Abstract
Cyanobacteria are eubacteria that perform oxygenic photosynthesis like plants. The initiation of transcription, mediated by the RNA polymerase holoenzyme, is the main determinant of gene regulation in eubacteria. The σ factor of the RNA polymerase holoenzyme is responsible for the recognition of a promoter sequence. In the cyanobacterium Synechocystis sp. PCC 6803, the primary σ factor, SigA, is essential for cell viability. The SigB, SigC, SigD, and SigE factors show significant sequence similarity with the SigA factor but are nonessential. In this study, we have used homology modeling to construct a three-dimensional model of Synechocystis RNA polymerase holoenzyme and all group 1 and 2 σ factors. According to the models, the overall three-dimensional structures of group 1 and 2 σ factors are similar, the SigB and SigD factors being the most similar ones. In addition, we have constructed a complete set of group 2 σ factor double inactivation strains, ΔsigBC, ΔsigBD, ΔsigBE, ΔsigCD, ΔsigCE, and ΔsigDE. All double mutants grow well under standard conditions, but differences are observed in stress conditions. The transition from lag phase to exponential growth is slow in the ΔsigBD strain, and all strains lacking the SigD factor were found to be sensitive to bright light. Furthermore, all group 2 σ factors were found to be involved in acclimation to salt- or sorbitol-induced osmotic stresses.
Cyanobacteria are evolutionarily ancient eubacteria that perform oxygenic photosynthesis and are known as the ancestors of chloroplasts (Rodríguez-Ezpeleta et al., 2005). Present-day cyanobacteria are found in most natural habitats. Growth and survival under a range of different environmental stress conditions make cyanobacteria valuable model organisms in studies of molecular mechanisms underlying the acclimation processes of autotrophs. The unicellular cyanobacterium Synechocystis sp. PCC 6803 (hereafter Synechocystis) has been used extensively in gene expression studies under a variety of different stress conditions (Hihara et al., 2001; Huang et al., 2002; Kanesaki et al., 2002; Shoumskaya et al., 2005; Foster et al., 2006; Singh et al., 2006; Summerfield and Sherman, 2007; Tuominen et al., 2008). These and other studies have demonstrated that acclimation to changing environmental conditions requires changes in gene expression over a wide range of different functions.
In eubacteria, the main determinant of gene regulation is the initiation of transcription mediated by the RNA polymerase holoenzyme. The eubacterial RNA polymerase holoenzyme is composed of a core enzyme (with the subunit composition α2, β, β′, ω) and a σ factor. The core enzyme of the RNA polymerase exhibits the RNA polymerase activity, while the σ factor is responsible for the recognition of promoter sequences. Most bacteria synthesize several σ factors that compete for the same RNA polymerase core (Maeda et al., 2000), and it is believed that replacement of the σ factor with another σ factor is a major switch for changing the global transcription pattern in eubacteria.
Nine genes encode σ factors in Synechocystis (Kaneko et al., 1996). The sigA gene encodes the principal (group 1) σ factor that is essential for cell viability. The sigB, sigC, sigD, and sigE genes encode group 2 σ factors (primary-like σ factors) that show extensive amino acid similarity with the SigA factor but are not essential for cell viability under optimal growth conditions (Imamura et al., 2003; Tuominen et al., 2003). Recent results have shown that group 2 σ factors are important under suboptimal conditions (Osanai et al., 2005; Singh et al., 2006; Tuominen et al., 2006, 2008; Summerfield and Sherman 2007). A complicated regulatory network between the group 1 and group 2 σ factors has been suggested to function in Synechocystis (Lemeille et al., 2005). The sigF, sigG, sigH, and sigI genes encode alternative σ factors that vary more considerably in amino acid sequence that those of group 1 and 2 σ factors.
The structure of the bacterial RNA polymerase holoenzyme from the thermophilic bacteria Thermus thermophilus (Vassylyev et al., 2002, 2005) and Thermus aquaticus (Murakami et al., 2002a, 2002b) has been determined by x-ray crystallography. In this study, we took advantage of the high sequence identity between bacterial RNA polymerases and used the crystal structure of the RNA polymerase of T. thermophilus to construct a three-dimensional model of the RNA polymerase holoenzyme with SigA factor in Synechocystis. In addition, we constructed three-dimensional models of all group 2 σ factors of Synechocystis. Based on the models, the overall three-dimensional structures of group 1 and 2 σ factors resemble each other; the SigB and SigD factors being the most similar ones. In addition to homolog models, we have constructed a complete set of double inactivation strains of group 2 σ factors, including ΔsigBC, ΔsigBD, ΔsigBE, ΔsigCD, ΔsigCE, and ΔsigDE. We show that although all double mutants grow well under standard conditions, the transfer from lag phase to exponential growth is slow in the ΔsigBD strain. All strains lacking the SigD factor were found to be sensitive to bright light. Furthermore, all group 2 σ factors were found to be involved in acclimation to osmotic stress.
RESULTS
Specific Features of the Synechocystis RNA Polymerase
We constructed the structural models of the Synechocystis RNA polymerase holoenzyme using the crystal structure of the RNA polymerase holoenzyme of T. thermophilus (Artsimovitch et al., 2005; Protein Data Bank [PDB] code 2A6E; chains A–F) as a template. The details of the different subunits of the RNA polymerase of T. thermophilus, including areas that are missing from the template structure, together with the RNA polymerase subunits of Synechocystis, are shown in Supplemental Table S1. The ω subunit was excluded from the model because its sequence identity (20%) with the template was too low to ensure reliable modeling. The other subunits were 40% to 50% identical (Supplemental Table S1). Ramachandran plots of the models showed that approximately 90% of amino acid residues were in the most favorable regions and less than 1% were in disallowed regions (the corresponding values for the template were 83.9% and 0.1%), indicating good overall reliability for the models.
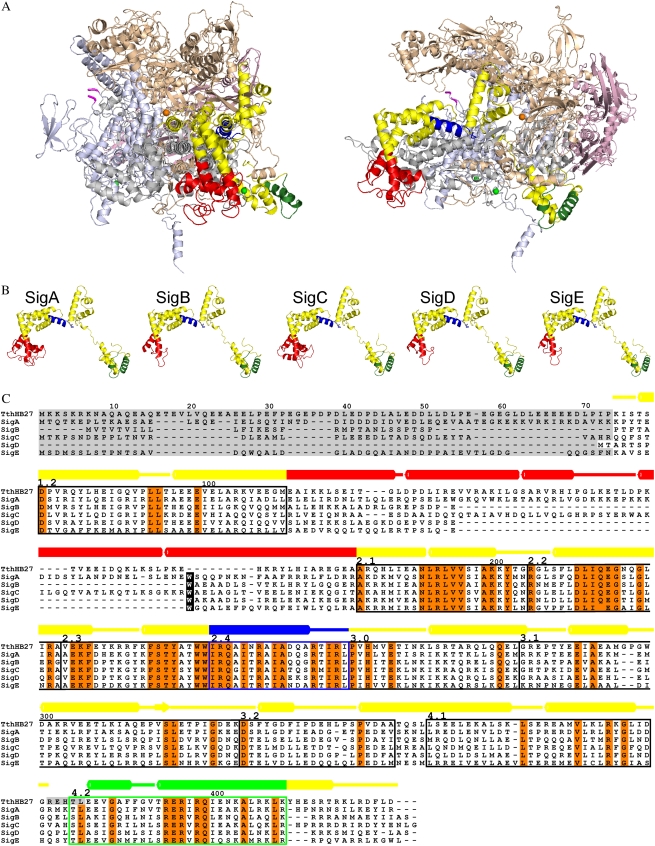
A specific feature of the cyanobacterial RNA polymerase is that the β′ subunit has been split into two different polypeptides (Schneider and Hasekorn, 1988). The γ subunit of the RNA polymerase in cyanobacteria is homologous with the N-terminal part of the β′ subunit of other eubacteria, and the cyanobacterial β′ subunit is homologous with the C-terminal part of the β′ subunit of other eubacteria. In Synechocystis, the γ subunit consists of 626 residues (light gray in Fig. 1) and the β′ subunit consists of 1,317 residues (light blue in Fig. 1). According to the model, the splitting has very little effect on the overall structure of the RNA polymerase of Synechocystis, as the last amino acid residue of the γ subunit and the first amino acid of the β′ subunit are located on the surface of RNA polymerase. Another special feature of the cyanobacterial β′ subunit is that it includes a large insertion (Iyer et al., 2004) containing 635 amino acid residues in Synechocystis. The first and last amino acid residues of the insertion are shown in magenta in Figure 1A, but the insertion is not included in the model because of the lack of template for modeling. Although the three-dimensional structure of the cyanobacterial insertion remains to be solved, our model suggests that the insertion can be accommodated in the three-dimensional structure without introducing dramatic changes in the other parts of the RNA polymerase holoenzyme (Fig. 1A).
