Abstract
In contrast to animals, where polyamine (PA) catabolism efficiently converts spermine (Spm) to putrescine (Put), plants have been considered to possess a PA catabolic pathway producing 1,3-diaminopropane, Δ1-pyrroline, the corresponding aldehyde, and hydrogen peroxide but unable to back-convert Spm to Put. Arabidopsis (Arabidopsis thaliana) genome contains at least five putative PA oxidase (PAO) members with yet-unknown localization and physiological role(s). AtPAO1 was recently identified as an enzyme similar to the mammalian Spm oxidase, which converts Spm to spermidine (Spd). In this work, we have performed in silico analysis of the five Arabidopsis genes and have identified PAO3 (AtPAO3) as a nontypical PAO, in terms of homology, compared to other known PAOs. We have expressed the gene AtPAO3 and have purified a protein corresponding to it using the inducible heterologous expression system of Escherichia coli. AtPAO3 catalyzed the sequential conversion/oxidation of Spm to Spd, and of Spd to Put, thus exhibiting functional homology to the mammalian PAOs. The best substrate for this pathway was Spd, whereas the N1-acetyl-derivatives of Spm and Spd were oxidized less efficiently. On the other hand, no activity was detected when diamines (agmatine, cadaverine, and Put) were used as substrates. Moreover, although AtPAO3 does not exhibit significant similarity to the other known PAOs, it is efficiently inhibited by guazatine, a potent PAO inhibitor. AtPAO3 contains a peroxisomal targeting motif at the C terminus, and it targets green fluorescence protein to peroxisomes when fused at the N terminus but not at the C terminus. These results reveal that AtPAO3 is a peroxisomal protein and that the C terminus of the protein contains the sorting information. The overall data reinforce the view that plants and mammals possess a similar PA oxidation system, concerning both the subcellular localization and the mode of its action.
Polyamines (PAs) are well-known, low-molecular-mass organic compounds, and the most abundant ones, putrescine (Put), spermidine (Spd), and spermine (Spm), exert a multifunctional role in plant development and defense (Thomas and Thomas, 2001; Mehta et al., 2002; Capell et al., 2004; Yamaguchi et al., 2006). Insights into the role of plant PA catabolism have been constantly gained in the last few years, and it has been correlated with diverse processes such as cell growth and development and programmed cell death (Ha et al., 1997; Yoda et al., 2003; Amendola et al., 2005; Paschalidis and Roubelakis-Angelakis, 2005a, 2005b; Moschou et al., 2008).
Oxidation is the main catabolic pathway of PAs. PA oxidation in mammals is exerted by multiple enzymatic activities with PA oxidase (PAO; EC 1.5.3.11), mainly localized in peroxisomes, and converting Spm to Put, via Spd producing hydrogen peroxide (H2O2; Schrader and Fahimi, 2004). The best substrates for this back-conversion process are the acetyl-derivatives of Spd and Spm (Schrader and Fahimi, 2004). On the contrary, in plants, PA oxidation is exerted mainly by apoplastic PAOs, oxidizing Spm and Spd to 1,3-diaminopropane, H2O2, and the corresponding aldehyde, thus being responsible for the terminal catabolism of PAs (Rea et al., 2004; Cona et al., 2006). Interestingly, none of the known plant PAOs has been reported to localize to peroxisomes (Angelini et al., 1995; Cona et al., 2005), while only the PAO from barley (Hordeum vulgare) has been reported to be localized in the vacuole (Cervelli et al., 2004). Also, Tavladoraki et al. (2006) reported that the Arabidopsis (Arabidopsis thaliana) gene AtPAO1 product could be localized in the cytoplasm, leaving an open window for novel PAO localization.
Biochemical evidence for the existence of a back-conversion pathway in plants was presented by Del Duca et al. (1995) and Tassoni et al. (2000), who found that exogenously supplied Spd to Helianthus tuberosus chloroplasts and Arabidopsis plants, respectively, was converted to Put, but the enzymatic activity(ies) responsible for this conversion could not be determined at that time; it was hypothesized that an enzyme similar to the animal PAO enzyme responsible for this pathway should exist in plants. Recently, Tavladoraki et al. (2006) identified the Arabidopsis gene AtPAO1 as a PAO able to convert Spm to Spd, thus being functionally similar to the mammalian Spm oxidase (SMO; EC 1.5.3.1), giving new insights into the plant PA catabolism and thus reinforcing the view that additional PAOs similar to animal PAOs should exist in plants.
In contrast to their mammalian counterparts, a correlation with PA oxidation has not been established yet for plant peroxisomes (Hayashi and Nishimura, 2006). On the other hand, plant peroxisomes possess a wide array of functions, exhibiting significant plasticity in response to changes in their cellular environment (Nyathi and Baker, 2006). Peroxisomes contain numerous enzymes involved in oxidative metabolism of a variety of substrates and also detoxifying enzymes to compensate for the high production rate of reactive oxygen species (ROS), especially from flavin-containing oxidases. Moreover, plant peroxisomes contribute to the defense against oxidative challenge, for which β-oxidation is the most conserved process. In addition to this role in plant defense, molecules with hormonal action, such as jasmonic acid (JA), auxin, and salicylic acid (SA), are all derived from peroxisomal β-oxidation in plants. Although mammalian peroxisomes possess a PA-derived ROS source via flavin-containing PAOs (Wu et al., 2003), the same process for plant peroxisomes remains obscure. However, a thorough knowledge of the oxidative sources of plant peroxisomes is required to further understand the physiological role(s) of ROS.
In this work, an in silico analysis of the Arabidopsis genome for putative PAO genes was performed, which revealed that AtPAO3 is a putative peroxisomal protein. To further explore this finding, heterologous expression of the gene encoding for AtPAO3 and purification of the respective protein was performed. Transient expression of the AtPAO3 protein fused to the C terminus of the GFP protein revealed that AtPAO3 is localized in organelles. The peroxisomal localization of the AtPAO3 was further documented by colocalization experiments in Nicotiana benthamiana leaves. Finally, its novel back-converting enzymatic activity able to convert Spm to Spd and Spd to Put was established. This is the second enzyme characterized so far that possesses back-converting activity, while the first was that reported by Tavladoraki et al. (2006), also in Arabidopsis. Our overall results presented herein support that plant peroxisomes possess PA catabolic activity and that PA catabolism in plants and mammals share critical features.
RESULTS
PAO3 Cloning and Sequence Comparison
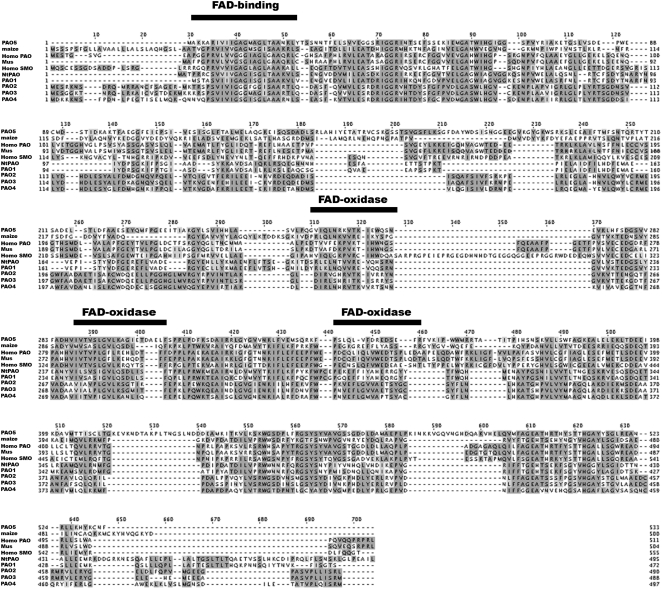
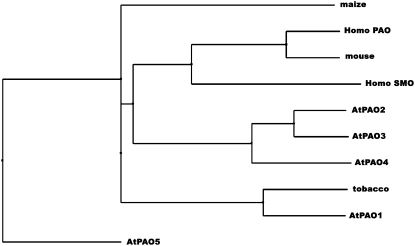
In silico analysis of the Arabidopsis genome showed that it contains five putative PAOs, AtPAO1 (At5g13700; GenBank accession no. NM_121373), AtPAO2 (At2g43020; GenBank accession no. AF364952), AtPAO3 (At3g59050; GenBank accession no. AY143905), AtPAO4 (At1g65840; GenBank accession no. AF364953), and AtPAO5 (At4g29720; GenBank accession no. AK118203), all of which encode proteins with a single amine oxidase domain (Tavladoraki et al., 2006). Irrespective of their origin, all PAO proteins contain four conserved motifs (Fig. 1). More specifically, PAOs in their N terminus (amino acids 30–60) contain a flavin adenine dinucleotide (FAD)-binding motif, while the positions 300 to 330, 380 to 410, and 440 to 460 approximately contain three specific PAO motifs (Fig. 1). All these motifs were present in AtPAO3 as indicated using the Simple Modular Architecture Research Tool software (Fig. 1; Letunic et al., 2006). Sequence comparison and phylogenetic analysis of the five putative genes indicated that PAO genes in Arabidopsis can be separated into three paraphyletic groups (Fig. 2). More specifically, the gene AtPAO1 can be classified with the well-characterized plant PAO genes from maize (Zea mays; ZmPAO; GenBank accession no. NM_001111636; Tavladoraki et al., 1998) and tobacco (Nicotiana tabacum; NtPAO; GenBank accession no. AB200262; Yoda et al., 2006). On the other hand, AtPAO2, AtPAO3, and AtPAO4 can be clustered together in one group, while AtPAO5 is the most distinct PAO gene (Fig. 2).
