Abstract
The zinc finger protein vascular endothelial zinc finger 1 (Vezf1) has been implicated in the development of the blood vascular and lymphatic system in mice, and has been characterized as a transcriptional activator in some systems. The chicken homolog, BGP1, has binding sites in the β-globin locus, including the upstream insulator element. We report that in a mouse embryonic stem cell line deletion of both copies of Vezf1 results in loss of DNA methylation at widespread sites in the genome, including Line1 elements and minor satellite repeats, some imprinted genes, and several CpG islands. Loss of methylation appears to arise from a substantial decrease in the abundance of the de novo DNA methyltransferase, Dnmt3b. These results suggest that naturally occurring mutations in Vezf1/BGP1 might have widespread effects on DNA methylation patterns and therefore on epigenetic regulation of gene expression.
Keywords: DNA methylation, BGP1, Vezf1, Dnmt3b
DNA methylation is an indispensable epigenetic modification that is directly connected to gene silencing. It is important for many cellular processes including silencing of repetitive elements, X inactivation, imprinting, and development. Methylated sites on promoters provide signals that result in the recruitment of repressive histone modifications and down-regulation of gene expression (Miranda and Jones 2007). Recent studies have suggested a role of DNA methylation during differentiation in repressing a subset of germline specific genes (Weber et al. 2007; Zilberman 2007). Anomalous DNA methylation has been linked to several forms of cancer and other genetic diseases (Esteller 2005).
DNA methylation is established in early development and maintained throughout the cell’s lifetime by de novo DNA methyltransferases Dnmt3a and Dnmt3b and the maintenance methyltransferase (MTase) Dnmt1 (Goll and Bestor 2005). Considering the scale at which genomic expression is regulated by DNA methylation, relatively few cofactors/modifiers have been shown to be involved in this process. A number of proteins interact directly with the MTases and regulate their activity. Recently, it has been shown that Dnmt3L interacts directly with Dnmt3a2 and Dnmt3b and targets these factors to promoters carrying unmethylated H3K4, promoting de novo DNA methylation at these sites (Ooi et al. 2007). Similarly, UHRF1 interacts with Dnmt1 and delivers it to hemimethylated CpG sites, helping in maintenance methylation (Bostick et al. 2007; Sharif et al. 2007). LSH, a chromatin remodeling factor, also interacts directly with Dnmt3a and Dnmt3b; it has been suggested that this increases the accessibility of the heterochromatic DNA to MTase enzymes (Muegge 2005; Zhu et al. 2006). Other mechanisms for regulation of DNA methylation act indirectly through the histone modification pathway: CGBP, a zinc finger protein, binds specifically to unmethylated DNA and associates with the Set 1 H3K4 MTase complex, restricting its activity and thereby influencing the DNA methylation pattern (Lee and Skalnik 2005). At the level of transcription, the MTases are known to be regulated by binding of the ubiquitous transcription factors Sp1 and Sp3 to MTase promoters (Ishida et al. 2003; Jinawath et al. 2005). However, the roles of any tissue-specific transcription factors or regulatory mechanisms controlling their expression in different cell types are unknown.
Here, we present evidence that the absence of a zinc finger protein, vascular endothelial zinc finger 1 (Vezf1)/BGP1, causes major loss of genomic methylation at specific sites, which include certain repeat elements, some imprinted loci, and many CpG islands. We identified BGP1 some years ago as a DNA-binding protein that recognizes poly dG sequences (Lewis et al. 1988; Clark et al. 1990). Recently, we began to reinvestigate the role of BGP1 as a regulatory factor because of our identification of BGP1-binding sites (A. West, M. Gaszner, and G. Felsenfeld, unpubl.) within the compound insulator element at the 5′ end of the chicken βA-globin locus, where they play an essential role in the functioning of the insulator (Recillas-Targa et al. 2002). Cloning of BGP1 has revealed that it is the chicken homolog of the mouse gene Vezf1 and the human gene DB1 (A. West, M. Gaszner, and G. Felsenfeld, unpubl.). Reports of Vezf1 and DB1 indicate that the protein might serve as a transcriptional regulatory factor (Aitsebaomo et al. 2001; Miyashita et al. 2004). However, our earlier studies of another BGP1 site in the cβA-globin promoter had shown that BGP1 does not have a strong regulatory effect on cβA-globin expression in transient assays, and we had suggested that it might function through its effects on chromatin structure (Lewis et al. 1988).
The availability of mouse embryonic stem cells in which both alleles of Vezf1 were deleted homozygously (Kuhnert et al. 2005) made it possible to investigate further the role of Vezf1/BGP1 in gene expression. We show that loss of the gene has genome-wide effects on DNA methylation, and that these can be explained by a preferential depletion of Dnmt3b1, which carries the catalytic domain responsible for de novo DNA methylation.
Results
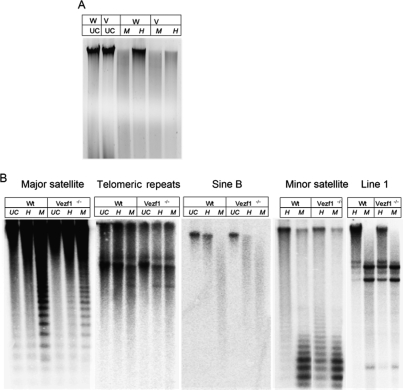
As part of a survey of gene expression patterns in mouse Vezf1−/− embryonic stem (ES) cells, we examined overall genomic levels of histone H3 acetylation and DNA methylation. Histone acetylation levels, determined by chromatin immunoprecipitation (ChIP), were lower in the null line than in wild-type controls (Supplemental Fig. S1), suggesting that the abundance of transcriptionally active chromatin might be reduced. DNA methylation levels were measured by digestion of DNA with the methylation sensitive restriction enzyme HpaII. This revealed, unexpectedly, a marked loss in DNA methylation in Vezf1−/− cells compared with wild-type ES cells (Fig. 1A).
Figure 1.
Loss of Vezf1 results in loss of DNA methylation. (A) Ethidium bromide stained 0.8% agarose gel loaded with genomic DNA from wild-type (W) and Vezf1−/− (V) ES cells. DNA was cleaved with HpaII (H), or MspI (M), or uncut (Schulz et al. 2002). (B) Methylation analysis by Southern blot; DNA was digested with HpaII and MspI and probed for different repetitive elements in the genome. MspI digestion was used as control to represent complete loss of DNA methylation. Line1 and Minor satellites show a distinct loss of methylation. A lesser loss of methylation is also seen at Sine B, telomeric, and major satellite repeats.
