Abstract
Our model of the native fatty acid synthase (FAS) depicts it as a dimer of two identical multifunctional proteins (Mr ≈ 272,000) arranged in an antiparallel configuration so that the active Cys-SH of the β-ketoacyl synthase of one subunit (where the acyl group is attached) is juxtaposed within 2 Å of the pantetheinyl-SH of the second subunit (where the malonyl group is bound). This arrangement generates two active centers for fatty acid synthesis and predicts that if we have two appropriate halves of the monomer, we should be able to reconstitute an active fatty acid-synthesizing site. We cloned, expressed, and purified catalytically active thioredoxin (TRX) fusion proteins of the NH2-terminal half of the human FAS subunit protein (TRX-hFAS-dI; residues 1–1,297; Mr ≈ 166) and of the C-terminal half (TRX-hFAS-dII-III; residues 1,296–2,504; Mr ≈ 155). Adding equivalent amounts of TRX-hFAS-dI and TRX-hFAS-dII-III to a reaction mixture containing acetyl-CoA, malonyl-CoA, and NADPH resulted in the synthesis of long-chain fatty acids. The rate of synthesis was dependent upon the presence of both recombinant proteins and reached a constant level when they were present in equivalent amounts, indicating that the reconstitution of an active fatty acid-synthesizing site required the presence of every partial activity associated with the subunit protein. Analyses of the product acids revealed myristate to be the most abundant with small amounts of palmitate and stearate, possibly because of the way the fused recombinant proteins interacted with each other so that the thioesterase hydrolyzed the acyl group in its myristoyl state. The successful reconstitution of the human FAS activity from its domain I and domains II and III fully supports our model for the structure–function relationship of FAS in animal tissues.
Keywords: subunit organization, active site recognition
The human fatty acid synthase (FAS; EC 2.3.1.85), like other animal synthases, catalyzes the synthesis of long-chain fatty acids from acetyl-CoA, malonyl-CoA, and NADPH. The native enzyme has a molecular weight of 544,000 and is composed of two identical subunit proteins (1, 2). Each subunit protein is multifunctional and contains the seven catalytic activities and a site for attachment of the prosthetic group, 4′-phosphopantetheine (acyl carrier protein). Earlier electron microscopic studies of the subunits of rat liver FAS (3) and guinea pig Harderian gland FAS (4) indicated that the subunit consists of a rod-like linear structure. Further refinement of the structure was achieved by small-angle neutron-scattering studies of the chicken liver FAS that showed that FAS has an overall appearance of two side-by-side cylinders with an overall dimension of 160 × 146 × 73 Å, with each cylinder being 160 Å in length and 73 Å in diameter (5). Moreover, each subunit cylinder is divided into three domains (I, II, and III) that are 32 Å, 82 Å, and 46 Å in length, respectively. Detailed proteolytic mapping of the chicken liver FAS by using various proteases and identification of the partial activities led us to propose the following order of the catalytic subdomains from the amino to the carboxyl termini of the multifunctional polypeptide: β-ketoacyl synthase (ketosynthase), acetyl-CoA and malonyl-CoA transacylases (AT/MT), β-hydroxyacyl dehydratase (dehydratase), enoyl reductase, β-ketoacyl reductase (ketoreductase), acyl carrier protein (ACP), and thioesterase (TE) (6–10). Based on the tryptic digestion of the FAS subunits, the partial activities were grouped into three domains: domain I (ketosynthase, AT/MT), domain II (dehydratase, enoyl reductase, ketoreductase, ACP), and domain III (TE). This order was later confirmed by comparisons of the amino acid sequences of the various animal FASs that had been deduced from the nucleotide sequences of the cDNA encoding the corresponding subunits (11–16). Moreover, the amino acid sequences of the N-terminal end of the major kallikrein cleavage peptides were determined and compared with the predicted amino acid sequences of the FAS subunit, thus establishing the exact locations of the partial activities along the FAS subunit (16). Further refinement of the order of the partial activities has recently been achieved by cloning and expressing the human FAS and its subdomains in Escherichia coli (17). Based on these results and others (16–18), the dehydratase was found to be located in domain I together with the ketosynthase and the transacylases (AT/MT).