Figure 1.
AtPAO3 sequence alignment with other known plant and mammalian PAOs. AtPAO3 (At3g59050; GenBank accession no. AY143905) amino acid sequence similarity with AtPAO1 (At5g13700; GenBank accession no. NM_121373), AtPAO2 (At2g43020; GenBank accession no. AF364952), AtPAO4 (At1g65840; GenBank accession no. AF364953), AtPAO5 (At4g29720; GenBank accession no. AK118203), maize (ZmPAO; GenBank accession no. NM_001111636), Homo PAO (H. sapiens PAO; GenBank accession no. NM_152911), Mus (GenBank accession no. NM_153783), Homo SMO (H. sapiens SMO; GenBank accession no. NM_175839), and tobacco (NtPAO; GenBank accession no. AB200262) PAOs. Conserved motifs spanning the protein sequences are also indicated (black bars). Alignment and (default) shading were accomplished using ClustalW version 1.8.
Figure 2.
Phylogenetic tree representing the evolutionary relationship of the AtPAO3 gene with other known plant and mammalian PAOs. AtPAO3 (At3g59050; GenBank accession no. AY143905) sequence similarity with AtPAO1 (At5g13700; GenBank accession no. NM_121373), AtPAO2 (At2g43020; GenBank accession no. AF364952), AtPAO4 (At1g65840; GenBank accession no. AF364953), AtPAO5 (At4g29720; GenBank accession no. AK118203), maize (ZmPAO; GenBank accession no. NM_001111636), Homo PAO (H. sapiens PAO; GenBank accession no. NM_152911), mouse (PAO; GenBank accession no. NM_153783), Homo SMO (H. sapiens SMO; GenBank accession no. NM_175839), and tobacco (NtPAO; GenBank accession no. AB200262) PAOs.
In mammals, PAOs able to back-convert PAs are localized in peroxisomes (Pledgie et al., 2005). From the above-mentioned Arabidopsis genes, AtPAO2, AtPAO3, and AtPAO4 are predicted from the AraPerox database to be localized in plant peroxisomes (Reumann et al., 2004), containing a peroxisomal targeting signal 1-type signal responsible for the localization of mammalian PAOs in peroxisomes. These triggered us to further investigate AtPAO3 as a starting point for the comparative analysis between plant and mammalian PA catabolism. AtPAO3 protein exhibits 84% identity with AtPAO2, 77% identity with an unknown protein from Vitis vinifera (GenBank accession no. CU459235.1), 72% identity with Os04g0623300 gene product from Oryza sativa (GenBank accession no. NM_001060458.1), and 61% identity with AtPAO4. The similarity to other PAOs, including those localized in peroxisomes of mammals, was very low.
The coding region of the AtPAO3 cDNA was obtained by performing PCR on a pUNI 51 vector containing the AtPAO3 cDNA (Arabidopsis Biological Resource Center [ABRC] Stock Center, clone no. U21196). The size of the AtPAO3 coding region is 1,467 bp. In addition, we checked the expression of the gene in both leaves and roots using reverse transcription (RT)-PCR and detected that the gene is mostly expressed in the leaves (Supplemental Fig. S1).
AtPAO3 Heterologous Expression and Protein Purification
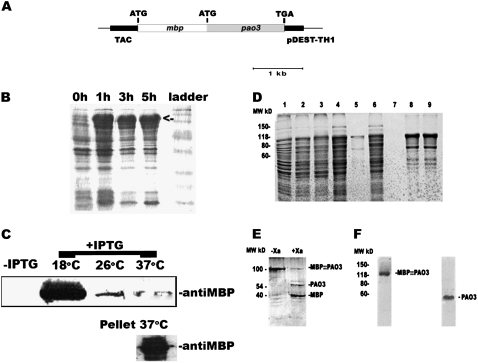
AtPAO3 encodes for a 488-amino acid protein, with a calculated molecular mass of 54.1 kD and pI 5.81. To overexpress the AtPAO3 protein in a heterologous system, the AtPAO3 cDNA containing the complete open reading frame was introduced into the pDEST-TH1 vector, to fuse AtPAO3 to the maltose-binding protein (MBP; MBP:PAO3) under the inducible tac promoter (fusion of the trp and lac promoters; Fig. 3A). This construct was introduced into Escherichia coli BL21 cells and induced with isopropyl β-d-thiogalactopyranoside (IPTG) for protein expression (Fig. 3B). AtPAO3 protein was detected at high levels in total cell lysates as soon as 1 h after induction (Fig. 3B). At 37°C, the accumulation of the MBP:PAO3 protein was found mostly in the pellet fraction of cell lysates, as indicated using an anti-MBP-specific polyclonal antibody (Fig. 3C). After decreasing the culture temperature from 37°C to 18°C and the inductive IPTG concentration from 1 mm to 0.05 mm, higher MBP:PAO3 protein accumulation was detected in the soluble fraction (Fig. 3C). Lower temperatures or IPTG concentrations did not have any significant effect on the amount of MBP:PAO3 protein accumulation (data not shown).
Figure 3.
AtPAO3 expression in E. coli and AtPAO3 protein digestion and chromatographic purification. A, The construct used to produce recombinant AtPAO3 in E. coli BL21 cells. B, Time course of AtPAO3 protein accumulation in the total cellular extracts from induced E. coli BL21 cells. C, Detection of the MBP:PAO3 protein fusion in the soluble fraction, in noninduced (−IPTG) and induced cells (+IPTG), using an anti-MBP polyclonal antibody at different temperatures. At higher temperatures (e.g. 37°C), the MBP:PAO3 protein accumulated mostly in the pellet fraction. D, First step protein expression and purification. 1, Cell lysate from noninduced culture; 2, supernatant of the cell lysate from induced culture; 3, pellet of the cell lysate from induced culture; 4, initial flow through the amylose resin; 5, part of the amylose resin before elution of the protein; 6, second flow through the amylose resin; 7, final flow through the amylose resin (10th); 8, elution fractions 4 and 5 from the amylose resin (recombinant MBP:PAO3 eluted between fractions 3–7). E, Digestion of the eluted MBP:PAO3 protein with the specific protease Xa factor for 24 h. As a control, MBP:PAO3 protein was used without addition of Xa factor (left). F, Gel filtration of the MBP:PAO3 protein purified with the amylose resin producing a single MBP:PAO3 band and AtPAO3 protein after digestion and purification with Xa factor. The data presented are from a single representative experiment that was repeated twice with similar results.
To purify the MBP:PAO3 protein, supernatants from cell lysates from induced cultures were passed through an amylose resin to bind the MBP tag, and AtPAO3 protein was purified to >80% (Fig. 3D). Furthermore, digestion of the MBP:PAO3 fusion with the Xa factor, a protease that specifically cleaves the junction between the MBP and PAO3 in the Ile-Leu-Glu-Gly-Arg sequence, resulted in two separate proteins, MBP with a molecular mass of 42 kD and AtPAO3 with a molecular mass of 54 kD (Fig. 3E). The MBP:PAO3 protein was further purified using gel filtration (Fig. 3F). More specifically, the MBP:PAO3 protein sample was applied again to an amylose resin and subsequently to a Sephadex gel filtration column equilibrated with MBP buffer, pH 7.0. Elution was performed with the same buffer, collecting fractions of 3.0 mL. The procedure was repeated twice, and the MBP:PAO3 protein was dialyzed in MBP buffer. More than 3 mg of recombinant MBP:PAO3 L−1 culture could be obtained after the gel filtration step at 18°C and inductive IPTG 0.05 mm (Fig. 3F).
