB-13, AB109562; REV/IFL1, AF188994; PHV/ATHB-9, AJ440967; PHB/ATHB-14, Y11122; CNA/ATHB-15, AJ439449; ATHB-8, Z50851; and PpHB10, AB032182. The RAP locus codes for OSHB1 to OSHB5 are Os03g0109400, Os10g0480200, Os12g0612700, Os03g0640800, and Os01g0200300, respectively. The TIGR locus codes are LOC_Os03g01890, LOC_Os10g33960, LOC_Os12g41860, LOC_Os03g43930, and LOC_Os01g10320, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of HD-ZIP III proteins, and nucleotide and amino acid sequences at the miR166-binding site of the OSHB1 to OSHB5 genes.
Supplemental Figure S2. Real-time RT-PCR analysis of OSHB genes in various tissues.
Supplemental Figure S3. In situ localization of OSHB transcripts in reproductive phase.
Supplemental Table S1. Primers and TaqMan probes used in this study.
Supplementary Material
This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (20248001 to Y.N. and 17780003 and 20061005 to J.-I.I.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Yasuo Nagato (anagato@mail.ecc.u-tokyo.ac.jp).
Some figures in this article are displayed in color online but in black and white in print.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Agalou A, Purwantomo S, Overnäs E, Johannesson H, Zhu X, Estiati A, de Kam RJ, Engström P, Slamet-Loedin IH, Zhu Z, et al (2008) A genome-wide survey of HD-Zip genes in rice and analysis of drought-responsive family members. Plant Mol Biol 66 87–103 [DOI] [PubMed] [Google Scholar]
- Baima S, Nobili F, Sessa G, Lucchetti S, Ruberti I, Morelli G (1995) The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 121 4171–4182 [DOI] [PubMed] [Google Scholar]
- Baima S, Possenti M, Matteucci A, Wisman E, Altamura MM, Ruberti I, Morelli G (2001) The Arabidopsis ATHB-8 HD-ZIP protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol 126 643–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Floyd SK (2008) Patterning and polarity in seed plant shoots. Annu Rev Plant Biol 59 67–88 [DOI] [PubMed] [Google Scholar]
- Chandler JW, Cole M, Flier A, Grewe B, Werr W (2007) The AP2 transcription factors DORNRÖSCHEN and DORNRÖSCHEN-LIKE redundantly control Arabidopsis embryo patterning via interaction with PHAVOLUTA. Development 134 1653–1662 [DOI] [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL (2003) Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol 13 1768–1774 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Perea JV, Bowman JL (2001) Establishment of polarity in lateral organs of plants. Curr Biol 11 1251–1260 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Izhaki A, Baum SF, Floyd SK, Bowman JL (2004) Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 131 2997–3006 [DOI] [PubMed] [Google Scholar]
- Floyd SK, Bowman JL (2004) Gene regulation: ancient microRNA target sequences in plants. Nature 428 485–486 [DOI] [PubMed] [Google Scholar]
- Floyd SK, Bowman JL (2006) Distinct developmental mechanisms reflect the independent origins of leaves in vascular plants. Curr Biol 16 1911–1917 [DOI] [PubMed] [Google Scholar]
- Floyd SK, Zalewski CS, Bowman JL (2006) Evolution of class III homeodomain-leucine zipper genes in streptophytes. Genetics 173 373–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM (2005) Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol 15 1899–1911 [DOI] [PubMed] [Google Scholar]
- Hemsley A, Arnheim N, Toney MD, Cortopassi G, Galas DJ (1989) A simple method for site-directed mutagenesis using the polymerase chain reaction. Nucleic Acids Res 17 6545–6551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson DC, Zhang X, Brooks L III, and Scanlon MJ (2006) RAGGED SEEDLING2 is required for expression of KANADI2 and REVOLUTA homologues in the maize shoot apex. Genesis 44 372–382 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6 271–282 [DOI] [PubMed] [Google Scholar]
- Itoh JI, Kitano H, Matsuoka M, Nagato Y (2000) SHOOT ORGANIZATION genes regulate shoot apical meristem organization and the pattern of leaf primordium initiation in rice. Plant Cell 12 2161–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh JI, Nonomura K, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, Kitano H, Nagato Y (2005) Rice plant development: from zygote to spikelet. Plant Cell Physiol 46 23–47 [DOI] [PubMed] [Google Scholar]
- Izhaki A, Bowman JL (2007) KANADI and class III HD-Zip gene families regulate embryo patterning and modulate auxin flow during embryogenesis in Arabidopsis. Plant Cell 19 495–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Kaur N, Garg R, Thakur JK, Tyagi AK, Khurana JP (2006) Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct Integr Genomics 6 47–59 [DOI] [PubMed] [Google Scholar]
- Juarez MT, Kui JS, Thomas J, Heller BA, Timmermans MCP (2004) microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428 84–88 [DOI] [PubMed] [Google Scholar]
- Kamiya N, Nagasaki H, Morikami A, Sato Y, Matsuoka M (2003) Isolation and characterization of a rice WUSCHEL-type homeobox gene that is specifically expressed in the central cells of a quiescent center in the root apical meristem. Plant J 35 429–441 [DOI] [PubMed] [Google Scholar]
- Kerstetter RA, Bollman K, Taylor RA, Bomblies K, Poethig RS (2001) KANADI regulates organ polarity in Arabidopsis. Nature 411 706–709 [DOI] [PubMed] [Google Scholar]
- Kouchi H, Hata S (1993) Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol Gen Genet 238 106–119 [DOI] [PubMed] [Google Scholar]