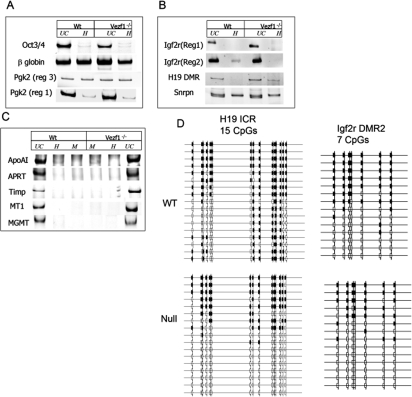
To determine in more detail the sites of altered methylation, specific regions of the genome were probed by Southern blot analysis of HpaII restriction digests. In the mutant cells, Line1 elements and minor satellite repeats showed a substantial loss of methylation. There was partial loss at Sine B and telomeric repeats, whereas major satellite repeat sequences were not affected (Fig. 1B). We also examined methylation at HpaII sites in the upstream regions of several individual genes by methylation-dependent PCR analysis (MDPCR; see Materials and Methods): β-globin and Pgk2, both single-copy, tissue-specific genes (Singer-Sam et al. 1990; Kafri et al. 1992; Ariel et al. 1994), were examined and the results show that whereas β-globin partially lost methylation in Vezf1−/− cells, sequences upstream of Pgk-2 showed no decrease in DNA methylation (Fig. 2A). Loss of Vezf1 also had varying effects at several imprinted loci. Those surveyed were the region upstream of the H19 gene, including the imprinted control region (ICR) with all four CTCF-binding sites (Bell and Felsenfeld 2000), the upstream regions 1 and 2 (reg1 and reg2) of the maternally imprinted Igf2r gene (Stoger et al. 1993), and the upstream region of Snrpn (Tada et al. 1997). As shown in Figure 2B, in wild-type cells the H19 ICR and reg2 of Igf2r had relatively few copies in which every site in the probed region was methylated (i.e., most copies were cut at least once by HpaII). However, these levels of methylation were further reduced in Vezf1−/− cells. There was no change in methylation at Igf2r reg1 which is unmethylated, or at the site upstream of Snrpn which is methylated in the wild-type cells. The PCR results were confirmed by Southern analysis of some of these sites after DNA had been cleaved with methylation-sensitive restriction enzymes (Supplemental Fig S2). CpG islands in the promoters of several housekeeping genes, which usually are maintained in an unmethylated state, did not show any gain of methylation as determined by MDPCR (Fig. 2C). To investigate in detail the methylation state of the individual CpG residues at the H19 ICR and reg2 of Igf2r, we used bisulfite modification of the DNA, followed by cloning and sequencing of the products (Warnecke et al. 1997). As shown in Figure 2D, individual CpGs were selectively reduced in methylation in Vezf1−/− cells, consistent with the results in Figure 2, B and C. At all but three sites there is a decrease in methylation. It should be noted that these changes are not uniformly distributed: Some CpG sites, especially at the H19 ICR sites 8–13, appear to be particularly sensitive to loss of Vezf1, while others are not. This may reflect differences in the relative contributions of different methylating enzymes to methylation of individual CpG sites within the imprinted loci.
Figure 2.
Methylation-dependent PCR analysis of sites at selected genes. Genomic DNA was cleaved with HpaII (H) and MspI (M). (A–C) Ethidium-bromide-stained 0.8% agarose gel loaded with PCR amplified cleaved genomic DNA from wild-type and Vezf1−/− ES cells. (A) Tissue-specific genes. (B) Imprinted genes. (C) Housekeeping genes. (D) Methylation analysis at H19 ICR and Igf2r DMR2 by bisulfite sequencing. The results show a significant loss of methylation at some sites (see the text).
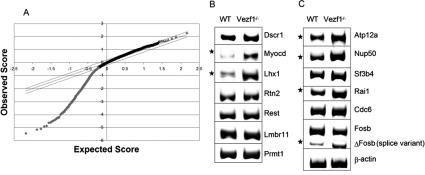
The above observations prompted us to investigate the global methylation of CpG islands in the Vezf1−/− cells, using methylated DNA immunoprecipitation (MeDIP) followed by hybridization of the immunoprecipitated product to a 4.6K mouse CpG island microarray. Using criteria described in the Materials and Methods, analysis of the data (significance analysis of microarray [SAM]) (Tusher et al. 2001) revealed genome-wide loss of methylation at >1300 CpG islands (CpGis); there were no sites of significantly increased methylation (Fig. 3A). Among these, 500 CpGis lost 20%–60% of their wild-type methylation (q value 0%–2%) of which 76 CpGis were located within 3 Kb upstream of the promoters and 71 Cpgis within 3 Kb downstream from genes (Supplemental Tables 4, 5). Many of these CpG islands were found to be associated with genes involved in tumorigenesis (Mcm7, Tceb1, Brf1, Mtmr11, Fev, and Tmc6), testis-specific genes (Abca14, Sart3, Tbx3), neuronal-specific genes involved in brain and craniofacial development (Abat, Tmem24, Dscr1, Tcfap2a, Foxb1, and Ppil3), homeobox genes (Lhx1 and Pbx1), and several tissue-specific genes (Supplemental Table 4). To investigate whether this loss of DNA methylation led to a change in expression, 14 genes were randomly selected (Fig. 3B,C), and the levels of transcript were compared in wild-type and Vezf1−/− cells using RT–PCR. A total of six genes showed an increase in levels of expression. Although this gene activation may reflect the reduced DNA methylation, a direct effect of the loss of Vezf1 on expression of some of these genes cannot be excluded.
Figure 3.
(A) MeDIP/CpG island microarray analysis, comparing genomic DNA from wild-type and Vezf1−/− ES cells. Observed score (see the Materials and Methods) is plotted against expected score (based on randomized sample). The limiting parallel lines are chosen to give a false discovery rate of 29.7%. Analysis shows that almost all the CpGis with significant change (points lying outside the parallel lines) have lost DNA methylation (gray diamonds). Gray triangles represent gain of methylation. (B,C) RT–PCR analysis of genes selected from the microarray that showed significant loss of DNA methylation at CpGis were tested for change in expression. The asterisks indicate the genes that show increased expression in the Vezf1−/− cells.