The AtPAO3 protein was further purified from the MBP tag after digestion with the Xa factor by applying it to an amylose resin to rebind the MBP. The Xa factor was removed through a hydroxyapatite column, resulting in a single band corresponding to the AtPAO3 protein (Fig. 3F). The final purified protein was subsequently used for the following experiments.
AtPAO3 Is a FAD-Binding Protein
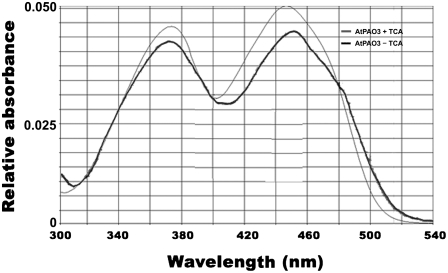
PAO proteins characterized so far are flavoproteins, able to bind via noncovalent bond(s) FAD molecule(s) that act as electron donors to molecular oxygen, necessary for PAO-oxidizing activity (Tavladoraki et al., 2006). Thus, in silico analysis of the AtPAO3 protein showed that it contains at the N terminus of the protein a domain responsible for the binding of a FAD molecule. To test whether AtPAO3 protein is able to bind to a FAD molecule in vitro, we disrupted the noncovalent bonds through addition of 5% TCA to the purified protein, which was subsequently precipitated. Supernatant absorbance was measured and exhibited the characteristic two-curve spectrum, typical for FAD-containing proteins, with absorption maxima at 380 and 460 nm, indicating that AtPAO3 is in an oxidized state (Fig. 4). A similar spectrum was obtained without disrupting the bonds between the FAD and AtPAO3, while the slight differences observed are most probably due to the protein microenvironment that surrounds the FAD molecule, which alters its absorbance characteristics (Fig. 4). No significant differences were observed between the AtPAO3 and MBP:PAO3 absorbance, suggesting that MBP protein does not affect the absorbance features of protein (data not shown). Each mole of protein could adhere approximately 0.8 mol of FAD, suggesting a close to 1:1 molecular ratio for FAD:PAO3 protein. In some cases, the supernatant exhibited an absorbance peak at 280 nm, indicating contamination of the supernatant from the precipitated protein.
Figure 4.
Absorption spectrum of the AtPAO3 purified protein between wavelengths from 300 to 540 from TCA-treated or nontreated protein. AtPAO3 exhibits the characteristic absorbance of FAD-containing enzymes. The data presented are from a single representative experiment that was repeated many times with similar results.
Reaction Catalyzed by AtPAO3
In plants, most characterized proteins encoded by PAO genes are responsible for the terminal catabolism of Spd and Spm. Only recently was the Arabidopsis gene AtPAO1 shown to encode for a FAD-binding PAO protein that exhibits a single step back-conversion activity, converting Spm to Spd (Tavladoraki et al., 2006). Analysis of the substrate specificity of the protein purified in this work showed that AtPAO3 is able to oxidize both Spd and Spm, producing H2O2, whereas no oxidizing activity was found when using Put (Fig. 5A). Both the crude bacterial lysate and the purified protein exhibited similar activities with respect to the oxidizing specificity (data not shown). Interestingly, Δ1-pyrroline was not a product of AtPAO3, as no yellowish product was observed after its reaction with o-aminobenzaldehyde, typical for most PAO enzymes that catalyze the terminal deamination of Spd and Spm.
Figure 5.
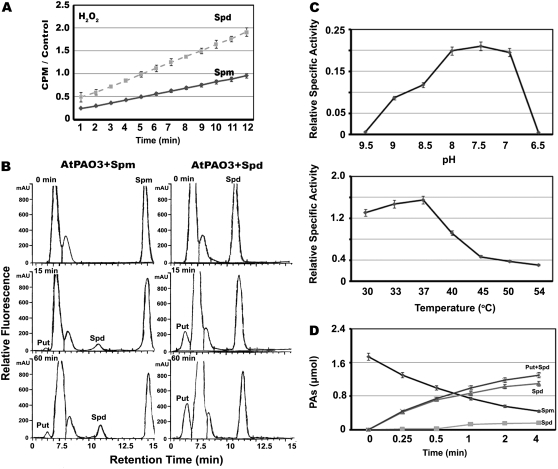
Biochemical properties of AtPAO3. A, Relative H2O2 production (CPM, counts per minute normalized to controls) using Spd and Spm as substrates (10 mm each). B, HPLC analysis of the AtPAO3-dependent Spm conversion to Spd (15 min) and Put (15 and 60 min; AtPAO3+Spm), and Spd conversion to Put (15 and 60 min; AtPAO3+ Spd). A total 1 mm substrate was used and 2 μg of AtPAO3 enzyme. The peaks with retention time varying between approximately 7 to 8 min correspond to traces of benzoyl chloride used as derivatization reagent. C, AtPAO3 dependence from pH and temperature. D, Conversion of Spm to Spd and Put by AtPAO3 as a fraction of time, using 2 μg of AtPAO3 enzyme and Spm as substrate. Data are the means from three independent experiments ± se.
The previous results prompted us to identify the reaction products of AtPAO3-oxidizing activity. To this end, an in vitro assay and HPLC were combined (Fig. 5B). Analysis of the reaction products revealed that both Spm (retention time, approximately 15 min) and Spd (retention time, approximately 10.9 min) are oxidized by AtPAO3, giving rise to Spd and Put, or Put alone, respectively, without any concomitant production of 1,3-diaminopropane (Fig. 5B). Production of Put from Spm could be detected after approximately 15 min in the in vitro assay using 2 μg of AtPAO3 protein, while when Spd was used as substrate, significantly higher amounts of Put at the same time points could be observed (Fig. 5B). These results reinforce the view that AtPAO3 encodes for an enzyme with PA-oxidizing activity, able to back-convert Spm and Spd to the final product Put in a two-step reaction producing H2O2 as end-product.
The pH optimum for the oxidation of the substrates was found to be 7.5 (Fig. 5C), similar to AtPAO1 (pH = 8; Tavladoraki et al., 2006) but different from that of ZmPAO (pH 6.5). Interestingly, the enzyme was quickly inactivated at lower pH values, and at pH > 9.5 or < 6.5, no significant activity was found (Fig. 5C). The pH dependence of the enzyme did not follow the bell-shaped dependency of AtPAO1, suggesting different ionization groups within the active site (Tavladoraki et al., 2006; results herein). Interestingly, the mouse (Mus musculus) peroxisomal PAO exhibits similar pH dependence (Wu et al., 2003).
The optimum temperature was found to be 37°C, similar to the Homo sapiens peroxisomal PAO (Wang et al., 2003), whereas at higher temperatures, the enzyme was quickly inactivated (Fig. 5C). Moreover, equimolar amounts of Spd and Put to Spm were produced via AtPAO3 oxidation activity (Fig. 5D). The enzyme exhibited a progressive inactivation with time, possibly due to end-product inhibition, either through H2O2 or Put (Fig. 5D).
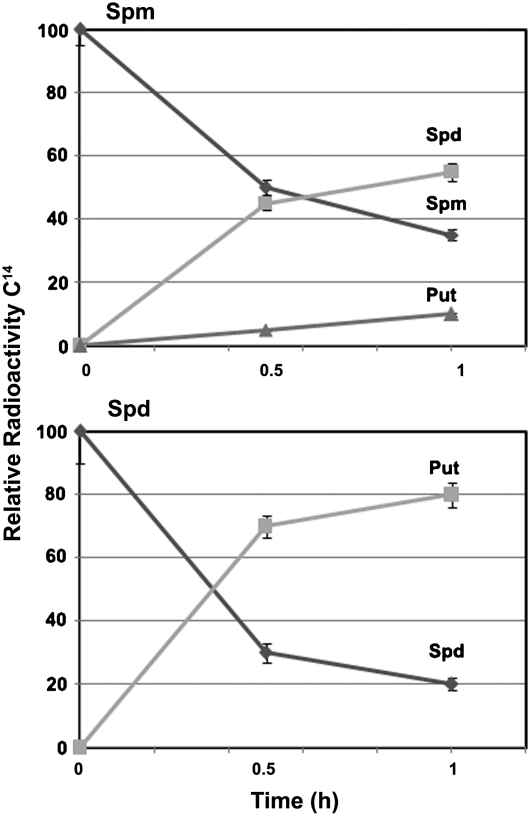
To further confirm the back-converting activity of AtPAO3, a radiometric method was employed using radiolabeled PAs as substrates, and the conversion efficiency to each product was estimated as a fraction of time (Fig. 6). Thus, 80% of the initial Spd was converted to Put within 1 h, while 65% of initial Spm was converted to Spd and Put at the same time point (Fig. 6; Spd and Spm, respectively). On the contrary, radiolabeled Put was not recognized as substrate by AtPAO3.