- McConnell JR, Barton MK (1998) Leaf polarity and meristem formation in Arabidopsis. Development 125 2935–2942 [DOI] [PubMed] [Google Scholar]
- McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK (2001) Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411 709–713 [DOI] [PubMed] [Google Scholar]
- Miyao A, Tanaka K, Murata K, Sawaki H, Takeda S, Abe K, Shinozuka Y, Onosato K, Hirochika H (2003) Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell 15 1771–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee K, Bürglin TR (2006) MEKHLA, a novel domain with similarity to PAS domains, is fused to plant homeodomain-leucine zipper III proteins. Plant Physiol 140 1142–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki H, Itoh J, Hayashi K, Hibara K, Satoh-Nagasawa N, Nosaka M, Mukouhata M, Ashikari M, Kitano H, Matsuoka M, et al (2007) The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proc Natl Acad Sci USA 104 14867–14871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardmann J, Ji JB, Werr W, Scanlon MJ (2004) The maize duplicate genes narrow sheath1 and narrow sheath2 encode a conserved homeobox gene function in a lateral domain of shoot apical meristems. Development 131 2827–2839 [DOI] [PubMed] [Google Scholar]
- Ochando I, Jover-Gil S, Ripoll JJ, Candela H, Vera A, Ponce MR, Martinez-Laborda A, Micol JL (2006) Mutations in the microRNA complementarity site of the INCURVATA4 gene perturb meristem function and adaxialize lateral organs in Arabidopsis. Plant Physiol 141 607–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi-Ito K, Fukuda H (2003) HD-Zip III homeobox genes that include a novel member, ZeHB-13 (Zinnia)/ATHB-15 (Arabidopsis), are involved in procambium and xylem cell differentiation. Plant Cell Physiol 44 1350–1358 [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Kubo M, Demura T, Fukuda H (2005) Class III homeodomain leucine-zipper proteins regulate xylem cell differentiation. Plant Cell Physiol 46 1646–1656 [DOI] [PubMed] [Google Scholar]
- Otsuga D, Deguzman B, Prigge M, Drews G, Clark S (2001) REVOLUTA regulates meristem initiation at lateral positions. Plant J 25 223–236 [DOI] [PubMed] [Google Scholar]
- Page RD (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12 357–358 [DOI] [PubMed] [Google Scholar]
- Ponting CP, Aravind L (1999) START: a lipid-binding domain in StAR, HD-ZIP and signalling proteins. Trends Biochem Sci 24 130–132 [DOI] [PubMed] [Google Scholar]
- Prigge MJ, Clark SE (2006) Evolution of the class III HD-Zip gene family in land plants. Evol Dev 8 350–361 [DOI] [PubMed] [Google Scholar]
- Prigge MJ, Otsuga D, Alonso JM, Ecker JR, Drews GN, Clark SE (2005) Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17 61–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP (2002) MicroRNAs in plants. Genes Dev 16 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D, Mandel T, Kuhlemeier C (2000) Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D, Pesce ER, Stieger P, Mandel T, Baltensperger K, Bennett M, Traas J, Friml J, Kuhlemeier C (2003) Regulation of phyllotaxis by polar auxin transport. Nature 426 255–260 [DOI] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP (2002) Prediction of plant microRNA targets. Cell 110 513–520 [DOI] [PubMed] [Google Scholar]
- Satoh N, Hong SK, Nishimura A, Matsuoka M, Kitano H, Nagato Y (1999) Initiation of shoot apical meristem in rice: characterization of four SHOOTLESS genes. Development 126 3629–3636 [DOI] [PubMed] [Google Scholar]
- Scanlon MJ, Freeling M (1997) Clonal sectors reveal that a specific meristematic domain is not utilized in the maize mutant narrow sheath. Dev Biol 182 52–66 [DOI] [PubMed] [Google Scholar]
- Sentoku N, Sato Y, Kurata N, Ito Y, Kitano H, Matsuoka M (1999) Regional expression of the rice KN1-type homeobox gene family during embryo, shoot, and flower development. Plant Cell 11 1651–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa G, Steindler C, Morelli G, Ruberti I (1998) The Arabidopsis Athb-8, -9 and -14 genes are members of a small gene family coding for highly related HD-ZIP proteins. Plant Mol Biol 38 609–622 [DOI] [PubMed] [Google Scholar]
- Steeves TA, Sussex IM (1989) Patterns in Plant Development, Ed 2. Cambridge University Press, Cambridge, UK
- Talbert PB, Adler HT, Parks DW, Comai L (1995) The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development 121 2723–2735 [DOI] [PubMed] [Google Scholar]
- Tang G, Reinhart BJ, Bartel DP, Zamore PD (2003) A biochemical framework for RNA silencing in plants. Genes Dev 17 49–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Taylor JJ, Ye ZH (1997) Disruption of interfascicular fiber differentiation in an Arabidopsis mutant. Plant Cell 9 2159–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH (1999) IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis, encodes a homeodomain-leucine zipper protein. Plant Cell 11 2139–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH (2001) Alteration of auxin polar transport in the Arabidopsis ifl1 mutants. Plant Physiol 126 549–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong RQ, Ye ZH (2004) amphivasal vascular bundle 1, a gain-of-function mutation of the IFL1/REV gene, is associated with alterations in the polarity of leaves, stems and carpels. Plant Cell Physiol 45 369–385 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.