Effects on DNA methylation mechanisms
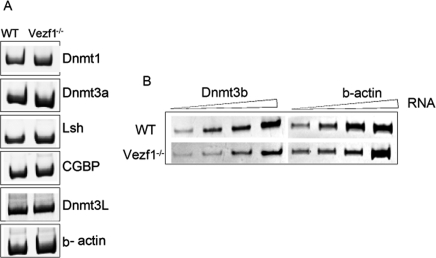
DNA methylation in ES cells involves both maintenance and de novo mechanisms. The above results suggested that levels of one or more of the DNA MTases might be affected in Vezf1−/− cells. As a first step, total DNA methylation activity in vitro was compared in nuclear extracts from wild-type and Vezf1−/− ES cells by methylating plasmid DNA in the presence of 3[H] S-adenosyl methionine. There was no significant difference in the methylation activity, indicating that the level of major MTase activity (predominantly Dnmt1) was essentially unaltered (Supplemental Fig. S3). To distinguish among the different methylating enzymes, we next determined the expression levels in wild-type and Vezf1−/− cells of the three known active DNA MTases. As shown in Figure 4, A and B, RT–PCR amplification of Dnmt1 and Dnmt3a transcripts revealed that Dnmt3a was increased slightly and Dnmt1 was unaffected. In contrast, Dnmt3b expression was considerably reduced in Vezf1−/− cells compared with the wild-type cells. These results were also confirmed at the protein level by Western blot analysis (Supplemental Fig. S4). RT–PCR analysis of transcripts for other proteins known to have an effect on DNA methylation, LSH and CGBP (Dennis et al. 2001; Carlone et al. 2005) and Dnmt3L also showed no change in their levels of expression (Fig. 4B).
Figure 4.
Effect of Vezf1 deletion on expression levels of DNA MTases and of genes coding for other proteins known to modulate DNA methylation levels in the genome. (A,B) One-step RT–PCR was performed with total RNA from wild-type and Vezf1−/− cells to amplify DNA MTases, LSH, CGBP, Dnmt3 L, and β-actin as a control for the amount of RNA template used in the RT–PCR reaction.
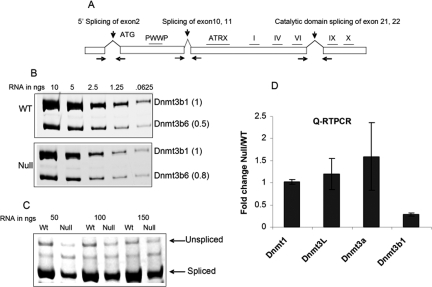
The results described above by RT–PCR reflected contributions from several Dnmt3b splice variants, and it was not clear which of these were changed in the mutant line. Dnmt3b transcripts are known to undergo alternative splicing; more than eight splice variants have been identified, and subsets of these have been shown to be expressed, in different cell types (Fig. 5A; Robertson et al. 1999; Saito et al. 2002; Ostler et al. 2007). Some of these isoforms are catalytically inactive since the splicing results in the loss of critical MTase motifs from the protein. Dnmt3b1 is the catalytically active form expressed in ES cells. The catalytically inactive forms Dnmt3b6 and weak amounts of Dnmt3b7 are also expressed; these lack the essential MTase motifs in the catalytic domain, which are lost by alternative splicing of exons 21 and 22 (Weisenberger et al. 2004). To identify these splice variants, primer pairs were constructed which spanned several exon–intron boundaries of the Dnmt3b gene allowing them to be visualized simultaneously. The results show that expression of both Dnmt3b1 and Dnmt3b6 is decreased in Vezf1−/− ES cells. Interestingly, semiquantitative analysis of RT–PCR products revealed a Dnmt3b6 to Dnmt3b1 ratio of 0.5:1 in wild-type cells but 0.8:1 in Vezf1−/− cells (Fig. 5B).
Figure 5.
Analysis of the splice variants of Dnmt3b; Dnmt3b transcript is decreased in Vezf1−/− cells. (A) Map of Dnmt3b gene showing three characterized splicing events. To analyze alternative splicing events, total RNA from wild-type and Vezf1−/− ES cells was reverse transcribed and amplified by PCR for 27 cycles. (B) Catalytic domain splicing. (C) 5′ end splicing. (D) Relative quantitative RT–PCR analysis for the measurement of DNA MTases using Taqman expression assay (Materials and Methods) The Ct values for all MTases are normalized to that of β-actin.
Splicing events known to take place on Dnmt3b transcripts were further investigated: 5′ splicing, which can remove exon 2, and splicing of exons 10 and 11. Semiquantitative RT–PCR measurements (Weisenberger et al. 2004) showed that Vezf1−/− ES cells expressed Dnmt3b mRNAs containing exons 10 and 11, which is also characteristic of the Dnmt3b1 and 3b6 variants found as the major components in normal ES cells (data not shown). Experiments designed to detect splicing at exon 2 at the 5′ end of the gene revealed both spliced and unspliced forms of Dnmt3b transcripts; at least 90% of the mRNA was spliced (Fig. 5C). However, the abundance of the spliced form was 2.5-3 fold lower in Vezf1−/− cells than in wild type. We also detected in small amounts in ES cells a previously unreported variant (see the Discussion; Supplemental Fig. S7).
We further determined the expression levels of DNA MTases by quantitative RT–PCR measurements. For Dnmt3b, the primers were designed to target the 3′ end of the transcript, which detect only the catalytically active full-length form Dnmt3b1. The analysis confirmed a fourfold reduction in the Dnmt3b1 transcript in Vezf1−/− ES cells (Fig. 5D), whereas the levels of Dnmt1 and Dnmt3L remained unchanged. There was an ∼1.5-fold increase in Dnmt3a transcript. However, this increase does not appear to compensate at the sites we observed for the loss of Dnmt3b activity.
The level of Dnmt3b protein was measured by Western blot, and appeared decreased in the mutant cells, consistent with the above finding (Supplemental Fig. S4). Both the splice variant Dnmt3b6 and the full-length variant Dnmt3b1 can be seen, and both are diminished in abundance, as compared with an actin control. No other Dnmt3b variants are significantly expressed in ES cells.
Vezf1 can rescue DNA methylation defects in Vezf1−/− ES cells
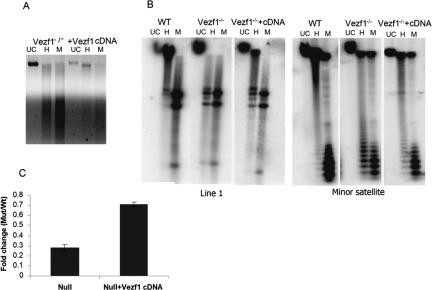
The Vezf1−/− ES cells were stably transfected with a plasmid carrying Vezf1 cDNA expressed under a CMV promoter. The expression of Vezf1 was checked by RT–PCR analysis (Supplemental Fig. S5A); a clone was selected in which Vezf1 was expressed at levels close to wild type. As shown in Figure 6A, the genomic DNA from the Vezf1-transfected clone is more resistant to HpaII digestion than is DNA from the Vezf1−/− cells. Southern blot analysis of DNA methylation of the Line1 and minor satellite repeats from the transfected clone clearly displayed a substantial rescue of DNA methylation at both repetitive elements when compared with Vezf1−/− (Fig. 6B). Quantitative RT–PCR showed that the level of Dnmt3b expression was also considerably restored (Fig. 6C). Measurements of both Vezf1 and Dnmt3b protein in the transfected Vezf1−/− cells were consistent with the above results (Supplemental Fig. S5B).