Figure 6.
Radiometric assay for the detection of AtPAO3 reaction products and conversion efficiency using Spm and Spd as substrates. Data are the means from three independent experiments ± se.
Kinetic Parameters of AtPAO3
Because the known mammalian PAOs oxidize N1-acetyl derivatives with higher affinity than the nonacetylated ones (Wang et al., 2003), the in vitro activity of AtPAO3 was determined not only using Spd and Spm as substrates, but also using N1-AcSpd, N8-AcSpd, and N1-AcSpm (Table I). AtPAO3 was found to oxidize not only Spd (Km = 0.204 mm) and Spm (Km = 0.588 mm) but also N1-AcSpd (Km = 1 mm) and N1-AcSpm (Km = 2 mm), converting them to AcSpd and AcPut, respectively (Table I; data not shown). The substrate specificity was Spd > Spm > N1-AcSpd > N1-AcSpm (Table I), suggesting that AtPAO3 differs with respect to the substrate specificity from the functionally homologous mammalian enzymes.
Table I.
Kinetics in the presence/absence of specific inhibitors for the PAO3 enzyme
Data are the means from a single representative experiment.
| Substrate | Km | Kcat | Kcat |
|---|---|---|---|
| mma | s−1b | Km−1 | |
| Agmatine | 0 | 0 | – |
| Cadaverine | 0 | 0 | – |
| Put | 0 | 0 | – |
| Spd | 0.204 | 1.250 | 6.12 |
| Spm | 0.588 | 0.188 | 0.31 |
| N1-AcSpd | 1 | 0.042 | 0.042 |
| N8-AcSpd | >5000 | 0 | – |
| N1-AcSpm | 2 | 0.020 | 0.01 |
| Inhibitor | Ki (μM)c | – | – |
| Guazatine | 0.028 | – | – |
| Aminoguanidine | 1.08 | – | – |
| Put | 61.5 | ||
| N8-AcSpd | 40.8 | – | – |
Substrate concentrations were <1.25 mm. Data are the means from three independent experiments ±se.
10 mm substrate was used for Kcat determination.
Inhibitors concentrations were <1.25 mm. Data are the means from three independent experiments ±se.
In addition, the inhibition constants of two well-known PAO inhibitors, guazatine and aminoguanidine, were determined using Spd as substrate (Bacchi et al., 2004; Tavladoraki et al., 2006). Guazatine, the most potent inhibitor characterized so far for PAO enzymes, including AtPAO1 (Tavladoraki et al., 2006), significantly inhibited AtPAO3 activity with a Ki of 0.028 μm (Table I). Aminoguanidine also inhibited PAO3 but with a higher Ki of 1.08 μm (Table I). In addition, Put, the end-product of PAO3 activity and N8-AcSpd, also inhibited the enzyme (Table I; Ki = 61.5 μm and Ki = 40.8 μm, respectively). Put similarly inhibited AtPAO3 when using Spm, N1-AcSpd, and N1-AcSpm as substrates (Ki = 60.1, 50.2, and 40.9 μm, respectively).
PAO3 Is Localized in Plant Peroxisomes
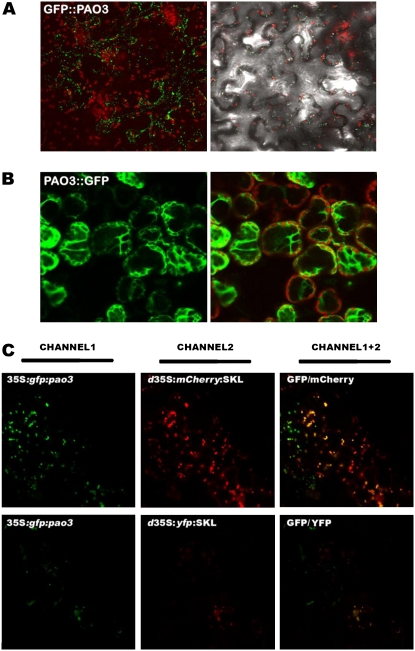
In silico protein localization prediction software failed to identify AtPAO3 as a putative peroxisomal protein. However, AtPAO3 contains a peroxisomal targeting signal 1, consisting of a Ser, an Arg, and a Met residue (SRM sequence; amino acids 486–488) at the C-terminal end of the protein, which has been shown to target proteins to peroxisomes in plants (Reumann et al., 2004). To investigate the in vivo AtPAO3 localization, we fused AtPAO3 cDNA to either the C-terminal (35S:GFP:PAO3) or N-terminal (35S:PAO3:GFP) part of the GFP. Agrobacterium-mediated transient expression of the 35S:GFP:PAO3 in N. benthamiana leaves showed a green fluorescent pattern in small spots distributed throughout the cytoplasm of the epidermal cells (Fig. 7A). In contrast, when 35S:PAO3:GFP construct was used, a diffuse fluorescence throughout the cytoplasm was found (Fig. 7B), suggesting that the fusion remained as a soluble protein in the cytosol. These results indicate that AtPAO3 has targeting information to small cytoplasmic organelles and that the sorting motifs are masked when GFP is fused to the C terminus.
Figure 7.
AtPAO3 protein localization and co-localization with plant peroxisomes in N. benthamiana. A, GFP fluorescence localization of the transiently expressed GFP:PAO3 in peroxisomes of N. benthamiana leaves, merged with the localization of chlorophyll fluorescence (left), and with inverse phase microscopy (right). B, GFP fluorescence localization of the transiently expressed PAO3:GFP into the cytoplasm of N. benthamiana leaves (left) and merged with the localization of chlorophyll fluorescence (right). C, GFP fluorescence localization of the GFP: PAO3 expressing binary vector (top left), mCherry fluorescence localization of the mCherry:SKL expressing binary vector (specific for plant peroxisomes; top middle), merged image of the GFP and mCherry fluorescence (top right), in peroxisomes of N. benthamiana leaves transiently expressing the two constructs. GFP fluorescence localization of the GFP:PAO3 expressing binary vector (bottom left), YFP fluorescence localization of the YFP:SKL construct (specific for plant peroxisomes; bottom middle), merged image of the GFP and YFP fluorescence (bottom right), in peroxisomes of N. benthamiana leaves transiently expressing the two constructs. The data presented are from a single representative experiment that was repeated twice with similar results. Data were obtained using a 40× immersion objective.
To determine where AtPAO3 is localized, we performed a colocalization experiment using yellow fluorescent protein (YFP) and Cherry markers of compartments that resembled the spots observed in 35S:GFP:PAO3 (d35S:YFP:SKL, d35S:mCherry:SKL for peroxisomal and d35S:mYFP for mitochondrial localization; SKL represents the motif Ser-Lys-Leu responsible for peroxisomal localization; Nelson et al., 2007; Fig. 6C). Agrobacterium-mediated transient expression of the 35S:GFP:PAO3 in N. benthamiana leaves with simultaneous cotransformation of the d35S:mCherry:SKL (Fig. 7C, top), or the d35S:YFP:SKL (Fig. 7C, bottom) showed that the signals colocalized, suggesting that AtPAO3 is a peroxisomal protein (Fig. 7). On the contrary, 35S:GFP:PAO3 and d35S:mYFP did not colocalize (data not shown).