Figure 1.
Model of Synechocystis RNA polymerase. A, Homology modeling of the RNA polymerase holoenzyme with the SigA factor was performed using the crystal structure of the RNA polymerase holoenzyme of T. thermophilus (Artsimovitch et al., 2005; PDB code 2A6E; chains A–F) as the template. Views are from the front toward the catalytic center (left) and from the side (right). The core enzyme consists of two α subunits (light pink), the β subunit (light brown), the β′ subunit (light blue), and the γ subunit (gray). The first and last residues of the cyanobacterial insertion in the β′ subunit are indicated with magenta. In the otherwise yellow σ factor, the 4.2 region is green, the 2.4 region is blue, and the NCR connecting the conserved regions 1.2 and 2.1 is red. The catalytic magnesium ion is presented as an orange sphere, and two zinc ions are presented as green spheres. B, Models of principal (SigA) and group 2 (SigB–SigE) σ factors of Synechocystis. The coloring is as in A. C, Sequence alignment of Synechocystis and T. thermophilus σ factors. Absolutely conserved residues are shown in orange, and conserved regions from 1.2 to 4.2 are indicated with boxes. Residues missing from the structures are shaded gray. The numbering of amino acid residues and the secondary structure assignment, with barrels denoting α-helices, are shown above the alignment for the template. The coloring of secondary structures is as in A. The cyanobacterial conserved Trp residue is indicated with a black background.
The three-dimensional models (Fig. 1B) and sequence alignment (Fig. 1C) are shown for the primary σ factor and for all group 2 σ factors. The SigA factor is included in the holoenzyme model (Fig. 1A). The basic structure of all σ factors is similar, consisting essentially of α-helices; the α-helices are shown as colored barrels above the sequence alignment (Fig. 1C). Based on amino acid sequence homology, four homologous regions that are further divided into subregions have been identified in group 1 and 2 σ factors (Lonetto et al., 1992). These subregions are indicated as boxes in Figure 1C. The 4.2 region (green) recognizes the −35 promoter element, and the 2.4 region (blue) recognizes the −10 promoter element. According to the three-dimensional models, these elements are similar in group 1 and group 2 σ factors. The most variable part of the group 1 and group 2 σ factors of Synechocystis is the nonconserved domain (NCD; red in Fig. 1) between the conserved domains 1.2 and 2.1. In primary σ factors of different bacteria, the length of the NCD region varies from two (Bacillus subtilis) to 315 (Bradyrhizobium japonicum) amino acids; in cyanobacteria, the variation is from 40 to 88 amino acids. In Synechocystis, the length of the NCD is 86 amino acids in SigA, 42 in SigB, 84 in SigC, 43 in SigD, and 44 in SigE. Based on secondary structure predictions (data not shown), the NCD is suggested to be helical in all group 1 and group 2 σ factors of Synechocystis. However, sequence identities in the NCDs are low, and thus this area in the models is less reliable than the other regions. Although the length of the NCD is similar in SigB, SigD, and SigE factors, only the NCD sequences of the SigB and SigD factors are similar (47% identity). The long NCDs of SigA and SigC do not show high sequence identity.
Single and Double Inactivation Strains of Group 2 σ Factors in Synechocystis
In order to study the role of each group 2 σ factor, we constructed single and double inactivation strains. The sigB (strain ΔsigB), sigC (strain ΔsigC), sigD (strain ΔsigD), and sigE (strain ΔsigE) genes were inactivated with a kanamycin (Kn) resistance cassette in Synechocystis. The constructs of all inactivation strains are shown in Supplemental Figure S1. The double inactivation strains were constructed by inactivating the second sig gene with a streptomycin/spectinomycin (Str/Spc) resistance cassette. The sigC gene was inactivated in the ΔsigB strain (resulting in ΔsigBC), the sigD gene was inactivated in the strains ΔsigB (resulting in ΔsigBD) and ΔsigC (resulting in ΔsigCD), and the sigE gene was inactivated in strains ΔsigB (resulting in ΔsigBE), ΔsigC (resulting in ΔsigCE), and ΔsigD (resulting in ΔsigDE). Two lines that were descendants of two independently raised colonies on the first selection plate were originally tested from each inactivation strain. Because the two lines behaved similarly, the results are shown only for one line. Testing of two independent lines minimizes the possibility that the results could be affected by secondary mutations. PCR verification confirmed that the strains were completely segregated (Supplemental Fig. S2).
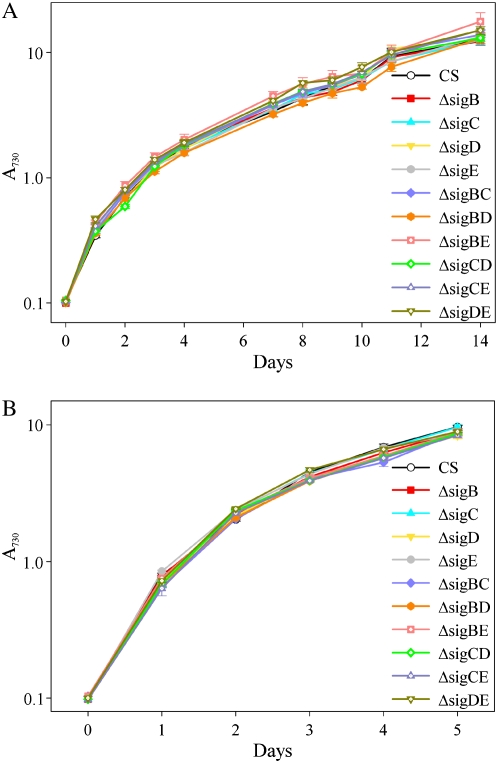
First we measured the growth rates of all single and double inactivation strains under standard growth conditions (32°C, continuous light of 40 μmol photons m−2 s−1, ambient CO2 concentration). A730 was set to 0.1, and the growth of a 50-mL cell culture in a 250-mL Erlenmeyer flask was followed for 14 d. All inactivation strains grew autotrophically with the same growth rate as the control strain (Fig. 2A). In our standard conditions, growth was exponential only at the very beginning of the growth experiment; thereafter, the growth was linear for a few days. Finally, growth of the cells slowed down, and by the 14th d the cells were hardly growing (Fig. 2A). As some researchers routinely grow Synechocystis cells under CO2-enriched atmosphere, we tested the growth of all inactivation strains under otherwise similar conditions as our standard conditions but supplemented the air of the growth chamber with 3% CO2. All strains grew faster under high-CO2 conditions than under ambient CO2 conditions, the doubling time being 8 h at high CO2 and 14 h at air level CO2 during the 1st d, but no differences were detected between the control and inactivation strains (Fig. 2B). The growth rate became slower throughout the whole experiment, both in 3% CO2 and in ambient CO2.
Figure 2.
Growth of single and double inactivation strains of group 2 σ factors of Synechocystis. The A730 of the cell culture was set to 0.1, and the cells were grown in BG-11 medium under the continuous illumination of 40 μmol photons m−2 s−1 at 32°C under air levels of CO2 (A) or under 3% CO2 (B). Each growth curve represents the mean of five independent experiments, and the error bars denote se.