Figure 6.
Transfection of Vezf1−/− ES cells with a Vezf1 expression construct restores DNA methylation. (A) Ethidium-bromide-stained gel of genomic DNA from Vezf1−/− ES cells transfected with a construct expressing Vezf1 and cleaved with HpaII (H) and MspI (M). Compared with untransfected Vezf1−/− cells, the transfected cells show less cleavage by HpaII, reflecting increased methylation of these sites. (B) DNA methylation analysis of repetitive elements in transfected cells: Southern blot of genomic DNA cleaved with HpaII and MspI and hybridized with probes against Line1 and minor satellite repeats. Both repeat elements show partial rescue of methylation in comparison with untransfected cells. (C) Relative quantitative RT–PCR analysis of Dnmt3b transcripts, normalized to β-actin as control using the TaqMan assay.
Vezf1-binding sites at the Dnmt3b locus
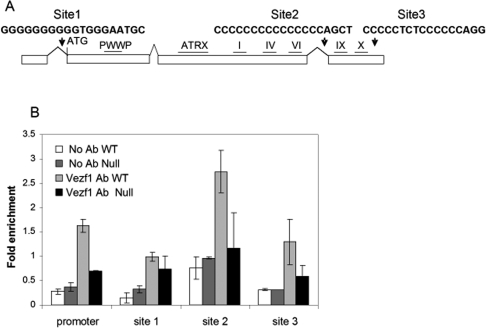
Vezf1/BGP1 is a C2H2 zinc finger protein that has been characterized biochemically and shown to bind to poly G tracts as well as to some bipartite sequences containing poly Cs or Gs. The sequence in the neighborhood of Dnmt3b contains several candidate Vezf1-binding sites; the three best potential sites were found within two introns near the 5′ and 3′ ends of the gene, and within the 3′ untranslated region (UTR) of the Dnmt3b gene (Fig. 7A; sites 1, 2, and 3, respectively). A ChIP/quantitative PCR assay was performed with primers and TaqMan probes covering all three potential binding sites, using a polyclonal antibody against chicken Vezf1/BGP1 (Supplemental Fig. S6). The comparative analysis of Vezf1 binding in wild type versus Vezf1−/− in Figure 7B shows that Vezf1 is present at site 2. There is little or no binding to other sites.
Figure 7.
ChIP analysis of Vezf1/BGP1 binding and histone acetylation in vivo at sites in the Dnmt3b gene. (A) Map of the Dnmt3b gene with positions of the sites studied, showing oligo-G sequences that are potential Vezf1-binding sites. (B) ChIP of formaldehyde cross-linked chromatin from wild-type and Vezf1−/− ES cells with anti-Vezf1 antibody. DNA from IP fractions was used for quantitative PCR analysis at the Dnmt3b promoter and at the three oligo-G sequences. Fold enrichment over input is compared for wild-type and Vezf1−/− cells (see the Materials and Methods).
ChIP assays using antibodies directed against several histone H3 modifications revealed about threefold reduced levels of acetylation, compared with wild-type cells, over the Dnmt3b promoter in Vezf1−/− cells (Supplemental Fig. S7). Small changes in acetylation were observed at downstream sites 1–3. Only small changes were seen at these sites for H3K4 dimethylation, H3K9 dimethylation, and H3K27 trimethylation, with the exception of site 3 in the 3′ UTR, where H3K4 dimethylation was somewhat increased in Vezf1−/− cells (Supplemental Fig. S7).
Discussion
Mouse Vezf1 was identified as a gene that is highly expressed in vascular endothelium during early embryonic development. Vezf1 is also expressed in neuronal and mesodermal tissues in embryos, and in endothelial and hematopoietic cell lines (Xiong et al. 1999). Mouse and human Vezf1 bind in a sequence-specific manner to GC-rich sequences of several genes, including IL-3, FLK-1, FLT-1, ET-1, and stathmin/OP18 (Koyano-Nakagawa et al. 1994; Aitsebaomo et al. 2001; Miyashita et al. 2004; D. Lemons and H. Stuhlmann, in prep.). However, transcriptional activation of at least some of the target gene promoters appears to be dependent on interactions between VEZF1 and components of the Rho GTPase machinery, including p68RacGAP (Aitsebaomo et al. 2004) and RhoB (Lebowitz and Prendergast 1998; Kuhnert and Stuhlmann 2007). The homologous protein in chicken, BGP1, has known binding sites in the chicken β-globin promoter (Lewis et al. 1988); and in the insulator at the 5′ end of the β-globin locus (A. West, M. Gaszner, and G. Felsenfeld, unpubl.).
Targeted inactivation of the Vezf1 gene in mice revealed that it is essential during embryogenesis, and that it acts in a dosage-dependent fashion to regulate the development of blood and lymphatic vascular systems (Kuhnert et al. 2005). Furthermore, embryoid bodies derived from in vitro differentiating ES cells with both Vezf1 alleles inactivated (Vezf1−/−) fail to form a well-organized vascular network and display vascular sprouting defects (Z. Zhou and H. Stuhlmann, in prep.).
Results from our present study show that Vezf1−/− ES cells display widespread loss of DNA methylation of the genome, and that this loss correlates with an approximately fourfold reduced expression of the de novo DNA MTase, Dnmt3b, compared with wild-type ES cells. Of interest, both Dnmt3b-null and Vezf1-null embryos show ectopic hemorrhage in the head region, subcutaneous edema, and die during midgestation stages (Kuhnert et al. 2005; Ueda et al. 2006); however, vascular defects have not been studied in detail in the Dnmt3b-null mice, and the mutant mice show additional defects in other tissues (Ueda et al. 2006). Therefore, it is plausible that at least some of the defects observed in the Vezf1 mutant mice are due to the considerable loss of Dnmt3b activity.
Complete loss of Dnmt3b (Dnmt3b−/−) causes fetal lethality in mice (Okano et al. 1999). In humans, hypomorphic mutations in the DNMT3B gene are found in individuals with ICF (immunodeficiency, centromere instability, facial anomalies) syndrome (Xu et al. 1999; Ehrlich et al. 2003). Patients with ICF syndrome display developmental defects that are characterized by region-specific losses of DNA methylation. These include satellite 2 and 3 repeats, other sporadic repeats like D4Z4, the inactive X chromosome, and cancer-testis (C-T) genes, suggesting that these sequences are bona fide targets of Dnmt3b (Hansen et al. 2000; Kondo et al. 2000). Of interest, mice carrying ICF-like hypomorphic missense mutations survive to term, but most of them die shortly after birth. The mice display phenotypes that are similar to those found in ICF patients, including hypomethylation of repetitive sequences (Ueda et al. 2006). We show here that the partial loss of Dnmt3b expression in Vezf1−/− ES cells causes methylation defects similar to those found in ICF patients, Dnmt3b−/− embryos, and ICF-like hypomorphic mutant mice. Our microarray analysis of CpG island methylation revealed a decrease in methylation levels surrounding the promoters of several genes, some of which were shown recently to have misregulated expression in ICF cells (Jin et al. 2008). Together, these results strongly suggest a role for Vezf1 in regulating genomic DNA methylation patterns.