AtPAO3 Is an Early Responsive Gene to Abscisic Acid, Jasmonate Treatments, and Mechanical Damage
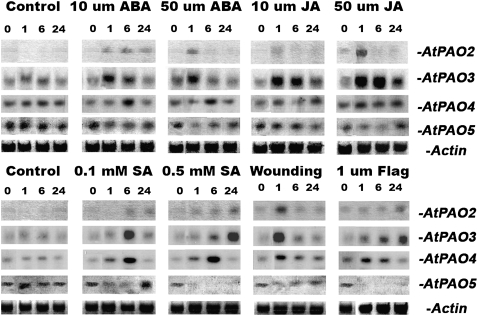
Because peroxisomes are organelles involved in various stress responses (Nyathi and Baker, 2006), we decided to monitor the abundance of AtPAO3 mRNA under various stress treatments. In addition, to gain a more comprehensive view of the regulation of PA catabolism and to compare to AtPAO3, we monitored the expression of the other members of the AtPAO gene family under the same conditions. Thus, the expression pattern of AtPAO1, AtPAO2, AtPAO3, AtPAO4, and AtPAO5 was followed after supplying 10-d-old Arabidopsis seedlings with various chemicals for 24 h (Fig. 8). Interestingly, AtPAO1 could not be detected either by RT-PCR or by northern blotting under the various treatments, consistent with the results obtained by Tavladoraki et al. (2006).
Figure 8.
AtPAO2, AtPAO3, AtPAO4, and AtPAO5 mRNA abundance under various chemical stimuli 1, 6, and 24 h after the corresponding treatment (each well contains 10 μg of total RNA and exposure time was 48 h). The data presented are from a single representative experiment that was repeated twice with similar results.
Among the various treatments performed, abscisic acid (ABA) induced AtPAO2 and AtPAO3 mRNA accumulation 1 h posttreatment, while induction of AtPAO4 took longer (6 h). On the other hand, AtPAO5 mRNA abundance was slightly reduced upon ABA treatment. Interestingly, AtPAO3 mRNA returned to basal levels after 24 h in ABA-treated plants (Fig. 8, ABA). JA-treated seedlings showed a marked and transient increase in AtPAO3 mRNA, constantly accumulated up to 6 h to later decrease at 24 h, while AtPAO2 mRNA accumulated only 1 h posttreatment. Treatment with JA did not provoke any significant increase of AtPAO4 or AtPAO5 mRNAs at all time points examined (Fig. 8; JA). SA treatment clearly induced AtPAO3 and AtPAO4 mRNA accumulation 6 h posttreatment, while AtPAO2 showed a slight increase 24 h posttreatment. Interestingly, AtPAO5 significantly accumulated after 24 h, at 0.1 mm SA (Fig. 8, SA). AtPAO2, AtPAO3, and AtPAO4 mRNA rapidly accumulated in wounded plants 1 h after wounding and returned to almost basal levels 6 h thereafter. In addition, seedlings treated with flagellin 22, a pathogen elicitor that activates the plant basal defense, exhibited AtPAO2 and AtPAO3 induction after the 6-h time point, with constant increase up to 24 h, while AtPAO4 was induced 1 h posttreatment returning to basal levels 24 h thereafter (Fig. 8, flagellin). On the other hand, AtPAO5 mRNA abundance showed a significant decrease in seedlings treated with flagellin.
DISCUSSION
PA oxidation has been considered to be a divergent process giving the notion that this pathway evolved separately in plants and animals, in contrast to PA biosynthesis, which is a conserved process among these kingdoms (Kaur-Sawhney et al., 2003; Kusano et al., 2007). On the other hand, mammalian PA catabolism has historically gained a respectful interest due to its implication in the tumor-genesis process (Toninello et al., 2006). Only recently plant PA catabolism has started to gain interest, as a consequence of two major facts, the complete analysis of the plant PA biosynthetic pathway, and the correlation of PA catabolism with programmed cell death processes, both at physiological and under stress conditions (Tiburcio et al., 1994; Yoda et al., 2003, 2006; Rea et al., 2004; Paschalidis and Roubelakis-Angelakis., 2005b; Moschou et al., 2008).
In an attempt to bridge the gap in our knowledge of PA oxidation between plants and animals, we strived to search the Arabidopsis genome with the scope to identify putative genes that could be functionally similar to the mammalian PAOs in terms of specific activity and localization. Using data obtained from combinatorial in silico analysis and the expression patterns of pao genes, and taking into account the possible localization predicted by AraPerox database (Reumann et al., 2004), we concluded that AtPAO3 gene could serve as a starting point for our research. In this work, we have described the purification of recombinant AtPAO3 using the heterologous expression system of E. coli (Fig. 3). In contrast to previous studies that either failed to express the protein in the soluble fraction (Tavladoraki et al., 2006) or introduced significant changes (deletion of the N terminus of the protein) in the structure of the PAO protein in order to obtain it in the soluble fraction (Yoda et al., 2006), we have managed to produce the protein in a soluble form using a fusion of the protein to MBP protein. The results have revealed that AtPAO3 exhibits the typical mammalian PAO back-conversion activity being able to produce Put as a final oxidation product (Figs. 5 and 6). A striking difference between the two counterparts is the substrate specificity.
More specifically, in mammals, Spd/Spm N1-acetyltransferase (SSAT) is the responsible enzyme for the acetylation of Spd and Spm, and the corresponding acetylated derivatives are orientated toward the catabolic pathway in peroxisomes, where di-acetyl-Spm and N1-acetyl-Spd serve as the optimum substrate for PAOs (SSAT-dependent PA catabolism). In contrast, AtPAO3 exhibits strong preference for nonacetylated Spd and Spm, indicating that AtPAO3 probably does not depend upon a yet-unrecognized SSAT activity (SSAT-independent catabolism; Table I). Moreover, AtPAO1 also showed preference for Spm and not its acetylated derivatives, suggesting that it is also SSAT independent (Tavladoraki et al., 2006). Although Spm can be oxidized by the mammalian peroxisomal PAO, it is a poor substrate when compared to N1-acetyl-Spm and N1-acetyl-Spd. In mammals, the necessity of Spm and Spd acetylation relates to the fact that both PAs are acetylated in order to be transported from the cells and eventually being excreted (Seiler, 1995; Wu et al., 2003). High PAO levels in mammals could prevent transport of N1-acetyl-Spm and N1-acetyl-Spd from the cell and increase the Spd and Put levels of the PA pool (Wu et al., 2003). On the other hand, these PA analogues in plants are of unknown physiological function and they can only be found in trace amounts in Arabidopsis, while in H. tuberosum relatively high titers of N1-acetyl-Spd were found in chloroplasts upon exogenous Put addition (Del Duca et al., 1995; Tassoni et al., 2000). Moreover, Del Duca et al. (1995) hypothesized that exogenous Put addition causes the inhibition of a PAO activity responsible for the conversion of Spd to Put. This is now reinforced by our results in which Put, the end-product of the full back-conversion pathway, exerted an inhibitory effect (Table I). The biochemical evidence for the existence of the PAO-dependent back-conversion pathway in the perennial dicot plant H. tuberosus indicates that the PA back-conversion is a common pathway in plants, rather than species-specific.
Interestingly, AtPAO1 and AtPAO3 exhibit a significant difference in their respective mRNA abundance in plant cells. More specifically, AtPAO3 is a rather constitutively expressed mRNA at relatively high levels in plant tissues (Supplemental Fig. S1), whereas AtPAO1 could only be detected using nested RT-PCR (Tavladoraki et al., 2006; results herein). In mammals, the peroxisomal PAO is also constitutively expressed at high levels, whereas the SMO is essentially an inducible gene (Pledgie et al., 2005). This striking similarity could imply a type of regulation of PA back-conversion with a yet-unknown role for both plants and animals. With respect to the SSAT dependence, AtPAO3 mimics AtPAO1 that is also involved in a pathway in which the SSAT probably does not participate. On the other hand, although both AtPAO1 and AtPAO3 follow a SSAT-independent pathway, their preference for substrates is radically different. AtPAO1 exhibits strong resemblance to the mammalian SMO, converting Spm to Spd but unable to convert Spd to Put (Tavladoraki et al., 2006), whereas AtPAO3 is able to oxidize both Spm and Spd, exhibiting preference to Spd (Table I; Fig. 6). In mammals, when PAO oxidizes N1-acetyl-Spm or N1-acetyl-Spd, FAD is reduced and subsequently oxidized by O2 to generate H2O2, and in our study, AtPAO3 also produced H2O2 (Fig. 5). In contrast to Fsm1, the PAO homolog from yeast (Saccharomyces cerevisiae), AtPAO3 is unable to oxidize N8-acetyl-Spd (Landry and Sternglanz, 2003). Thus, Spd produced by AtPAO1 from Spm might be of physiological significance, because this can be further used by AtPAO3 to efficiently produce Put.