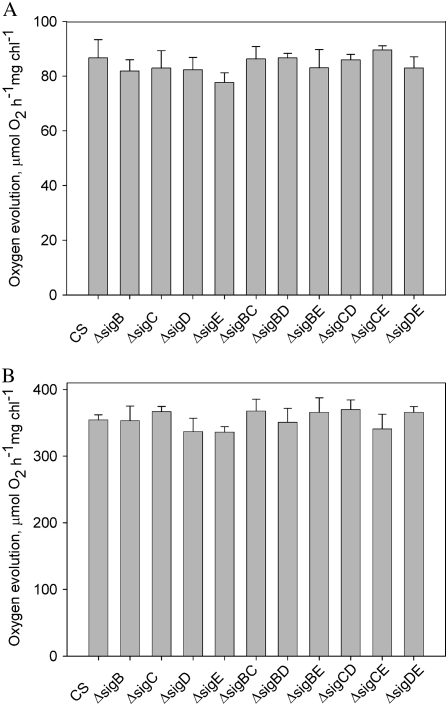
In accordance with similar growth rates, also photosynthetic activity, measured as light-saturated oxygen evolution, was found to be similar in all strains (Fig. 3A). Furthermore, similar PSII electron transport capacity was measured from the control and all inactivation strains (Fig. 3B). These findings indicate that two group 2 σ factor genes can be inactivated simultaneously in any combination without affecting the growth rate or photosynthesis under standard growth conditions.
Figure 3.
Light-saturated rates of photosynthesis (A) and PSII electron transport (B) in the σ factor inactivation strains. The rate of in vivo photosynthetic oxygen evolution was measured with an oxygen electrode in BG-11 medium supplemented with 10 mm NaHCO3 under saturating light (500 μmol photons m−2 s−1) at 32°C. The light-saturated rate of PSII electron transport was measured similarly as photosynthesis, except that 0.7 mm 2,6-dichlorobenzoquinone was used as an artificial electron acceptor. The measurements were repeated three times using independent liquid cultures each time. The error bars denote se.
All Strains with an Inactivated sigD Gene Grow Slowly under Bright Light, and the Transition from Lag to Exponential Phase Is Slow in the ΔsigBD Strain
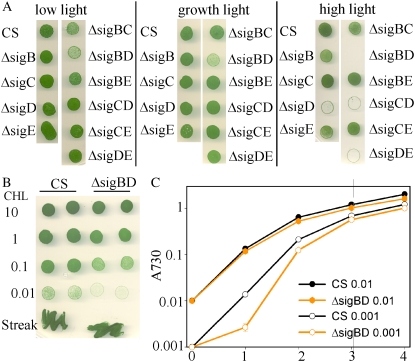
To follow the growth of the inactivation strains under different light conditions, dilute Synechocystis cultures (A730 was 0.1, corresponding to 3.6 × 106 cells mL−1) were spotted on BG-11 plates and the plates were grown under constant irradiance of 20, 40, or 80 μmol photons m−2 s−1 at 32°C. Under low light (20 μmol photons m−2 s−1), the growth of the ΔsigBD strain was delayed, and also the ΔsigBC strain grew slightly more slowly than the control strain or the other inactivation strains (Fig. 4A).
Figure 4.
Growth of the inactivation strains under different light intensities. A, The A730 of each strain was set to 0.1, and 5 μL of each cell culture was spotted on BG-11 plates. The plates were grown under continuous illumination of 20 (low light), 40 (growth light), or 80 (high light) μmol photons m−2 s−1 at 32°C for 7 d (growth and high light) or 8 d (low light). The data shown are representative of three independent experiments showing similar results. B, The A730 of the control (CS) and ΔsigBD strains was set to 10, 1, 0.1, or 0.01, and 5 μL of each dilution was spotted on BG-11 plates or cells were streaked directly from an old plate. The data shown are representative of three independent experiments showing similar results. C, Growth of the control and ΔsigBD strains in liquid culture. The A730 of the cell culture was set to 0.01 (closed symbols) or 0.001 (open symbols), and the cells were grown in BG-11 medium under continuous illumination of 40 μmol photons m−2 s−1 at 32°C for 4 d. Each growth curve represents the mean of three independent experiments, and the error bars denote se.
Under standard growth conditions, the inactivation strains grew like the control strain, except that the ΔsigBD strain grew slowly in the spot test (Fig. 4A). This was a surprise for us, as similar growth rates were measured for the control and ΔsigBD strains in liquid cultures (Fig. 2) and we did not see any difference between the control and ΔsigBD strains when we streaked a new plate using cells directly from the old plate (Fig. 4B). The difference in growth rates between the ΔsigBD and control strains became larger when more dilute cultures were used as starting material in the spot experiments, but the difference disappeared completely if dense cultures (10-fold concentration after adjusting A730 to 1.0) were spotted on the plates (Fig. 4B). Because slow growth of the ΔsigBD strain on plates was clearly dependent on cell density at the beginning of the experiment, we next tested the cell density dependence of growth in liquid culture. At initial A730 of 0.01, both control and ΔsigBD strains grew rapidly. When A730 was 0.001, 100 times more dilute than was used in the beginning of our standard growth curve measurements, the doubling time of the control strain was only 6 h during the 1st d, indicating very fast growth. The ΔsigBD strain, in turn, grew slowly during the 1st d, the doubling time being 17 h (Fig. 4C). During the next 2 d, the growth of the control strain slowed down as the density of the culture became higher, but the ΔsigBD strain grew faster, and finally the growth difference between the strains almost disappeared. The finding that only the double mutant ΔsigBD, not ΔsigB or ΔsigD, showed slow growth in dilute culture indicates that the presence of either the SigB or the SigD factor is sufficient for normal efficient transfer of the cells from the lag growth phase to the exponential growth phase. Partial redundancy of the functions of the SigB and SigD factors may be related to the fact that these are the two most homologous σ factors of Synechocystis.
The dependence of the growth of the ΔsigBD strain on cell density suggests that cell-to-cell communication might be important for growth. We tested the possibility that cells of the control and ΔsigBD strains secrete different chemical signals to the growth medium. The A730 was set to 0.001 and the control and ΔsigBD strains were grown for 3 d. Thereafter, the cells were removed from the growth medium and new dilute batch cultures were started so that cells of the control strain grew in used ΔsigBD strain-BG-11 medium and the new ΔsigBD culture was started in used control strain medium. We compared the growth of control and ΔsigBD strains in these and in fresh BG-11 media; at the beginning of the experiment, the A730 was set to 0.001. The ΔsigBD strain always grew more slowly than the control strain, despite the used growth medium (data not shown), indicating that a secreted chemical signal is unlikely to explain the slow growth of the ΔsigBD strain in dilute culture or that the chemical signal is short-lived.
In the spot test experiments, the clearest phenotypes were seen under high-light conditions. Strains with an inactivated sigD gene (ΔsigD, ΔsigCD, and ΔsigDE) hardly grew at all at 80 μmol photons m−2 s−1, and the double inactivation strain ΔsigBD died (Fig. 4A). These results indicate that the SigD factor is extremely important for acclimation to bright light and further underline the redundancy of the functions of the SigB and SigD factors. The results also show that the antibiotic resistance cassette used to inactivate the gene does not interfere with the results, as the sigD gene was inactivated with a Kn cassette in strains ΔsigD and ΔsigDE and with a Str/Spc cassette in strains ΔsigBD and ΔsigCD (Supplemental Fig. S1).