In an attempt to understand the mechanism by which Vezf1 regulates the expression of Dnmt3b, we identified three binding motifs for Vezf1, all in the introns or 3′ UTR of the Dnmt3b gene. Data from ChIP experiments (Fig. 7B) shows that Vezf1 is present at one of these sites, and may be present at a second, in wild-type cells. The mechanism by which loss of Vezf1 from binding sites within the Dnmt3b gene results in a decrease of its expression is not yet clear. Our results, however, show that the Dnmt3b6 transcript, which is an inactive splice variant of Dnmt3b1, is also decreased in abundance in Vezf1−/− ES cells, but to a lesser extent than Dnmt3b1, perhaps due to improper alternative splicing. This hypothesis is also supported by a number of independent observations. First, a primer set designed across exons 4 and 7 of the Dnmt3b gene revealed a new isoform in which exon 6 is spliced out (Supplemental Fig. S8). This expressed isoform was present in wild-type ES cells, and was not detected in the Vezf1−/− ES cells. The novel splice product would result in removal of 42 amino acids N-terminal of the PHD domain of Dnmt3b protein. Second, in surveying effects of the absence of Vezf1 on expression of tissue specific genes (Fig. 3B) we noted another example of an effect on splicing: Fosb showed decreased expression of full-length transcript and increased expression of an alternatively spliced shorter isoform, ΔFosb, in Vezf1−/− cells compared with the wild-type ES cells. It remains to be determined whether Vezf1/BGP1 plays a more widespread role in regulation of splicing. Long uninterrupted poly-G sequences (n > 10), binding sites for BGP1 (Lewis et al. 1988), can be found in introns of many genes, including Fosb. One possibility is that changes in rates of transcription affect the relative probabilities of following different splicing pathways. It is interesting to note that the zinc finger-binding domain of Vezf1 has considerable homology with that of the DNA-binding factor Maz, which, it has been suggested, can slow transcript elongation (Ashfield et al. 1994), and could, in principle, alter the relative proportion of splice variants.
Whatever the mechanism, it is clear that Vezf1/BGP1 has a significant effect on Dnmt3b expression in ES cells, and consequently a major but selective effect on patterns of DNA methylation, which in turn has the potential to affect gene expression throughout the genome. We note that Vezf1 protein does not appear to interact directly with Dnmt3a, Dnmt3b, or Dnmt3L, as judged by the negative results obtained in attempts to immunoprecipitate these MTases with an antibody to Vezf1 (data not shown).
We suggest also that the possibility of defects in Vezf1 function might be explored in connection with certain genetic disorders like ICF syndrome. Sixty percent to 90% of ICF patients have a missense mutation in the catalytic domain of DNMT3B, causing a partial loss of its activity (Hansen et al. 1999; Xu et al. 1999). There are, however, known ICF patients who lack DNMT3B mutations but have a distinctive set of DNA methylation defects, pointing to the existence of distinct ICF subtypes and the possible involvement of another gene (Jiang et al. 2005). Alterations in DNA methylation are also seen in almost all cancers. Satellite 2 and 3 hypomethylation is very common in cancer cells (Narayan et al. 1998), and can cause chromosomal instability (Wong et al. 2001; Schulz et al. 2002; Tsuda et al. 2002; Ehrlich et al. 2003). It has also been proposed that cancer-associated hypomethylation of satellite DNA sequences might function in altering gene expression to favor carcinogenesis (Ehrlich 2002). It is therefore plausible that Vezf1 may be a candidate oncogene whose altered expression can result in transformation at least mediated by changes in expression/splicing of Dnmt3b.
Materials and methods
Maintenance and transfection of ES cells
Wild-type ES cells and Vezf1−/− cells were cultured in the absence of MEFs in 1× GMEM medium (Sigma, G5154) supplemented with 2 mM glutamine (GIBCO-BRL), 1 mM sodium pyruvate (GIBCO-BRL), 1× nonessential amino acids, 10% (v/v) fetal bovine serum (characterized; Hyclone), 50 μM β-mercaptoethanol, and 500–1000 U/mL leukocyte inhibitory factor (Chemicon catalog no. ESG1107). The Vezf1−/− ES cells were maintained in 600 μg/mL Geneticin.
Stable transfection of Vezf1−/− ES cells was performed in Nucleofector II using Mouse ES cell Nucleofector Kit from Amaxa Biosystems. The human cDNA for Vezf1 cloned in mammalian expression vector pME18S was obtained from Dr. Cam Patterson (University of North Carolina, Chapel Hill, NC). The Vezf1−/− ES cells were cotransfected with a hygromycin resistance marker. The clones resistant to the antibiotic were selected, and the presence of a Vezf1 transcript was detected by RT–PCR analysis.
Methylation analysis
The DNA methylation activity of the wild-type and Vezf1−/− ES cells was determined by an in vitro DNA MTase activity assay. Twenty-five micrograms of wild-type and Vezf1−/− ES nuclear extract as determined by Bio-Rad protein assay (catalog no. 500-0006) were incubated with 1 μg of λ-phage DNA in the presence of 2 μCi of 3H-methyl-S-adenosyl L-methionine (15 Ci/mmol; Perkin-Elmer) in 120 mM NaCl, 10 mM Tris-HCl (pH 7.9), 10 mM EDTA, and 1 mM dithiothreitol (DTT) for 1 h at 30°C. In vitro methylated DNA was spotted on a Whatman DE-81 ion-exchange filter, washed five times with 0.2 M ammonium bicarbonate, and incorporated radioactivity was measured by scintillation counting.
For Southern blots, genomic DNA was digested, separated by electrophoresis on 1% agarose gels, and transferred on a Zeta-Probe GT membrane (Bio-Rad). The rest of the procedure was according to the manufacturer’s protocol. The oligonucleotide probes used for repetitive elements have been described (Dennis et al. 2001) A 330-bp fragment was used to analyze the MluI site of region 2 of the Igf2r gene (Tada et al. 1997). The H19 imprinted region was analyzed by using a 1.5-kb probe corresponding to positions 1282–2808 bp (GenBank accession no. U19619). The HhaI site at the 3′ end of the β- globin gene was analyzed for methylation by using a 1-kb probe corresponding to positions 3841–4921 bp (GenBank acession no. J00413) against an 8-kb BamHI fragment containing the HhaI site (Tada et al. 1997).