Most known animal PAOs are localized in peroxisomes; AtPAO3 is so far the second organellar PAO recognized in plants in addition to PAO from barley, which is sorted to vacuoles (Cervelli et al., 2004). The localization of AtPAO3 in peroxisomes gives direct evidence for the implication of this organelle in PA oxidation and back-conversion in plant tissues (Fig. 7). This compartmentalization could be of physiological importance considering the oxidative nature of PAO, because the produced H2O2 could be efficiently scavenged through the abundant peroxisomal catalase. Moreover, the apparent nonorganellar localization of AtPAO1, which functionally resembles the mammalian SMO and the AtPAO3 sorting to peroxisomes, could suggest a similar system to the mammalian oxidation system but lacking the SSAT.
The strong induction of the AtPAO3 by JA (early), wounding (early), SA (middle to late), and flagellin (constant; Fig. 8) suggests the relevance of the enzyme to stress defense responses (Alvarez et al., 1998; Asai et al., 2000). The H2O2 produced by the AtPAO-oxidizing activity could act as a second messenger that signals downstream events under biotic stress, as described previously (Orozco-Cardenas et al., 2001; Takahashi et al., 2004; Vacca et al., 2004; Sasaki-Sekimoto et al., 2005; Skopelitis et al., 2006; Moschou et al., 2008). Moreover, PAs have been implicated in the wound response in Arabidopsis (Perez-Amador et al., 2002). In mammals, another product of the oxidation of the N1-acetylated PAs, 3-acetamidopropanal, may be enzymatically deacetylated to yield cytotoxic 3-aminopropanal that is thought to contribute, either alone or in concert with H2O2, to tissue damage following traumatic injury (Wu et al., 2003). What is worth noticing is the pattern of flagellin-induced AtPAO3 increase, which in contrast to the other treatments produces a constantly increasing pattern rather than a transient increase (Fig. 8). The previous implies that the differential signal perception between biotic and abiotic stress produces alternative patterns of AtPAO3 activation. In the case of abiotic stress, PA oxidation from AtPAO3 could have a signaling role, whereas in the case of biotic stress, AtPAO3 could produce H2O2 for a prolonged period of time to participate in this way in the plant's basal defense against pathogenic bacteria.
Our overall results support that AtPAO3 is a peroxisomal PAO, able to back-convert Spm to Spd and subsequently Spd to Put, and provides plant peroxisomes with a new role in PA oxidation/back-conversion. Moreover, the correlation of AtPAO3 with stress responses involving oxidative burst gives evidence for a PA-peroxisomal interplay under (a)biotic stress conditions.
MATERIALS AND METHODS
Chemicals
All chemicals were purchased from Sigma unless stated otherwise, while radiolabeled PAs were purchased from Amersham.
In Silico Analysis and cDNA Acquisition
For the multiple protein alignment and the phylogenetic tree construction, the Expasy Database tools were used (http://au.expasy.org/tools/). For motif and domain scan, the Simple Modular Architecture Research Tool database was used (http://smart.embl.de). The cDNA encoding for the AtPAO3 (clone no. U21196) was provided by the ABRC stock center (http://www.arabidopsis.org).
Plant Material and Treatments
Seeds of Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 were surface sterilized and sown in 24-well (10 seeds per well) tissue culture clusters, containing 1 mL/well of sterile Murashige and Skoog medium (Murashige and Skoog salts; Duchefa) supplemented with 0.5% Suc and grown with shaking (150 rpm) in a culture room under 16-h-d (26°C)/8-h night (22°C) diurnal cycles. Fresh medium, 500 μL/well, was added 9 d after sowing, and experiments were conducted 1 d later. Prior to treatment initiation, the medium remaining in the wells was removed and 1 mL of fresh medium with the different compounds tested was added.
Gene Constructs
For the AtPAO3 cDNA cloning into vector pDONR207, the primer pairs GGGGACAAGTTTGTACAAAAAAGCAGGCTTGATGGAGTCCGGAGGCAAC (PAO3-FOR) and GGGGACCACTTTGTACAAGAAAGCTGGGTATTACATACGGGAGATCAGAAG (PAO3-REV) were used, and inserts were cloned into the pDEST-TH1 (Hammrastrom et al., 2002) and pGWB6 (GFP:PAO3) vectors using the GATEWAY recombination system (Invitrogen). To eliminate the stop codon of PAO3 cDNA, the PAO3-REV-no stop primer GGGGACCACTTTGTACAAGAAAGCTGGGTATTACATACGGGAGATCAGCG was used in combination with PAO3-FOR, and AtPAO3 was cloned into a second pDONR207 vector and further transferred into the pGWB5 (PAO3:GFP) vector using the GATEWAY recombination system. The vector pDEST-TH1 was used for protein expression, whereas vectors pGWB6 and pGWB5 correspond to the N-terminal and C-terminal GFP binary fusion vectors, respectively. All vectors prepared were direct sequenced.
PCR amplifications were performed using a thermal PTC-200 PCR cycler and the Pfu (Promega) or Taq (Minotech) polymerase following standard PCR protocols.
Protein Expression and Purification
Escherichia coli strain BL21 cells were transformed with standard transformation protocols and were induced for protein expression by adding 0.05 to 1 mm IPTG in the Luria Nutrient Broth culture media supplemented with 100 μg mL−1 ampicillin and 2 g L−1 Glc after reaching OD600 = 0.5. The cells were harvested at 2,500g for 20 min at RT and frozen overnight at −80°C. Cells were dissolved in 10 mL of MBP buffer (20 mm Tris, pH 7.4, 400 mm NaCl, and 1 mm EDTA, 0.5 mm PMSF) and lysed at 4°C in a sonicator using four cycles of 15 to 20 s. Cell debris (pellet fraction) was removed after a centrifugation step at 10,000g at 4°C, and supernatant was filtrated using Miracloth and passed through an amylose resin (Biolabs) at 4°C. Protein was eluted using MBP buffer supplemented with 20 mm maltose, and collected fractions were electrophoretically resolved.
Protein fusion was digested using the protease Xa factor as described by the manufacturer (Biolabs). The Xa factor was removed using a hydroxyapatite column as described by the manufacturer (Biolabs).
In Vitro Activity Assays
In vitro activity was determined using four different assays. To determine directly the H2O2 production, a modified method of luminol measurement was used. Thus, 2 μg of purified protein were incubated in a 690-μL H2O2 lumination buffer (10 mm Tris, pH 7.0, 1 mm CaCl2, 0.1 mm KCl) supplemented with 100 μL 2.5 mg mL−1 luminol, 10 μL of 0.1 unit mL−1 peroxidase, and 1 to 10 mm final concentration of Put 2HCl, Spd 3HCl, or Spm 4HCl. Counts were measured in a scintillation counter (Beckman) for 10 min, and values were taken in the linear part of the assay. For determination of the kinetic parameters, the method of the oxidation of 4-aminopterine was used (Tavladoraki et al., 2006). Analysis of AtPAO3 oxidation products was performed by incubating saturating amounts of substrate with 4 μg of purified protein, in a final volume of 200 μL, buffered with 100 mm Tris, pH 7.5. Reaction was stopped by the addition of equal volume of 5% (v/v) perchloric acid. The samples were centrifuged for 20 min at 12,000g at 4°C, the supernatant was derivatized, and PAs were determined as previously described, using an HP 1100 High Performance Liquid Chromatographer (Hewlett-Packard; Kotzabasis et al., 1993). Additionally, reaction products were estimated using radiolabeled Spm by a modification of the radiometric method of Paschalidis and Roubelakis-Angelakis (2005a), using [1,4-14C]Put, [1,4-14C]Spd, [1,4-14C]Spm (Amersham; specific activities, 4.37 GBq mmol−1) as labeled substrates and 4 μg of AtPAO3 enzyme. The assay mixture contained 0.2 mL of Tris, pH 7.5, 1 mm unlabeled Put, Spd, and Spm and 3.7 KBq of [1,4-14C]Put, [1,4-14C]Spd, [1,4-14C]Spm. After incubation on a shaker at 37°C for 60 min, the reaction was terminated by adding 150 μL of saturated sodium carbonate. Products were separated on a thin layer chromatography plate using as a mobile phase Chlorophorm/Triethylamine 4:1 (v/v). The plate was dried out for 10 min at 60°C, the radioactivity was estimated after scraping off the spots from the plate, and the powder from each spot was measured using a scintillation counter (Beckman). To identify the retention factor of each spot, 1 mm standards of PAs were also used and were visualized under UV light following dansylation as described by Flores and Galston (1982). Additionally, radiolabeled products were dansylated to maintain the Rr accuracy.