All Group 2 σ Factors Are Involved in Osmotic Acclimation of Synechocystis
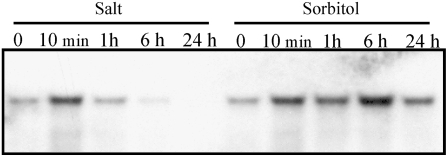
Our earlier experiments indicated activation of the sigB gene by a short osmotic shock (Tuominen et al., 2003). We studied the expression of the sigB gene in more detail by following the amount of sigB transcripts under salt- and sorbitol-induced osmotic stress in the control strain. Addition of 0.7 m NaCl induced an 8-fold increase in the amount of sigB transcripts within 10 min, and after a 1-h treatment, the amount of sigB transcripts was still twice as high as that measured under the standard growth conditions (Fig. 5). Thereafter, the amount of sigB transcripts decreased below the amount measured under the standard growth conditions, and only traces of sigB transcripts were detected after 24 h of salt treatment (Fig. 5). Sorbitol-induced osmotic stress, in turn, caused more permanent up-regulation of sigB transcripts, and three times as high levels of sigB transcripts, compared with the amount measured under standard growth conditions, were measured even after 24 h of sorbitol-induced osmotic stress (Fig. 5). In addition to the SigB factor, the SigD factor also has been suggested to be involved in signal transduction of short high-salt and high-sorbitol shock treatments (Shoumskaya et al., 2005). To get a more comprehensive picture of the importance of the different σ factors under osmotic stress, we grew cells of all inactivation strains for 5 d in BG-11 medium supplemented with either 0.7 m NaCl or 0.5 m sorbitol. These concentrations of salt and sorbitol were chosen because they slowed down the growth of the control strain by approximately 30% in our test experiments.
Figure 5.
Accumulation of sigB mRNA in the control strain under osmotic stress. The growth medium was supplemented with 0.7 m NaCl or 0.5 m sorbitol, as indicated, and samples were withdrawn before the addition (0) and after 10 min and 1, 6, and 24 h of incubation. Thereafter, total RNA was isolated and the amount of sigB mRNA was detected with the northern-blot technique. The data shown are representative of three independent northern-blot experiments showing similar results.
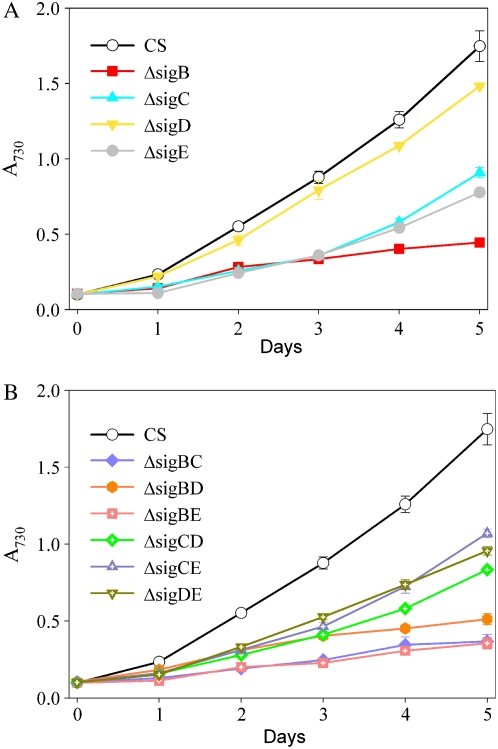
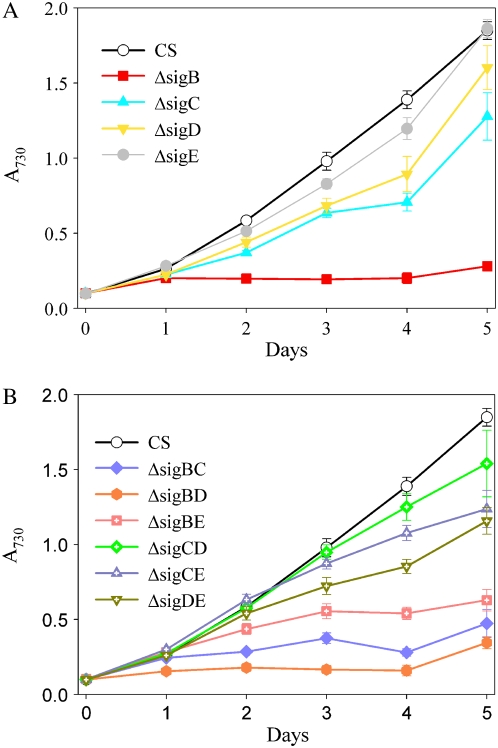
The growth of the ΔsigB strain was seriously retarded in high-salt conditions and practically ceased after 2 d (Fig. 6A), while the ΔsigD strain grew almost as well as the control strain, and only after a prolonged salt stress did the growth of the ΔsigD strain decrease slightly compared with the control strain. Inactivation strains ΔsigC and ΔsigE grew slowly under salt stress throughout the experiment (Fig. 6A). From the double mutants, the ΔsigBC, ΔsigBD, and ΔsigBE strains grew as slowly as the ΔsigB strain (Fig. 6B), indicating that inactivation of another group 2 σ factor in addition to the SigB factor did not cause a more severe phenotype under high-salt stress. The other double mutant strains, ΔsigCD, ΔsigCE, and ΔsigDE, grew similarly as the single inactivation strains ΔsigC and ΔsigE. These results suggest that SigB is the most crucial σ factor for acclimation to high salt. In addition, the SigC and SigE factors are required for optimal acclimation to high-salt stress. The SigD factor, in turn, has only a minor role, if any, for salt acclimation. Furthermore, these results indicate that the redundancy of the SigD and SigB factors does not extend to all functions of these two σ factors.
Figure 6.
Growth of single (A) and double (B) inactivation strains under high-salt stress. The A730 of the cell culture was set to 0.1, and the cells were grown in BG-11 medium supplemented with 0.7 m NaCl under continuous illumination of 40 μmol photons m−2 s−1 at 32°C. Each growth curve represents the mean of three independent experiments, and the error bars denote se.
In accordance with the results of the high-salt experiments, all inactivation strains with inactivated sigB gene were susceptible to sorbitol-induced osmotic stress (Fig. 7, A and B). Contrary to salt-induced osmotic stress, inactivation of the SigE factor had only a minor effect on growth when osmotic stress was induced with sorbitol (Fig. 7A). The ΔsigC strain showed reduced growth, but the effect of the SigC factor was milder under sorbitol-induced stress than under salt-induced stress. The SigD factor, in turn, was more important for acclimation to high sorbitol than for high-salt stress. Some double mutants showed peculiar behavior, as both ΔsigBE and ΔsigBC actually grew slightly better than the ΔsigB strain. The reasons for this behavior remain to be solved.
Figure 7.
Growth of single (A) and double (B) inactivation strains under high-sorbitol stress. The A730 of the cell culture was set to 0.1, and the cells were grown in BG-11 medium supplemented with 0.5 m sorbitol under continuous illumination of 40 μmol photons m−2 s−1 at 32°C. Each growth curve represents the mean of at least five independent experiments, and the error bars denote se.
DISCUSSION
Cyanobacterial genomes typically code for several group 2 σ factors. Studies of single inactivation strains of different group 2 σ factors in Synechocystis (Asayama et al., 2004; Tuominen et al., 2006, 2008; Summerfield and Sherman, 2007), Anabaena sp. PCC 7120 (Khudyakov and Golden, 2001), Synechococcus sp. PCC 7002 (Caslake et al., 1997), and Synechococcus sp. PCC 7942 (Goto-Seki et al., 1999) as well as double inactivation strains in Synechocystis (Fig. 2) indicate that these σ factors have little or no effect on growth under optimal conditions. Evidence accumulated in recent years strongly suggests that group 2 σ factors are required for acclimation under stress conditions (Table I).
Table I.