Methylation-dependent PCR analysis was performed by digesting genomic DNA with PvuII (β-globin, Pgk2, and Igf2r) or HindIII (ApoA1, Aprt, Timp, MT1, MGMT, and H19) alone and with HpaII restriction enzymes. Two-hundred-base-pair to 500-bp regions in the promoters of the genes, or as described in the results for the imprinted genes, were amplified from the 20 ng of digested genomic DNA using primers flanking the HpaII sites (Supplemental Table 2).
Bisulphite sequencing was carried out as described in Warnecke et al. (1997). Briefly, 1 μg of genomic DNA from wild-type or null cells was treated with sodium bisulphate and the converted DNA was amplified at H19 ICR and Igf2r DMR regions by nested PCR. The primer sequences are given in Supplemental Table 2. The amplified DNA was cloned in pGEM T Easy system (Promega) and individual clones were sequenced.
MeDIP and microarray analysis
Genomic DNA was isolated from both the wild-type and the Vezf1−/− ES cells, and equal amounts were used for MeDIP. MeDIP assay was performed as described (Weber et al. 2005). The immunoprecipitation (IP) was carried out in triplicate both for wild-type and the Vezf1−/− genomic DNA and the IP fraction was amplified by random PCR using the following primers: Primer A, GTTTCCCAGTCACGATCNNNNNNNN: Primer B, GTTTCCCAGTCACGATC, using the protocol published by University Health Network Microarray Center (UHNMC; http://www.microarrays.ca) to generate data that were tested for statistically significant changes in methylation. The IP products and inputs were sent to UHNMC for hybridization on a mouse CpG-island 4.6K array (MCGI4.6K). This is a single-spotted DNA array containing 4642 CpG-island clones. The data analysis was also performed by UHNMC using the SAM program (Tusher et al. 2001) to detect changes in methylation levels. The SAM analysis calculates a score by dividing the difference between the signal-to-input ratios of the null and the wild type by the average standard deviation (Tusher et al. 2001). The program yielded 1302 genes with decreases ranging from 5% to 60% (q value ranging from 0% to 64%) at a false discovery rate of 29.7%. We further sorted the data manually and reduced the list to 500 genes with fold change of 20%–60% and q values of 0%–2%.
RT–PCR and quantitative PCR analysis
The specified amount of DNase (Roche)-treated RNA purified using RNeasy Mini Kit (Qiagen) was reverse transcribed using the 1 Step RT–PCR kit (Invitrogen). The primers were designed at the exon–intron boundaries (Supplemental Table 1). All the RT–PCR products were in the size range of 200–500 bp. The primer sequences are provided in the Supplemental Material. Quantitative RT–PCR procedures used TaqMan One-Step RT–PCR Master Mix and TaqMan Gene Expression Assays in the ABI PRISM 7900 Sequence Detection System (Applied Biosystems). Quantitative PCR for ChIP studies was carried out on the same instrument with primer-probe sets designed using the software Primer Express. The analysis was done as described (Litt et al. 2001).
Antibody production and ChIP
Antibody against Vezf1 was produced in rabbits at Spring Valley Laboratories, Inc. Chicken Vezf1/BGP1, which shares >95% homology with the mouse protein, was cloned and expressed as a His-tagged recombinant protein in Escherichia coli. The overexpressed protein was purified in the presence of 7 M urea on Ni NTA Agarose (Qiagen) using the manufacturer’s protocol. The Vezf1 protein was separated by PAGE. Coomassie-stained bands were excised and used as an antigen to raise a polyclonal antibody. The antibody was able to immunoprecipitate His-tagged recombinant Vezf1 from E. coli cell extracts. The specificity of the antibody was tested by using it to detect Vezf1 from wild-type and Vezf1−/− nuclear extracts in Western blot (Supplemental Fig. S6).
For chromatin preparation, 10 confluent T175 flasks containing wild-type or Vezf1−/− ES cells were treated with 1% formaldehyde for 5 min at room temperature and the reaction was stopped by adding 125 mM glycine. The cells were washed twice and manually dislodged in chilled PBS. The pelleted cells were washed with 15 mL of 10 mM Tris (pH 8.0) buffer containing 10 mM EDTA, 0.5 mM EGTA, and 0.25% Triton X, and centrifuged at 2500 rpm for 10 min. The pellet was then washed with 15 mL of 10 mM Tris (pH 8.0) buffer containing 200 mM NaCl, 10 mM EDTA, and 0.5 mM EGTA, and again centrifuged at 3000 rpm for 10 min. The pellet was washed twice and resuspended in 5 mL of 10mM Tris (pH 8.0), 10 mM EDTA, and 0.5 mM EGTA buffer. This was followed by sonication and immunoprecipitation using a ChIP Assay kit (Upstate Biotechnologies/Millipore) according to the manufacturer’s protocol. One-hundred microliters of chromatin (0.2 μg/μL measured at A260) and 30 μg of anti-acetyl Histone H3 antibody, anti-H3K4 dimethyl, anti-K9 dimethyl, and anti-H3K27 trimethyl antibodies (Upstate Biotechnologies), or 90 μg of anti-Vezf1 antibody were used in each immunoprecipitation reaction.
Acknowledgments
We thank members of the Felsenfeld laboratory for helpful discussions and a critical reading of the manuscript, and Dr. Frank Kuhnert for generating the Vezf1−/− cell line. This work was supported by the intramural research program of the NIH, National Institute of Diabetes and Digestive and Kidney Diseases, and by NIH grant R01 HL 068648 to H.S.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1658408.