Physicochemical studies, including active enzyme centrifugation (3), of rat liver FAS demonstrated a requirement of the dimeric form of the synthase for fatty acid synthesis. The monomeric form of the enzyme tested positively for all the partial activities except that of the ketosynthase, the catalytic activity required for the condensation of the acyl with the malonyl group to affect chain elongation. The reason for this requirement was investigated by Stoops and Wakil (19–21), who found that the two FAS subunits are arranged in a head-to-tail fashion such that the active Cys-SH of the ketosynthase partial domain of one subunit is juxtaposed within 2 Å of the pantetheine-SH of the second subunit. In this arrangement, the acetyl-S-Cys- of the ketosynthase domain of one subunit is within the atomic distance needed to react with the malonyl-S-pantetheinyl derivative to spontaneously release CO2 and form the β-ketoacyl-S-pantetheinyl derivative. The latter acyl derivative is then sequentially reduced, dehydrated, and reduced again to form the saturated acyl derivative, which is then transferred to the Cys-SH of the ketosynthase for further elongation (2). Based on these results, a functional map and an organizational model for animal FAS was proposed in which the two subunits depicted in domains I, II, and III were arranged in an antiparallel fashion, thereby generating two sites for palmitate synthesis (2, 9, 10). Evidence presented by Singh et al. (22) indicated that both sites are active in palmitate synthesis. Recently, Chirala et al. (16) determined the boundaries between the various partial activities and their most probable location on the polypeptide subunits and showed the presence of an interdomain peptide containing more than 600 amino acids that span the central region of the FAS subunit. The role of this interdomain peptide in the structure–function relationship of the synthase is not clear at this time, but the peptide is presumed to be involved in the association and maintenance of the dimer. In the human FAS cDNA, there exists a BamHI site in the interdomain region (nucleotide 3,886) that was conveniently used to cut the cDNA molecule in half (1, 17). The 5′-half of the cDNA encodes the ketosynthase, AT/MT, the dehydratase, and about half of the interdomain region (domain I), whereas the 3′-half of the cDNA encodes the second half of the interdomain, the enoyl reductase, the ketoreductase, ACP, and TE (domains II and III) (16). We recently cloned, using this BamHI site, and expressed human FAS domain I in E. coli as a fusion protein with maltose-binding protein and reported the expression and purification of human FAS (hFAS) domains II and III as an N-terminal fusion protein with thioredoxin-6His (TRX-hFAS-dII-III) (17). In this report, we describe the cloning of human FAS domain I as a TRX-6His fusion protein (TRX-hFAS-dI), the expression of the protein in E. coli, and purification of the protein from the soluble extracts. In addition, we report the reconstitution of the FAS activity by assembling the two recombinant human FAS proteins (TRX-hFAS-dI and TRX-hFAS-dII-III).
MATERIALS AND METHODS
Materials.
The E. coli strains and the plasmid vectors used were as described (17). All the reagents and chemicals were purchased from the vendors reported earlier (1, 17).
Enzyme Assay.
The FAS activity was assayed spectrophotometrically by measuring the rate of oxidation of NADPH or by the incorporation of radiolabeled acetyl-CoA or malonyl-CoA into long-chain fatty acids (23). The radiolabeled fatty acid products derived from [1-14C]acetyl-CoA or [2-14C]malonyl-CoA were extracted, and the radioactivity was measured as discussed (1). The fatty acids were further analyzed as phenacyl ester derivatives by HPLC as described (17). The partial activities of FAS were assayed as described (1).
Expression and Purification of Recombinant TRX-hFAS-dI and TRX-hFAS-dII-III Proteins.
The construction of the TRX-hFAS-dI expression plasmid by using pET-32b(+) is described in Results and Discussion. The recombinant TRX-hFAS-dI and TRX-hFAS-dII-III proteins were expressed in E. coli BL21 (DE3) cells and were partially purified by affinity chromatography using TALON resin as described (17).
RESULTS AND DISCUSSION
Construction, Expression, and Purification of hFAS-dI as a TRX-6His Fusion Protein (TRX-hFAS-dI).