Growth of group 2 σ factor inactivation strains under different environmental conditions
Growth is compared with the growth of the control strain under the same conditions. +++, Similar growth as in the control strain; ++(+), slightly slower growth; ++, slower growth; +, only very slow growth; +/−, slow growth for a short time; −, no growth; ND, not determined.
| Growth Conditions | Inactivated sig Gene
|
Reference | |||
|---|---|---|---|---|---|
| sigB | sigC | sigD | sigE | ||
| Standard | +++ | +++ | +++ | +++ | This study |
| High CO2 | +++ | +++ | +++ | +++ | This study |
| Heat | ++(+) | +/− | ++(+) | ND | Tuominen et al. (2006, 2008) |
| Salt stress | +/− | + | ++(+) | + | This study |
| Sorbitol | +/− | ++ | ++ | ++(+) | This study |
| High light | +++ | +++ | + | +++ | This study |
| Light-activated heterotrophic growth | ND | ND | ND | − | Osanai et al. (2005) |
| Mixotrophic growth, 8 h of light/16 h of dark | +++ | ND | +++ | + | Summerfield and Sherman (2007) |
The SigB factor can be considered as a general stress-responsive σ factor. Rapid transient increase of SigB transcripts has been detected in high-salt stress (Fig. 5), in heat stress (Imamura et al., 2003; Shoumskaya et al., 2005; Tuominen et al., 2006), and after a dark-to-light shift (Tuominen et al., 2003). Furthermore, up-regulation of the sigB gene has been found to occur under oxidative stress induced by hydrogen peroxide treatment (Kanesaki et al., 2007) and under nitrogen starvation (Imamura et al., 2006). SigB factor has also been implicated to be involved in the regulation of many genes in light-dark transitions (Summerfield and Sherman, 2007) and in exponential or linear growth phases (Foster et al., 2006), but in these studies the growth of the control and ΔsigB strains was similar, suggesting that in these cases SigB can be complemented by other σ factors.
We show in this study that the SigB factor is rapidly up-regulated by osmotic stress induced by salt and by sorbitol and that cells do not grow without SigB under these conditions. On the other hand, similar rapid transient up-regulation of the sigB gene occurs at mild heat stress at 43°C, but the ΔsigB strain grows almost as well as the control strain (Tuominen et al., 2006). These results indicate that there is no simple direct correlation between the increase in sigB transcripts and the importance of the SigB factor under a particular stress condition.
It is well documented that the SigB factor is involved in the up-regulation of heat shock genes, especially the hspA gene, at high temperatures (Imamura et al., 2003; Singh et al., 2006; Tuominen et al., 2006). Many heat shock proteins, particularly HspA, are important chaperones in acclimation to osmotic stress (Asadulghani et al., 2004). We compared the expression of the hspA gene in the control and ΔsigB strains in osmotic stresses. In salt stress, the amount of hspA mRNA remained lower in ΔsigB than in the control strain (Supplemental Fig. S3), but the expression kinetics was similar in both strains. However, when osmotic stress was induced with sorbitol, the induction of the hspA gene was slower in the ΔsigB strain than in the control strain, but after 1 h the amount of hspA mRNA in ΔsigB exceeded that in the control strain (Supplemental Fig. S3). Interestingly, an hspA inactivation strain has been found to tolerate well mild (0.5 m NaCl) salt-induced osmotic stress (Asadulghani et al., 2004), and we have noticed that the ΔsigB strain grows well in BG-11 medium supplemented with 0.5 m NaCl (data not shown). These results indicate that inactivation of the sigB gene affects the expression of the hspA gene not only in heat stress but also in salt- and sorbitol-induced stresses.
In this study, we show that the SigC factor is involved in acclimation to salt stress and also to a lesser extent to sorbitol stress. Previously, SigC of Synechocystis and SigE, its closest homolog in Synechococcus sp. PCC 7002, were assigned roles in stationary phase-related gene expression and growth (Gruber and Bryant, 1998; Asayama et al., 2004; Imamura et al., 2006) and in Synechocystis also in acclimation to heat stress (Tuominen et al., 2008). We did not see differences in growth rate between the control and ΔsigC strains during the studied 14 d in standard conditions (Fig. 2), but Asayama et al. (2004) noticed a slight delay of growth in a sigC inactivation strain after 3 weeks. A unique feature of the SigC factor is that the sigC gene is not specifically up-regulated at high temperature (Tuominen et al., 2008) or under osmotic stress (Tuominen et al., 2003, Shoumskaya et al., 2005) or stationary phase (Asayama et al., 2004), yet SigC factor is required for acclimation to those conditions. These results suggest that the sigC gene might be regulated posttranscriptionally. One possible explanation is that the RNA polymerase core has an increased tendency to recruit the SigC factor under these stress conditions. In Escherichia coli, the small signaling molecule ppGpp (for guanosine 5′-diphosphate 3′-diphosphate) directly binds to the RNA polymerase core (Artsimovitch et al., 2004), and active transcription from promoters that depend on σS, the only group 2 σ factor in E. coli, requires ppGpp (Kvint et al., 2000). A similar system might explain why SigC is important in conditions in which the sigC gene is not up-regulated.
All strains with inactivated sigD gene were unable to grow under high light. DNA microarray analysis has shown that the expression of the sigD gene increases in response to high light in Synechocystis (Hihara et al., 2001; Huang et al., 2002). Furthermore, Imamura et al. (2003) reported an increase in the amount of SigD protein after high-light treatment. In another cyanobacterium, Synechococcus elongatus PCC 7942, one of the five group 2 σ factors, RpoD3, was recognized as a high-light-responsive σ factor (Seki et al., 2007). The amino acid sequences of the RpoD3 factor of Synechococcus elongatus PCC 7942 and the SigD factor of Synechocystis suggest that these factors are closely related (Seki et al., 2007).
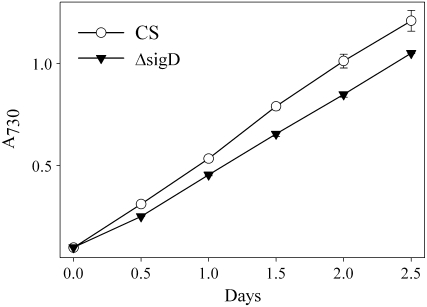
Enhancement of the expression of the sigD gene has been detected in both sorbitol- and salt-induced stresses (Shoumskaya et al., 2005). However, inactivation of the sigD gene only slightly slowed down the growth of Synechocystis under high-salt or high-sorbitol conditions (Figs. 6 and 7), and our earlier experiments did not show induction of sigD under salt stress (Tuominen et al., 2003). Shoumskaya et al. (2005) used more bright light in their experiments than we used in our experiments; thus, the amount of light during osmotic stress might explain the differences in the results. Interestingly, the SigD factor was suggested to be part of a signaling cascade in which signals are transduced by His kinase (Hik) 33 and the cognate response regulator (Rre) 31 during osmotic acclimation (Marin et al., 2003; Shoumskaya et al., 2005). We noticed that genes (all 10 genes under salt stress and 11 of 12 genes under sorbitol stress) belonging to the Hik33-Rre31 cascade were among those genes that were up-regulated under UV light and high-light stress conditions (Huang et al., 2002). Furthermore, almost all of these same genes were less induced in the ΔHik33 strain than in the wild-type strain under oxidative stress caused by hydrogen peroxide treatment (Kanesaki et al., 2007). Because UV light and high-intensity visible light, as well as treatment with high salt, can induce the production of reactive oxygen species, it is possible that the SigD factor is required for acclimation to conditions that enhance the production of reactive oxygen species. We tested the effect of mild oxidative stress by growing the ΔsigD and control strains in the presence of 0.1 μm methyl viologen. Methyl viologen accepts an electron from PSI and reduces oxygen to O2−, which is converted to hydrogen peroxide by superoxide dismutase. Synechocystis can grow in the presence of 0.5 μm methyl viologen in moderate light (Tichy and Vermaas, 1999). Figure 8 shows that the ΔsigD strain grew slightly more slowly than the control strain. This finding supports our idea that the SigD factor is involved in acclimation to oxidative stress.
Figure 8.
Growth of the ΔsigD and control strains under mild oxidative stress. The A730 of the cell culture was set to 0.1, 0.1 μm methyl viologen was added, and the cells were grown under continuous irradiance of 40 μmol photons m−2 s−1 at 32°C under air levels of CO2. Each growth curve represents the mean of three independent experiments, and the error bars denote se.