References
- Aitsebaomo J., Kingsley-Kallesen M.L., Wu Y., Quertermous T., Patterson C. Vezf1/DB1 is an endothelial cell-specific transcription factor that regulates expression of the endothelin-1 promoter. J. Biol. Chem. 2001;276:39197–39205. doi: 10.1074/jbc.M105166200. [DOI] [PubMed] [Google Scholar]
- Aitsebaomo J., Wennerberg K., Der C.J., Zhang C., Kedar V., Moser M., Kingsley-Kallesen M.L., Zeng G.Q., Patterson C. p68RacGAP is a novel GTPase-activating protein that interacts with vascular endothelial zinc finger-1 and modulates endothelial cell capillary formation. J. Biol. Chem. 2004;279:17963–17972. doi: 10.1074/jbc.M311721200. [DOI] [PubMed] [Google Scholar]
- Ariel M., Cedar H., McCarrey J. Developmental changes in methylation of spermatogenesis-specific genes include reprogramming in the epididymis. Nat. Genet. 1994;7:59–63. doi: 10.1038/ng0594-59. [DOI] [PubMed] [Google Scholar]
- Ashfield R., Patel A.J., Bossone S.A., Brown H., Campbell R.D., Marcu K.B., Proudfoot N.J. MAZ-dependent termination between closely spaced human complement genes. EMBO J. 1994;13:5656–5667. doi: 10.1002/j.1460-2075.1994.tb06904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell A.C., Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- Bostick M., Kim J.K., Esteve P.O., Clark A., Pradhan S., Jacobsen S.E. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- Carlone D.L., Lee J.H., Young S.R., Dobrota E., Butler J.S., Ruiz J., Skalnik D.G. Reduced genomic cytosine methylation and defective cellular differentiation in embryonic stem cells lacking CpG binding protein. Mol. Cell. Biol. 2005;25:4881–4891. doi: 10.1128/MCB.25.12.4881-4891.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S.P., Lewis C.D., Felsenfeld G. Properties of BGP1, a poly(dG)-binding protein from chicken erythrocytes. Nucleic Acids Res. 1990;18:5119–5126. doi: 10.1093/nar/18.17.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis K., Fan T., Geiman T., Yan Q., Muegge K. Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes & Dev. 2001;15:2940–2944. doi: 10.1101/gad.929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M. DNA hypomethylation, cancer, the immunodeficiency, centromeric region instability, facial anomalies syndrome and chromosomal rearrangements. J. Nutr. 2002;132 (Suppl.):2424S–2429S. doi: 10.1093/jn/132.8.2424S. [DOI] [PubMed] [Google Scholar]
- Ehrlich M., Hopkins N.E., Jiang G., Dome J.S., Yu M.C., Woods C.B., Tomlinson G.E., Chintagumpala M., Champagne M., Dillerg L., et al. Satellite DNA hypomethylation in karyotyped Wilms tumors. Cancer Genet. Cytogenet. 2003;141:97–105. doi: 10.1016/s0165-4608(02)00668-4. [DOI] [PubMed] [Google Scholar]
- Esteller M. Aberrant DNA methylation as a cancer-inducing mechanism. Annu. Rev. Pharmacol. Toxicol. 2005;45:629–656. doi: 10.1146/annurev.pharmtox.45.120403.095832. [DOI] [PubMed] [Google Scholar]
- Goll M.G., Bestor T.H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Hansen R.S., Wijmenga C., Luo P., Stanek A.M., Canfield T.K., Weemaes C.M., Gartler S.M. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc. Natl. Acad. Sci. 1999;96:14412–14417. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen R.S., Stöger R., Wijmenga C., Stanek A.M., Canfield T.K., Luo P., Matarazzo M.R., D’Esposito M., Feil R., Gimelli G., et al. Escape from gene silencing in ICF syndrome: Evidence for advanced replication time as a major determinant. Hum. Mol. Genet. 2000;9:2575–2587. doi: 10.1093/hmg/9.18.2575. [DOI] [PubMed] [Google Scholar]
- Ishida C., Ura K., Hirao A., Sasaki H., Toyoda A., Sakaki Y., Niwa H., Li E., Kaneda Y. Genomic organization and promoter analysis of the Dnmt3b gene. Gene. 2003;310:151–159. doi: 10.1016/s0378-1119(03)00545-6. [DOI] [PubMed] [Google Scholar]
- Jiang Y.L., Rigolet M., Bourc’his D., Nigon F., Bokesoy I., Fryns J.P., Hulten M., Jonveaux P., Maraschio P., Megarbane A., et al. DNMT3B mutations and DNA methylation defect define two types of ICF syndrome. Hum. Mutat. 2005;25:56–63. doi: 10.1002/humu.20113. [DOI] [PubMed] [Google Scholar]
- Jin B., Tao Q., Peng J., Soo H.M., Wu W., Ying J., Fields C.R., Delmas A.L., Liu X., Qiu J., et al. DNA methyltransferase 3B (DNMT3B) mutations in ICF syndrome lead to altered epigenetic modifications and aberrant expression of genes regulating development, neurogenesis and immune function. Hum. Mol. Genet. 2008;17:690–709. doi: 10.1093/hmg/ddm341. [DOI] [PubMed] [Google Scholar]
- Jinawath A., Miyake S., Yanagisawa Y., Akiyama Y., Yuasa Y. Transcriptional regulation of the human DNA methyltransferase 3A and 3B genes by Sp3 and Sp1 zinc finger proteins. Biochem. J. 2005;385:557–564. doi: 10.1042/BJ20040684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafri T., Ariel M., Brandeis M., Shemer R., Urven L., McCarrey J., Cedar H., Razin A. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes & Dev. 1992;6:705–714. doi: 10.1101/gad.6.5.705. [DOI] [PubMed] [Google Scholar]
- Kondo T., Bobek M.P., Kuick R., Lamb B., Zhu X., Narayan A., Bourc’his D., Viegas-Péquignot E., Ehrlich M., Hanash S.M. Whole-genome methylation scan in ICF syndrome: Hypomethylation of non-satellite DNA repeats D4Z4 and NBL2. Hum. Mol. Genet. 2000;9:597–604. doi: 10.1093/hmg/9.4.597. [DOI] [PubMed] [Google Scholar]
- Koyano-Nakagawa N., Nishida J., Baldwin D., Arai K., Yokota T. Molecular cloning of a novel human cDNA encoding a zinc finger protein that binds to the interleukin-3 promoter. Mol. Cell. Biol. 1994;14:5099–5107. doi: 10.1128/mcb.14.8.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhnert F., Stuhlmann H. Vezf1: A transcriptional regulator of the endothelium. In: Aird W.C., editor. Endothelial biomedicine. Cambridge University Press; Cambridge, UK: 2007. pp. 855–861. [Google Scholar]
- Kuhnert F., Campagnolo L., Xiong J.W., Lemons D., Fitch M.J., Zou Z., Kiosses W.B., Gardner H., Stuhlmann H. Dosage-dependent requirement for mouse Vezf1 in vascular system development. Dev. Biol. 2005;283:140–156. doi: 10.1016/j.ydbio.2005.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebowitz P.F., Prendergast G.C. Functional interaction between RhoB and the transcription factor DB1. Cell Adhes. Commun. 1998;6:277–287. doi: 10.3109/15419069809010787. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Skalnik D.G. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J. Biol. Chem. 2005;280:41725–41731. doi: 10.1074/jbc.M508312200. [DOI] [PubMed] [Google Scholar]
- Lewis C.D., Clark S.P., Felsenfeld G., Gould H. An erythrocyte-specific protein that binds to the poly(dG) region of the chicken β-globin gene promoter. Genes & Dev. 1988;2:863–873. doi: 10.1101/gad.2.7.863. [DOI] [PubMed] [Google Scholar]