Standard calibration curves for N1-AcSpm, N1-AcSpd, and N8-AcSpd were additionally prepared. The Δ1-aminopyrroline production was assayed by using the o-aminobenzaldehyde method and the formation of a yellowish product, as described in Moschou et al. (2008).
Protein Extraction, Western Blotting, RNA Extraction, and Northern Blotting
Proteins were extracted and treated as described in Papadakis and Roubelakis-Angelakis (2005). Total protein extracts were electrophoretically resolved using SDS-PAGE, transferred to membranes, and hybridized against an anti-MBP polyclonal antibody. Total RNA was extracted with the optimized hot-phenol method according to Rojo et al. (1998), transferred to nylon membranes, and hybridized against 32P-labeled probes. All hybridizations were performed at 67°C and the appropriate controls were used, including RNA ladders, for the correct size of each transcript to be estimated.
Templates for probes were obtained using the following primers and standard PCR conditions: for PAO1, GGTAGTGGTGAAGACAGAGG (PAO1-FOR) and AGCTTGTGCCTGAGATAAA (PAO1-REV); PAO2, ATGGAGTCCAGGAAAACTC (PAO2-FOR) and GAGACGAGATATAAGAAG (PAO2-REV); PAO3, GGGGACAAGTTTGTACAAAAAAGCAGGCTTGATGGAGTCCGGAGGCAAC (PAO3-FOR) and GGGGACCACTTTGTACAAGAAAGCTGGGTATTACATACGGGAGATCAGAAG (PAO3-REV); PAO4, GGTCACCAAACCCTTCATC (PAO4-FOR) and TTGAGCAAACGATGGAGAAG (PAO4-REV); PAO5 ATGGCGAAGAAAGCAAGA (PAO5-FOR) and AAAATTACATTTGTAAT (PAO5-REV).
RT-PCR Analysis
For semiquantitative RT-PCR reactions, total mRNA from leaves and roots was extracted and subjected to DNase I RNase-free treatment (Invitrogen) for 45 min at 37°C. After checking the RNA in agarose gel, the samples were subjected to RT-PCR reactions using polyT as primer and the Super RT enzyme according to the manufacturer's instructions (HT Biotechnology). The samples were normalized according to actin and the primers TTGAGCACATTCTGGAAGAGGTG (forward PAO3) and ACACTTTGCCGAGATGGTTTCAG (reverse PAO3) were used for AtPAO3-specific 30 cycle (exponential phase) quantitative RT-PCR reactions. Reactions with samples treated with DNase I but not subjected to RT were also used as negative controls.
Transient Expression and Colocalization
Agrobacterium tumefaciens strain C58C1 cells were transformed with the appropriate binary vectors using the freeze-thaw method and PCR confirmed. A. tumefaciens positive clones were grown in Luria-Bertani until OD600 = 0.6 and were pelleted after a centrifugation step at 3,000g for 20 min. Cells were resuspended in MMA (Murashige and Skoog salts, 10 mm MES, pH 5.7, 0.2 mm acetosyringone, 3% Suc) until OD600 = 0.5, incubated at RT for 1 h, and infiltrated in Nicotiana benthamiana leaves, using a 1-mL syringe. Leaves were analyzed after 48 h using a Leica TCS-NT confocal microscope, using the 40× objective. The excitation/emission wavelength was 480/508 nm for GFP, 513/527 nm for YFP, and 587/610 nm for mCherry. The YFP-PRX (PRX: peroxisomal) and mCherry-PRX constructs were purchased from the ABRC Stock Center (http://www.arabidopsis.org).
Colocalization was performed by mixing the MMA-resuspended A. tumefaciens GFP:PAO3 with A. tumefaciens YFP-PRX or mCherry-PRX in a ratio 1.2:1 (v/v). Merge of the images was performed using the Leica Confocal software version 2.61.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NM_121373 (AtPAO1), AF364952 (AtPAO2), NM_115767 (AtPAO3), AF364953 (AtPAO4), and AK118203 (AtPAO5).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression of AtPAO3 in leaves and roots. cDNAs were equalized according to actin.
Supplementary Material
Acknowledgments
The authors are grateful to Dr. Helena Berglund (Karolinska Institute, Sweden) for the generous gift of the pDEST-TH1 vector.
This work was supported by the National and European resources (EPEAKII-Pythagoras), Greek-Spain Bilateral Agreement, and COST858, COST FA065 Actions; by the Consejo Superior de Investigaciones Científicas I3P Programme cofinanced by the European Social Fund (to M.S.); and by the Onassis Foundation (scholarship to A.H.A.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Kalliopi A. Roubelakis-Angelakis (poproube@biology.uoc.gr).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alvarez ME, Pennell RI, Meijer PJ, Ishikawa A, Dixon RA, Lamb C (1998) Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92 773–784 [DOI] [PubMed] [Google Scholar]
- Amendola R, Bellini A, Cervelli M, Degan P, Marcocci L, Martini F, Mariottini P (2005) Direct oxidative DNA damage, apoptosis and radio sensitivity by spermine oxidase activities in mouse neuroblastoma cells. Biochim Biophys Acta 1755 15–24 [DOI] [PubMed] [Google Scholar]
- Angelini R, Federico R, Bonfante P (1995) Maize polyamine oxidase: antibody production and ultrastructural localization. J Plant Physiol 145 686–692 [Google Scholar]
- Asai T, Stone JM, Heard JE, Kovtun Y, Yorgey P, Sheen J, Ausubel FM (2000) Fumonisin B1-induced cell death in Arabidopsis protoplasts requires jasmonate-, ethylene-, and salicylate-dependent signaling pathways. Plant Cell 12 1823–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacchi CJ, Rattendi D, Faciane E, Yarlett N, Weiss LM, Frydman B, Woster P, Wei B, Marton LJ, Wittner M (2004) Polyamine metabolism in a member of the phylum Microspora (Encephalitozoon cuniculi): effects of polyamine analogues. Microbiology 150 1215–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capell T, Bassie L, Christou P (2004) Modulation of the polyamine biosynthetic pathway in transgenic rice confers tolerance to drought stress. Proc Natl Acad Sci USA 101 9909–9914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervelli M, Caro O, Penta A, Angelini R, Federico R, Vitale A, Mariottini P (2004) A novel C-terminal sequence from barley polyamine oxidase is a vacuolar sorting signal. Plant J 40 410–418 [DOI] [PubMed] [Google Scholar]
- Cona A, Moreno S, Cenci F, Federico R, Angelini R (2005) Cellular re-distribution of flavin-containing polyamine oxidase in differentiating root and mesocotyl of Zea mays L. seedlings. Planta 221 265–276 [DOI] [PubMed] [Google Scholar]
- Cona A, Rea G, Angelini R, Federico R, Tavladoraki P (2006) Functions of amine oxidases in plant development and defence. Trends Plant Sci 11 80–88 [DOI] [PubMed] [Google Scholar]
- Del Duca S, Beninati S, Serafini-Fracassini D (1995) Polyamines in chloroplasts: identification of their glutamyl and acetyl derivatives. Biochem J 305 233–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores HE, Galston AW (1982) Analysis of polyamines in higher plants by high performance liquid chromatography. Plant Physiol 69 701–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha HC, Woster PM, Yager JD, Casero RA (1997) The role of polyamine catabolism in polyamine analogue-induced programmed cell death. Proc Natl Acad Sci USA 94 11557–11562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarstrom M, Hellgren N, Van der Berg S, Berglund H, Hard T (2002) Rapid screening for improved solubility of small human proteins produced as fusion proteins in Escherichia coli. Protein Sci 11 313–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Nishimura M (2006) Arabidopsis thaliana: a model organism to study plant peroxisomes. Biochim Biophys Acta 1763 1382–1391 [DOI] [PubMed] [Google Scholar]
- Kaur-Sawhney R, Tiburcio AF, Atabella T, Galston AW (2003) Polyamines in plants: an overview. J Cell Mol Biol 2 1–12 [Google Scholar]
- Kotzabasis K, Christakis-Hampsas MD, Roubelakis-Angelakis KA (1993) A narrow bore HPLC method for the identification and quantitation of free, conjugated and bound polyamines. Anal Biochem 214 484–487 [DOI] [PubMed] [Google Scholar]
- Kusano T, Yamaguchi K, Berberich T, Takahashi Y (2007) Advances in polyamine research in 2007. J Plant Res 120 345–350 [DOI] [PubMed] [Google Scholar]