The SigE factor was important for acclimation to salt stress, but growth of the ΔsigE strain was only slightly affected under sorbitol stress (Figs. 6 and 7). Cyanobacteria accumulate the osmoprotective solute glucosylglycerol in response to salt stress, whereas sorbitol itself accumulates in cyanobacterial cells (Marin et al., 2006). Sorbitol was also shown to induce a more pronounced efflux of water from cells than salt (Kanesaki et al., 2002). DNA microarray analysis has revealed that although many genes are up-regulated or down-regulated in both salt-induced and sorbitol-induced osmotic stress, many differences are also found (Kanesaki et al., 2002; Shoumskaya et al., 2005). According to our results, SigE has an important role only in acclimation to salt stress. Earlier studies have also shown that the SigE factor is involved in the regulation of sugar catabolic pathways (Osanai et al., 2005). In addition, inactivation of the SigE gene was found to affect the expression of photosynthetic genes in light-dark transition (Yoshimura et al., 2007), but in our continuous light experiments the ΔsigE strain grew well in all tested light intensities.
The fact that the three-dimensional models of the SigB and SigD factors (Fig. 1) are the most similar among Synechocystis σ factors, and the finding that only the ΔsigBD strain (not ΔsigB or ΔsigD) shows slow transition from lag phase to exponential growth (Fig. 4), suggest partial redundancy in the functions of these two σ factors. Furthermore, the ΔsigBD strain is more sensitive to heat stress (Tuominen et al., 2006) and to high light (Fig. 4) than ΔsigB or ΔsigD. However, under osmotic stress, the ΔsigBD strain was not more sensitive than the ΔsigB strain. Of the other double mutants, the ΔsigBΔsigE strain was shown to grow badly under mixotrophic conditions in a 12-h-light/12-h-dark rhythm, although ΔsigB and ΔsigE strains grew well (Summerfield and Sherman, 2007). Both ΔsigB and ΔsigC strains are sensitive to heat stress (Tuominen et al., 2006, 2008). These two σ factors regulate completely different sets of genes at high temperature, which makes the double mutant ΔsigBC extremely sensitive to heat stress (Tuominen et al., 2008).
In conclusion, the overall three-dimensional structures of all group 1 and 2 σ factors in Synechocystis resemble each other, despite the NCD, which lies right next to the −10 promoter region in the three-dimensional structure (Fig. 1A) and thus might play a regulatory role in promoter binding. The overall structural similarity suggests that the different σ factors may have overlapping functions. The physiological results obtained with the single and double inactivation strains support this suggestion. In particular, functional redundancy of group 2 σ factors is confirmed by the finding that all possible combinations of group 2 σ factor double inactivation mutants are viable in Synechocystis. Functional redundancy, found in transition from lag phase to exponential growth, is obvious between the SigB and SigD factors that show significant sequence identity even within the NCD connecting the conserved domains 1.2 and 2.1. However, treatments under different stress conditions also revealed that each group 2 σ factor is important under a specific set of conditions. The SigD factor is involved in high-light responses in Synechocystis, and all group 2 σ factors have a role in acclimation to salt- or sorbitol-induced osmotic stress, the SigB factor being the most important one.
MATERIALS AND METHODS
Structural Modeling of Synechocystis RNA Polymerase Holoenzyme
The amino acid sequences of the subunits of RNA polymerase of Synechocystis (Table I) were obtained from CyanoBase (www.kazusa.or.jp/cyanobase) and aligned with the sequences of their respective counterparts from the Thermus thermophilus RNA polymerase structure (Artsimovitch et al., 2005; PDB code 2A6E; chains A–F were used as the structural template) with the program MALIGN (Johnson and Overington, 1993) in the Bodil visualization and modeling package (Lehtonen et al., 2004). The secondary structure predictions for the sequences were made with PredictProtein (Rost and Liu, 2003). As the sequence identities between the subunits of T. thermophilus and Synechocystis RNA polymerases were reasonably high (Supplemental Table S1), alignment was fairly straightforward. The only exceptions were the σ factors, which were aligned as follows: First, SigA and SigC were aligned with the template, the alignment was fixed, and then SigB, SigD, and SigE were aligned with the previously fixed alignment. Finally, the alignment was modified so that the Synechocystis σ factors have a conserved Trp at the end of the NCD before region 2.1 (Fig. 1C). Structural modeling was done with MODELLER (Šali and Blundell, 1993) using the very thorough variable target function optimization method. Ten models were generated for each of the σ factors, and those with lowest objective function, as given by MODELLER, were chosen for further investigation. The stereochemical quality of the models was assessed with PROCHECK (Laskowski et al., 1993).
Construction of Group 2 σ Factor Inactivation Strains in Synechocystis
The Glc-tolerant strain of Synechocystis (Williams, 1988) was used as a control strain. The sigB (sll0306), sigC (sll0184), sigD (sll2012), and sigE (sll1689) genes were amplified by PCR with primers specific for sigB (5′-ATGGTAACAGTGACAGTTAT-3′ and 5′-TAGCTCTTGGCCATCGTTA-3′), sigC (5′-ATGACTAAACCAAGCAACGA-3′ and 5′-AATCTAGCAAAATTTCCTGC-3′), sigD (5′-ATGACTGCCAGAACCAGCCC-3′ and 5′-GCCTCCCTACAGTTGGATCT-3′), and sigE (5′-ATGAGCGATATGTCTTCCCT-3′ and 5′-CTATAACCAACCTTTGAGGC-3′). The PCR products were cloned into the pCR-Blunt II-TOPO vector (Invitrogen). The pCR-Blunt II-TOPO-sigB was digested with KpnI and PstI, and the sigB fragment was ligated into the KpnI and PstI double-digested pUC19. The pUC19-sigB was digested with SmaI, and BamHI polylinker (New England Biolabs) was added. The inactivation plasmid pUC19-sigB-Kn was constructed by ligating the BamHI fragment of pUC4K (Amersham Biosciences), carrying the Kn resistance cassette, into the BamHI site. The pCR-Blunt II-TOPO-sigC was digested with SpeI and EcoRV, and the sigC fragment was ligated into the XbaI and SmaI double-digested pUC19. To construct the inactivation plasmid pUC19-sigC-Kn, the pUC19-sigC was digested with BglII and the BamHI fragment of pUC4K was then ligated into the BglII restriction site. To construct the inactivation plasmid pUC19-sigC-Ω, the BamHI fragment (the Ω fragment that confers resistance for Spc and Str) of pHP45Ω (Prentki and Krisch, 1984) was ligated into BglII-digested pUC19-sigC. The pCR-Blunt II-TOPO-sigD was digested with SpeI and EcoRV, and the sigD fragment was ligated into the XbaI and SmaI double-digested pUC19. The pUC19-sigD-Kn was constructed by ligating the BamHI fragment of pUC4K into BamHI-digested pUC19-sigD. To construct the inactivation plasmid pUC19-sigD-Ω, the BamHI fragment of pHP45Ω was ligated into BamHI-digested pUC19-sigD. The pCR-Blunt II-TOPO-sigE was digested with PstI and EcoRI, and the sigE fragment was ligated into the PstI and EcoRI double-digested pUC19. The inactivation plasmids pUC19-sigE-Kn and pUC19-sigE-Ω were constructed by ligating the BamHI fragment of pUC4K and the BamHI fragment of pHP45Ω, respectively, into BamHI-digested pUC19-sigD.
The control strain was transformed with the vector pUC19-sigB-Kn, pUC19-sigC-Kn, pUC19-sigD-Kn, or pUC19-sigE-Kn according to Williams (1988). Transformants were isolated on selective BG-11 agar plates containing Kn (50 μg mL−1). For double mutants, the ΔsigB and ΔsigC inactivation strains were transformed with pUC19-sigD-Ω; the ΔsigB, ΔsigC, and ΔsigD inactivation strains were transformed with pUC19-sigE-Ω; and the ΔsigB strain was transformed with pUC19-sigC-Ω. Transformants were isolated on BG-11 agar plates containing Kn (25 μg mL−1), Spc (20 μg mL−1), and Str (10 μg mL−1).
The complete replacement of the native gene with the inactivated gene was confirmed by PCR analysis of corresponding genomic DNA. Genomic DNA was isolated (Williams, 1988), and the sigB, sigC, sigD, or sigE gene, depending on the strain, was amplified by PCR using the same primers as in the cloning procedure.