- Litt M.D., Simpson M., Recillas-Targa F., Prioleau M.N., Felsenfeld G. Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. EMBO J. 2001;20:2224–2235. doi: 10.1093/emboj/20.9.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda T.B., Jones P.A. DNA methylation: The nuts and bolts of repression. J. Cell. Physiol. 2007;213:384–390. doi: 10.1002/jcp.21224. [DOI] [PubMed] [Google Scholar]
- Miyashita H., Kanemura M., Yamazaki T., Abe M., Sato Y. Vascular endothelial zinc finger 1 is involved in the regulation of angiogenesis: Possible contribution of stathmin/OP18 as a downstream target gene. Arterioscler. Thromb. Vasc. Biol. 2004;24:878–884. doi: 10.1161/01.ATV.0000126373.52450.32. [DOI] [PubMed] [Google Scholar]
- Muegge K. Lsh, a guardian of heterochromatin at repeat elements. Biochem. Cell Biol. 2005;83:548–554. doi: 10.1139/o05-119. [DOI] [PubMed] [Google Scholar]
- Narayan A., Ji W., Zhang X.Y., Marrogi A., Graff J.R., Baylin S.B., Ehrlich M. Hypomethylation of pericentromeric DNA in breast adenocarcinomas. Int. J. Cancer. 1998;77:833–838. doi: 10.1002/(sici)1097-0215(19980911)77:6<833::aid-ijc6>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Okano M., Bell D.W., Haber D.A., Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Ooi S.K., Qiu C., Bernstein E., Li K., Jia D., Yang Z., Erdjument-Bromage H., Tempst P., Lin S.P., Allis C.D., et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostler K.R., Davis E.M., Payne S.L., Gosalia B.B., Exposito-Cespedes J., Le Beau M.M., Godley L.A. Cancer cells express aberrant DNMT3B transcripts encoding truncated proteins. Oncogene. 2007;26:5553–5563. doi: 10.1038/sj.onc.1210351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recillas-Targa F., Pikaart M.J., Burgess-Beusse B., Bell A.C., Litt M.D., West A.G., Gaszner M., Felsenfeld G. Position-effect protection and enhancer blocking by the chicken β-globin insulator are separable activities. Proc. Natl. Acad. Sci. 2002;99:6883–6888. doi: 10.1073/pnas.102179399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson K.D., Uzvolgyi E., Liang G., Talmadge C., Sumegi J., Gonzales F.A., Jones P.A. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: Coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999;27:2291–2298. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y., Kanai Y., Sakamoto M., Saito H., Ishii H., Hirohashi S. Overexpression of a splice variant of DNA methyltransferase 3b, DNMT3b4, associated with DNA hypomethylation on pericentromeric satellite regions during human hepatocarcinogenesis. Proc. Natl. Acad. Sci. 2002;99:10060–10065. doi: 10.1073/pnas.152121799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz W.A., Elo J.P., Florl A.R., Pennanen S., Santourlidis S., Engers R., Buchardt M., Seifert H.H., Visakorpi T. Genomewide DNA hypomethylation is associated with alterations on chromosome 8 in prostate carcinoma. Genes Chromosomes Cancer. 2002;35:58–65. doi: 10.1002/gcc.10092. [DOI] [PubMed] [Google Scholar]
- Sharif J., Muto M., Takebayashi S., Suetake I., Iwamatsu A., Endo T.A., Shinga J., Mizutani-Koseki Y., Toyoda T., Okamura K., et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- Singer-Sam J., Grant M., LeBon J.M., Okuyama K., Chapman V., Monk M., Riggs A.D. Use of a HpaII-polymerase chain reaction assay to study DNA methylation in the Pgk-1 CpG island of mouse embryos at the time of X-chromosome inactivation. Mol. Cell. Biol. 1990;10:4987–4989. doi: 10.1128/mcb.10.9.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoger R., Kubicka P., Liu C.G., Kafri T., Razin A., Cedar H., Barlow D.P. Maternal-specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell. 1993;73:61–71. doi: 10.1016/0092-8674(93)90160-r. [DOI] [PubMed] [Google Scholar]
- Tada M., Tada T., Lefebvre L., Barton S.C., Surani M.A. Embryonic germ cells induce epigenetic reprogramming of somatic nucleus in hybrid cells. EMBO J. 1997;16:6510–6520. doi: 10.1093/emboj/16.21.6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda H., Takarabe T., Kanai Y., Fukutomi T., Hirohashi S. Correlation of DNA hypomethylation at pericentromeric heterochromatin regions of chromosomes 16 and 1 with histological features and chromosomal abnormalities of human breast carcinomas. Am. J. Pathol. 2002;161:859–866. doi: 10.1016/S0002-9440(10)64246-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher V.G., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda Y., Okano M., Williams C., Chen T., Georgopoulos K., Li E. Roles for Dnmt3b in mammalian development: A mouse model for the ICF syndrome. Development. 2006;133:1183–1192. doi: 10.1242/dev.02293. [DOI] [PubMed] [Google Scholar]
- Warnecke P.M., Stirzaker C., Melki J.R., Millar D.S., Paul C.L., Clark S.J. Detection and measurement of PCR bias in quantitative methylation analysis of bisulphite-treated DNA. Nucleic Acids Res. 1997;25:4422–4426. doi: 10.1093/nar/25.21.4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M., Davies J.J., Wittig D., Oakeley E.J., Haase M., Lam W.L., Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Weber M., Hellmann I., Stadler M.B., Ramos L., Paabo S., Rebhan M., Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- Weisenberger D.J., Velicescu M., Cheng J.C., Gonzales F.A., Liang G., Jones P.A. Role of the DNA methyltransferase variant DNMT3b3 in DNA methylation. Mol. Cancer Res. 2004;2:62–72. [PubMed] [Google Scholar]
- Wong N., Lam W.C., Lai P.B., Pang E., Lau W.Y., Johnson P.J. Hypomethylation of chromosome 1 heterochromatin DNA correlates with q-arm copy gain in human hepatocellular carcinoma. Am. J. Pathol. 2001;159:465–471. doi: 10.1016/S0002-9440(10)61718-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J.W., Leahy A., Lee H.H., Stuhlmann H. Vezf1: A Zn finger transcription factor restricted to endothelial cells and their precursors. Dev. Biol. 1999;206:123–141. doi: 10.1006/dbio.1998.9144. [DOI] [PubMed] [Google Scholar]
- Xu G.L., Bestor T.H., Bourc’his D., Hsieh C.L., Tommerup N., Bugge M., Hulten M., Qu X., Russo J.J., Viegas-Pequignot E. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402:187–191. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- Zhu H., Geiman T.M., Xi S., Jiang Q., Schmidtmann A., Chen T., Li E., Muegge K. Lsh is involved in de novo methylation of DNA. EMBO J. 2006;25:335–345. doi: 10.1038/sj.emboj.7600925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D. The human promoter methylome. Nat. Genet. 2007;39:442–443. doi: 10.1038/ng0407-442. [DOI] [PubMed] [Google Scholar]