- Landry J, Sternglanz R (2003) Yeast Fms1 is a FAD-utilizing polyamine oxidase. Biochem Biophys Res Commun 303 771–776 [DOI] [PubMed] [Google Scholar]
- Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P (2006) SMART 5: domains in the context of genomes and networks. Nucleic Acids Res 34 D257–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta RA, Cassol T, Li N, Ali N, Handa AK, Mattoo AK (2002) Engineered polyamine accumulation in tomato enhances phytonutrient content, juice quality, and vine life. Nat Biotechnol 20 613–618 [DOI] [PubMed] [Google Scholar]
- Moschou P, Dellis I, Paschalidis K, Roubelakis-Angelakis KA (2008) Transgenic tobacco plants over-expressing polyamine oxidase are not able to cope with oxidative burst generated by abiotic factors. Physiol Plant 133 140–156 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multi-color set of in vivo organelle markers for colocalization studies in Arabidopsis and other plants. Plant J 51 1126–1136 [DOI] [PubMed] [Google Scholar]
- Nyathi Y, Baker A (2006) Plant peroxisomes as a source of signaling molecules. Biochim Biophys Acta 1763 1478–1495 [DOI] [PubMed] [Google Scholar]
- Orozco-Cardenas ML, Narvaez-Vasquez J, Ryan CA (2001) Hydrogen peroxide acts as a second messenger for the induction of defence genes in tomato plants in response to wounding, systemin and methyl jasmonate. Plant Cell 13 179–191 [PMC free article] [PubMed] [Google Scholar]
- Papadakis AK, Roubelakis-Angelakis KA (2005) Polyamines inhibit NADPH oxidase-mediated superoxide generation and putrescine prevents programmed cell death induced by polyamine oxidase-generated hydrogen peroxide. Planta 220 826–837 [DOI] [PubMed] [Google Scholar]
- Paschalidis KA, Roubelakis-Angelakis KA (2005. a) Spatial and temporal distribution of polyamine levels and polyamine anabolism in different organs/tissues of the tobacco plant. Correlations with age, cell division/expansion, and differentiation. Plant Physiol 138 142–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschalidis KA, Roubelakis-Angelakis KA (2005. b) Sites and regulation of polyamine catabolism in the tobacco plant. Correlations with cell division/expansion, cell cycle progression, and vascular development. Plant Physiol 138 2174–2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Amador MA, Leon J, Green PJ, Carbonell J (2002) Induction of the arginine decarboxylase ADC2 gene provides evidence for the involvement of polyamines in the wound response in Arabidopsis. Plant Physiol 130 1454–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pledgie A, Huang Y, Hacker A, Zhang Z, Woster PM, Davidson NE, Casero RA (2005) Spermine oxidase SMO (PAOh1), not N1-acetylpolyamine oxidase PAO, is the primary source of cytotoxic H2O2 in polyamine analogue-treated human breast cancer cell lines. J Biol Chem 280 39843–39851 [DOI] [PubMed] [Google Scholar]
- Rea G, de Pinto MC, Tavazza R, Biondi S, Gobbi V, Ferrante P, De Gara L, Federico R, Angelini R, Tavladoraki P (2004) Ectopic expression of maize polyamine oxidase and pea copper amine oxidase in the cell wall of tobacco plants. Plant Physiol 134 1414–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Ma C, Lemke S, Babujee L (2004) AraPerox. A database of putative Arabidopsis proteins from plant peroxisomes. Plant Physiol 136 2587–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo E, Titarenko E, León J, Berger S, Vancanneyt G, Sánchez-Serrano JJ (1998) Reversible protein phosphorylation regulates jasmonic acid-dependent and -independent wound signal transduction pathways in Arabidopsis thaliana. Plant J 13 153–165 [DOI] [PubMed] [Google Scholar]
- Sasaki-Sekimoto Y, Taki N, Obayashi T, Aono M, Matsumoto F, Sakurai N, Suzuki H, Hirai MY, Noji M, Saito K, et al (2005) Coordinated activation of metabolic pathways for antioxidants and defence compounds by jasmonates and their roles in stress tolerance in Arabidopsis. Plant J 44 653–668 [DOI] [PubMed] [Google Scholar]
- Schrader M, Fahimi DH (2004) Mammalian peroxisomes and reactive oxygen species. Histochem Cell Biol 122 383–393 [DOI] [PubMed] [Google Scholar]
- Seiler N (1995) Polyamine oxidase, properties and functions. Prog Brain Res 106 333–344 [DOI] [PubMed] [Google Scholar]
- Skopelitis DS, Paranychianakis NV, Paschalidis KA, Pliakonis ED, Delis ID, Yakoumakis DI, Kouvarakis A, Papadakis AK, Stephanou EG, Roubelakis-Angelakis KA (2006) Abiotic stress generates ROS that signal expression of anionic glutamate dehydrogenases to form glutamate for proline synthesis in tobacco and grapevine. Plant Cell 18 2767–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Uehara Y, Berberich T, Ito A, Saitoh H, Miyazaki A, Terauchi R, Kusano T (2004) A subset of hypersensitive response marker genes, including HSR203J, is the downstream target of a spermine signal transduction pathway in tobacco. Plant J 40 586–595 [DOI] [PubMed] [Google Scholar]
- Tassoni A, Van Buuren M, Franceschetti M, Fornale ÌS, Bagni N (2000) Polyamine content and metabolism in Arabidopsis thaliana and effect of spermidine on plant development. Plant Physiol Biochem 38 383–393 [Google Scholar]
- Tavladoraki P, Rossi MN, Saccuti G, Perez-Amador MA, Polticelli F, Angelini R, Federico R (2006) Heterologous expression and biochemical characterization of a polyamine oxidase from Arabidopsis involved in polyamine back conversion. Plant Physiol 149 1519–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavladoraki P, Schinina ME, Cecconi F, Di Agostino S, Manera F, Rea G, Mariottini P, Federico R, Angelini R (1998) Maize polyamine oxidase: primary structure from protein and cDNA sequencing. FEBS Lett 426 62–66 [DOI] [PubMed] [Google Scholar]
- Thomas T, Thomas TJ (2001) Polyamines in cell growth and cell death: molecular mechanism and therapeutic applications. Cell Mol Life Sci 58 244–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiburcio AF, Besford RT, Borrell A (1994) Posttranslational regulation of arginine decarboxylase synthesis by spermine in osmotically-stressed oat leaves. Biochem Soc Trans 22 455S. [DOI] [PubMed] [Google Scholar]
- Toninello A, Pietrangeli P, De Marchi U, Salvi M, MondovÌ B (2006) Amine oxidases in apoptosis and cancer. Biochim Biophys Acta 1765 1–13 [DOI] [PubMed] [Google Scholar]
- Vacca RA, de Pinto MC, Valenti D, Passarella S, Marra E, Gara DL (2004) Production of reactive oxygen species, alteration of cytosolic ascorbate peroxidase, and impairment of mitochondrial metabolism are early events in heat shochinduced programmed cell death in tobacco bright-yellow 2 cells. Plant Physiol 134 1100–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Murray-Stewart T, Devereux W, Hacker A, Frydman B, Woster PM, Casero RA Jr (2003) Properties of purified human polyamine oxidase, PAOh1/SMO. Biochem Biophys Res Commun 304 605–611 [DOI] [PubMed] [Google Scholar]
- Wu T, Yankovskaya V, McIntire WS (2003) Cloning, sequencing, and heterologous expression of the murine peroxisomal flavoprotein N1-acetylated polyamine oxidase. J Biol Chem 278 20514–20525 [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Takahashi Y, Berberich T, Imai A, Miyazaki A, Takahashi T, Michael A, Kusano T (2006) The polyamine spermine protects against high salt stress in Arabidopsis thaliana. FEBS Lett 22 6783–8 [DOI] [PubMed] [Google Scholar]
- Yoda H, Hiroi Y, Sano H (2006) Polyamine oxidase is one of the key elements for oxidative burst to induce programmed cell death in tobacco cultured cells. Plant Physiol 142 193–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda H, Yamaguchi Y, Sano H (2003) Induction of hypersensitive cell death by hydrogen peroxide produced through polyamine degradation in tobacco plants. Plant Physiol 132 1973–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.