Growth Conditions and Measurements
Synechocystis was grown in BG-11 medium (Rippka et al., 1979) supplemented with 20 mm HEPES-NaOH, pH 7.5, under the continuous photosynthetic photon flux density (PPFD) of 40 μmol m−2 s−1 and ambient CO2 levels at 32°C. Liquid cultures were shaken at 90 rpm. These are referred to as standard growth conditions. For high-CO2 conditions, air was supplemented with 3% CO2. The BG-11 agar plates for strains ΔsigB, ΔsigC, ΔsigD, and ΔsigE were supplemented with Kn (50 μg mL−1), and plates for strains ΔsigBD, ΔsigBC, ΔsigBE, ΔsigCD, ΔsigCE, and ΔsigDE were supplemented with Kn (25 μg mL−1), Spc (20 μg mL−1), and Str (10 μg mL−1). For the experiments, cells were grown without antibiotics in liquid BG-11 medium.
The A730 of liquid cultures was set to 0.1, and the growth of the cells (50 mL of cell culture in a 250-mL Erlenmeyer flask) was monitored under standard growth conditions (PPFD of 40 μmol m−2 s−1, 32°C, shaking at 90 rpm) by measuring A730 for 14 d. Samples of dense cultures were diluted with BG-11 before the absorbance was measured, so that A730 did not exceed 0.4, and the dilutions were taken into account when the final results were calculated. In addition, growth of the control and ΔsigBD strains was followed so that A730 was set to 0.01 and 0.001 at the beginning of the growth experiment. Osmotic stress was induced by supplementing BG-11 medium with either 0.7 m NaCl or 0.5 m sorbitol, as indicated. The A730 was set to 0.1, and the growth of the cells was monitored under standard growth conditions (PPFD of 40 μmol m−2 s−1, 32°C) for 5 d. Oxidative stress was induced by supplementing BG-11 medium with 0.1 μm methyl viologen. The A730 was set to 0.1, and the growth of the cells was monitored under standard growth conditions (PPFD of 40 μmol m−2 s−1, 32°C) for 2.5 d.
The growth of the inactivation strains was screened on plates under various light conditions. The A730 of each strain was set to 0.1, and 5 μL of each cell suspension was spotted onto a BG-11 plate. The plates were then kept under continuous light at the PPFD of 20, 40, or 80 μmol m−2 s−1, as indicated, and photographed at the indicated times. The effect of dilution on growth on solid medium was tested in ΔsigBD and control strains. The A730 of the cell suspension was first set to 10, 1, 0.1, 0.01, and 0.001, and 5 μL of the cell suspensions was spotted onto BG-11 plates. The plates were kept in the standard growth conditions and photographed after 1 week.
Determination of Photosynthetic and PSII Capacity
In vivo photosynthetic activity of the control and inactivation strains (1 mL of cell suspension containing 10 μg chlorophyll mL−1) was measured in BG-11 medium supplemented with 10 mm NaHCO3 under saturating light (500 μmol photons m−2 s−1) with a Clark-type oxygen electrode (Hansatech) at 32°C. PSII capacity was measured similarly as photosynthetic capacity, except that 0.7 mm 2,6-dichloro-p-benzoquinone was used as an artificial electron acceptor and 0.7 mm ferricyanide was added to keep the quinone in an oxidized form.
RNA Isolation and Northern Blotting
Total RNA was isolated as described by Tyystjärvi et al. (2001). The RNAs were separated on 1.2% agarose-glyoxal gels and subsequently transferred to Hybond-N+ membranes (Amersham Biosciences) according to standard procedures (Sambrook and Russell, 2001). A 7-μg aliquot of total RNA was loaded per lane. Equal loading of the gels was confirmed by methylene blue staining (Sambrook and Russell, 2001). The gene-specific probes were amplified by PCR with primers specific for sigB (5′-TGGTAACAGTGACAGTTAT-3′ and 5′-GCTTCAATCATTTTCCGTTT-3′) and for hspA/sll1514 (5′-GTCTCTCATTCTTTACAATCC-3′ and 5′-TTAGGAAAGCTGAACTTTCAC-3′). The probes were labeled, hybridized, and detected using the DIG High Prime DNA Labeling and Detection Starter Kit II (Roche) according to the instruction manual of the kit.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The σ factor inactivation strains of Synechocystis.
Supplemental Figure S2. PCR analysis of inactivation strains.
Supplemental Figure S3. The amount of hspA transcripts in the control and ΔsigB strains under salt or sorbitol stress.
Supplemental Table S1. Similarity of RNA polymerase subunits in Synechocystis and T. thermophilus.
Supplementary Material
This work was supported by the Academy of Finland.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Taina Tyystjärvi (taityy@utu.fi).
The online version of this article contains Web-only data.
References
- Artsimovitch I, Patlan V, Sekine S, Vassylyeva MN, Yokoyama S, Vassylyev DG (2004) Structural basis for transcription regulation by alarmone ppGpp. Cell 117 299–310 [DOI] [PubMed] [Google Scholar]
- Artsimovitch I, Vassylyeva MN, Svetlov D, Svetlov V, Perederina A, Igarashi N, Matsugaki N, Wakatsuki S, Tahirov TH, Vassylyev DG (2005) Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycin. Cell 122 351–363 [DOI] [PubMed] [Google Scholar]
- Asadulghani, Nitta K, Kaneko Y, Kojima K, Fukuzawa H, Kosaka H, Nakamoto H (2004) Comparative analysis of the hspA mutant and wild-type Synechocystis sp. strain PCC 6803 under salt stress: evaluation of the role of hspA in salt stress management. Arch Microbiol 182 487–497 [DOI] [PubMed] [Google Scholar]
- Asayama M, Imamura S, Yoshihara S, Miyazaki A, Yoshida N, Sazuka T, Kaneko T, Ohara O, Tabata S, Osanai T, et al (2004) SigC, the group 2 sigma factor of RNA polymerase, contributes to the late-stage gene expression and nitrogen promoter recognition in the cyanobacterium Synechocystis sp. strain PCC 6803. Biosci Biotechnol Biochem 68 477–487 [DOI] [PubMed] [Google Scholar]
- Caslake LF, Gruber TM, Bryant DA (1997) Expression of two alternative sigma factors of Synechococcus sp. strain PCC 7002 is modulated by carbon and nitrogen stress. Microbiology 143 3807–3818 [DOI] [PubMed] [Google Scholar]
- Foster JS, Singh AK, Rothschild LJ, Sherman LA (2006) Growth-phase dependent differential gene expression in Synechocystis sp. strain PCC 6803 and regulation by a group 2 sigma factor. Arch Microbiol 187 265–279 [DOI] [PubMed] [Google Scholar]
- Goto-Seki A, Shirokane M, Masuda S, Tanaka K, Takahashi H (1999) Specificity crosstalk among group 1 and group 2 sigma factors in the cyanobacterium Synechococcus sp. PCC7942: in vitro specificity and a phylogenetic analysis. Mol Microbiol 34 473–484 [DOI] [PubMed] [Google Scholar]
- Gruber TM, Bryant DA (1998) Characterization of the alternative σ-factors SigD and SigE in Synechococcus sp. strain PCC 7002: SigE is implicated in transcription of post-exponential-phase-specific genes. Arch Microbiol 169 211–219 [DOI] [PubMed] [Google Scholar]
- Hihara Y, Kamei A, Kanehisa M, Kaplan A, Ikeuchi M (2001) DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell 13 793–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, McCluskey MP, Ni H, LaRossa RA (2002) Global gene expression profiles of the cyanobacterium Synechocystis sp. strain PCC 6803 in response to irradiation with UV-B and white light. J Bacteriol 184 6845–6858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura S, Tanaka K, Shirai M, Asayama M (2006) Growth phase-dependent activation of nitrogen-related genes by a control network of group 1 and group 2 σ factors in a cyanobacterium. J Biol Chem 281 2668–2675 [DOI] [PubMed] [Google Scholar]
- Imamura S, Yoshihara S, Nakano S, Shiozaki N, Yamada A, Tanaka K, Takahashi H, Asayama M, Shirai M (2003) Purification, characterization, and gene expression of all sigma factors of RNA polymerase in a cyanobacterium. J Mol Biol 325 857–872 [DOI] [PubMed] [Google Scholar]