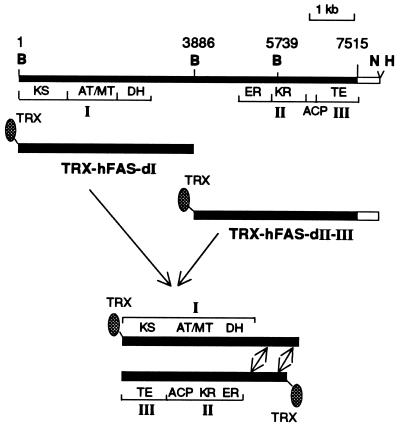
Recently, we described (17) the successful expression of human FAS domain I as a fusion protein with maltose-binding protein (pMALc2-hFAS-dI) and human FAS domains II and III as a TRX-6His fusion protein (TRX-hFAS-dII-III). In our experience, TRX fusion proteins give consistently better yields and higher levels of purity (16, 17). Consequently, we cloned human FAS domain I in pET-32b(+) vector so that we could express this domain also as a TRX fusion protein and express and purify both halves of the hFAS subunit in a similar manner. The hFAS domain I expression plasmid was constructed by first cloning the 8.3-kb EcoRI–NotI fragment from clone C6 (17) into the pFASTBAC1-generating clone pFASTBAC2, which was then used to isolate the 3.9-kb BamHI fragment for cloning into pET-32b(+) vector. Expression of the resulting clone was expected to yield a 166-kDa recombinant protein, TRX-hFAS-dI (Fig. 1).
Figure 1.
Schematic representation of the construction of TRX-hFAS-dI and TRX-hFAS-dII-III expression clones and the in vitro assembly of both recombinant proteins. The top bar represents the full-length human FAS cDNA in pFastBac1 vector. The arrows indicate the putative interaction between the two interdomains. B, BamHI; N, NotI; H, HindIII; KS, β-ketoacyl synthase; DH, β-hydroxyacyl dehydratase; ER, enoyl reductase; KR, β-ketoacyl reductase.
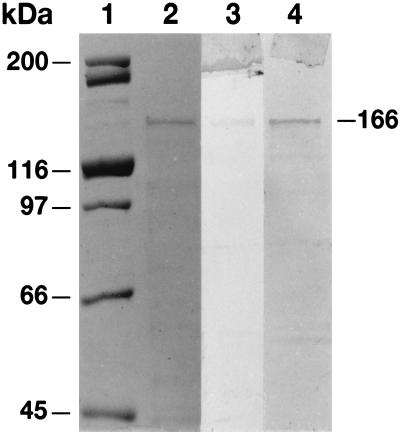
The recombinant TRX-hFAS-dI protein was purified from the crude soluble extracts of E. coli by affinity chromatography using TALON resin in a procedure similar to that reported earlier for TRX-hFAS-dII-III (17). Relative to dehydratase activity, the activities of the transacylases (AT/MT) were high in the crude extracts because of endogenous E. coli transacylases (24, 25). However, the affinity chromatography step removed the E. coli enzymes and yielded a preparation that had high specific activities for not only the transacylases but also the dehydratase (Table 1). As judged by SDS/PAGE analysis, the preparation of the recombinant TRX-hFAS-dI protein exhibited a single band with an estimated molecular mass of 166 kDa (Fig. 2, lane 2). Moreover, the protein band reacted positively with anti-human FAS antibodies (Fig. 2, lane 3) and S-protein alkaline phosphatase conjugate (Fig. 2, lane 4) in Western blot analysis.
Table 1.
Purification of recombinant TRX-hFAS-dI from E. coli
| Step | Total protein, mg | Specific activities, nmol/min/mg of protein
|
||
|---|---|---|---|---|
| AT | MT | DH | ||
| Cell-free supernatant | 110.0 | 42.5 | 60.8 | 0.38 |
| TALON column | 0.8 | 1,811.0 | 748.5 | 50.80 |
DH, β-hydroxyacyl dehydratase.
Figure 2.
SDS/PAGE and Western blot analyses of partially purified recombinant TRX-hFAS-dI. Protein standards of the indicated masses (lane 1) and of the TRX-hFAS-dI (5 μg) (lane 2) were subjected to SDS/PAGE analysis (8%), and the gel was then stained with Coomassie blue (lanes 1 and 2) or subjected to Western blot analysis by using anti-human FAS antibodies (lane 3) or S-protein alkaline phosphatase conjugate (lane 4).