- Iyer LM, Koonin EV, Aravind L (2004) Evolution of bacterial RNA polymerase: implications for large-scale bacterial phylogeny, domain accretion, and horizontal gene transfer. Gene 335 73–88 [DOI] [PubMed] [Google Scholar]
- Johnson MS, Overington JP (1993) A structural basis for sequence comparisons: an evaluation of scoring methodologies. J Mol Biol 233 716–738 [DOI] [PubMed] [Google Scholar]
- Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, et al (1996) Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res 3 109–136 [DOI] [PubMed] [Google Scholar]
- Kanesaki Y, Suzuki I, Allakhverdiev SI, Mikami K, Murata N (2002) Salt stress and hyperosmotic stress regulate the expression of different sets of genes in Synechocystis sp. PCC 6803. Biochem Biophys Res Commun 290 339–348 [DOI] [PubMed] [Google Scholar]
- Kanesaki Y, Yamamoto H, Paithoonrangsarid K, Shoumskaya M, Suzuki I, Hayashi H, Murata N (2007) Histidine kinases play important roles in the perception and signal transduction of hydrogen peroxide in the cyanobacterium, Synechocystis sp. PCC 6803. Plant J 49 313–324 [DOI] [PubMed] [Google Scholar]
- Khudyakov IY, Golden JW (2001) Identification and inactivation of three group 2 sigma factor genes in Anabaena sp. strain PCC 7120. J Bacteriol 183 6667–6675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvint K, Farewell A, Nyström T (2000) RpoS-dependent promoters require guanosine tetraphosphate for induction even in the presence of high levels of σs. J Biol Chem 275 14795–14798 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst 26 283–291 [Google Scholar]
- Lehtonen JV, Still DJ, Rantanen VV, Ekholm J, Bjöklund D, Iftikhar Z, Huhtala M, Repo S, Jussila A, Jaakkola J, et al (2004) BODIL: a molecular modeling environment for structure-function analysis and drug design. J Comput Aided Mol Des 18 401–419 [DOI] [PubMed] [Google Scholar]
- Lemeille S, Geiselmann J, Latifi A (2005) Crosstalk regulation among group 2-sigma factors in Synechocystis PCC6803. BMC Microbiol 5 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonetto M, Gribskov M, Gross CA (1992) The σ70 family: sequence conservation and evolutionary relationships. J Bacteriol 174 3843–3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H, Fujita N, Ishihama A (2000) Competition among seven Escherichia coli σ subunits: relative binding affinities to the core RNA polymerase. Nucleic Acids Res 28 3497–3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin K, Stirnberg M, Eisenhut M, Krämer R, Hagemann M (2006) Osmotic stress in Synechocystis sp. PCC 6803: low tolerance towards nonionic osmotic stress results from lacking activation of glucosylglycerol accumulation. Microbiology 152 2023–2030 [DOI] [PubMed] [Google Scholar]
- Marin K, Suzuki I, Yamaguchi K, Yamamoto H, Kanesaki Y, Hagemann M, Murata N (2003) Identification of histidine kinases that act as sensors in the perception of salt stress in Synechocystis sp. PCC 6803. Proc Natl Acad Sci USA 100 9061–9066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA (2002. a) Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science 296 1285–1290 [DOI] [PubMed] [Google Scholar]
- Murakami KS, Masuda S, Darst SA (2002. b) Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 Å resolution. Science 296 1280–1284 [DOI] [PubMed] [Google Scholar]
- Osanai T, Kanesaki Y, Nakano T, Takahashi H, Asayama M, Shirai M, Kanehisa M, Suzuki I, Murata N, Tanaka K (2005) Positive regulation of sugar catabolic pathways in the cyanobacterium Synechocystis sp. PCC 6803 by the group 2 σ factor SigE. J Biol Chem 280 30653–30659 [DOI] [PubMed] [Google Scholar]
- Prentki P, Krisch HM (1984) In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29 303–313 [DOI] [PubMed] [Google Scholar]
- Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY (1979) Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol 111 1–61 [Google Scholar]
- Rodríguez-Ezpeleta N, Brinkmann H, Burey SC, Roure B, Burger G, Loffelhardt W, Bohnert HJ, Philippe H, Lang BF (2005) Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr Biol 15 1325–1330 [DOI] [PubMed] [Google Scholar]
- Rost B, Liu J (2003) The ProdictProtein server. Nucleic Acids Res 31 3300–3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234 779–815 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell T (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schneider GJ, Hasekorn R (1988) RNA polymerase subunit homology among cyanobacteria, other eubacteria and archaebacteria. J Bacteriol 170 4136–4140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki A, Hanaoka M, Akimoto Y, Masuda S, Iwsaki H, Tanaka K (2007) Induction of a group 2 σ factor, RPOD3, by high light and the underlying mechanism in Synechococcus elongatus PCC 7942. J Biol Chem 282 36887–36894 [DOI] [PubMed] [Google Scholar]
- Shoumskaya MA, Paithoonrangsarid K, Kanesaki Y, Los DA, Zinchenko VV, Taticharoen M, Suzuki I, Murata N (2005) Identical Hik-Rre systems are involved in perception and transduction of salt signals and hyperosmotic signals but regulate the expression of individual genes to different extents in Synechocystis. J Biol Chem 22 21531–21538 [DOI] [PubMed] [Google Scholar]
- Singh AK, Summerfield TC, Li H, Sherman LA (2006) The heat shock response in the cyanobacterium Synechocystis sp. strain PCC 6803 and regulation of gene expression by HrcA and SigB. Arch Microbiol 186 273–286 [DOI] [PubMed] [Google Scholar]
- Summerfield TC, Sherman LA (2007) Role of sigma factors in controlling global gene expression in light/dark transitions in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol 189 7829–7840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichy M, Vermaas W (1999) In vivo role of catalase-peroxidase in Synechocystis sp. strain PCC 6803. J Bacteriol 181 1875–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuominen I, Pollari M, Aguirre von Wobeser E, Tyystjärvi E, Ibelings BW, Matthijs HCP, Tyystjärvi T (2008) Sigma factor SigC is required for heat acclimation of the cyanobacterium Synechocystis sp. strain PCC 6803. FEBS Lett 582 346–350 [DOI] [PubMed] [Google Scholar]
- Tuominen I, Pollari M, Tyystjärvi E, Tyystjärvi T (2006) The SigB sigma factor mediates high-temperature responses in the cyanobacterium Synechocystis sp. PCC6803. FEBS Lett 580 319–323 [DOI] [PubMed] [Google Scholar]
- Tuominen I, Tyystjärvi E, Tyystjärvi T (2003) Expression of primary sigma factor (PSF) and PSF-like sigma factors in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol 185 1116–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyystjärvi T, Herranen M, Aro EM (2001) Regulation of translation elongation in cyanobacteria: membrane targeting of the ribosome nascent-chain complexes controls the synthesis of D1 protein. Mol Microbiol 40 476–484 [DOI] [PubMed] [Google Scholar]
- Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S (2002) Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature 417 712–719 [DOI] [PubMed] [Google Scholar]
- Vassylyev DG, Svetlov V, Vassylyeva MN, Perederina A, Igarashi N, Matsugaki N, Wakatsuki S, Artsimovitch I (2005) Structural basis for transcription inhibition by tagetitoxin. Nat Struct Mol Biol 12 1086–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JGK (1988) Construction of specific mutations in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Methods Enzymol 167 766–778 [Google Scholar]
- Yoshimura T, Imamura S, Tanaka K, Shirai M, Asayama M (2007) Cooperation of group 2 σ factors, SigD and SigE for light-induced transcription in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett 581 1495–1500 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.