As reported earlier (17), the partially purified TRX-hFAS-dII-III protein had the expected molecular mass of 155 kDa and reacted positively in Western blot analyses with anti-hFAS antibodies and S-protein alkaline phosphatase conjugate. The preparation had the expected partial catalytic activities of FAS, namely the enoyl reductase, ketoreductase, and TE with specific activities of 95, 2,750, and 51 nmol of substrate utilized/min/mg of protein, respectively (17).
Reconstitution of FAS Activity from TRX-hFAS-dI and TRX-hFAS-dII-III.
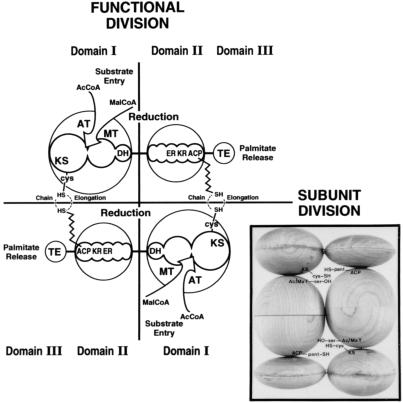
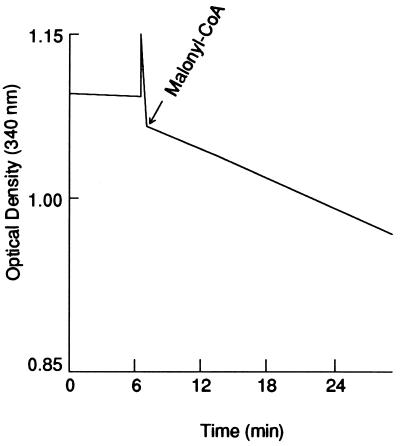
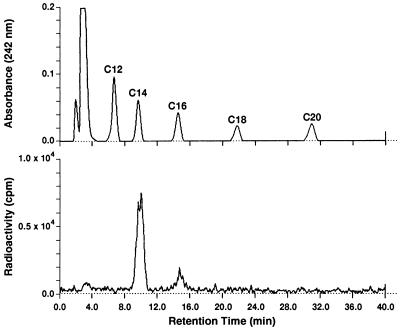
As stated earlier, the antiparallel arrangement of the two identical multifunctional subunit proteins generates two active sites for palmitate synthesis (Fig. 3). Each site contains the requisite seven partial activities and the 4′-phosphopantetheine prosthetic group and are separated by about 660 amino acids of the interdomain region of both subunits (16) (Figs. 1 and 3). As predicted from our earlier model of FAS (2), which is shown in Fig. 3, an active site for palmitate synthesis could be constituted from the N-terminal and C-terminal halves of the FAS subunits. The way in which human FAS domain I and human FAS domains II and III were expressed, the C-terminal sequences of the domain I protein contain 340 amino acids of the N-terminal half of the interdomain region, and the N-terminal end of the domain II-III protein contains the remaining 320 amino acids of the interdomain peptide region. Consequently, we predicted that mixing these two purified proteins could reconstitute an active site of FAS (Figs. 1 and 3) through possible interaction of the interdomain and the active site components of these proteins. As shown in Fig. 4, mixing the N-terminal half (TRX-hFAS-dI) with the C-terminal half (TRX-hFAS-dII-III) of the FAS subunit and adding it to a reaction mixture containing NADPH, acetyl-CoA, and malonyl-CoA resulted in rapid oxidation of NADPH. When [1-14C]acetyl-CoA or [2-14C]malonyl-CoA was present in the reaction mixture, the fatty acid products were radiolabeled. The absence of either domain I or domain II-III from the reaction mixture did not result in oxidation of NADPH nor did it lead to the formation of radiolabeled fatty acids. In a typical reaction carried out at 25°C for 30 min, the presence of 0.3 nmol each of TRX-hFAS-dI and TRX-hFAS-dII-III resulted in the oxidation of 16.5 nmol of NADPH and the incorporation of 1.2 nmol of acetyl-CoA and 8.8 nmol of malonyl-CoA into fatty acids, thus giving a stoichiometric relationship between the three substrates (acetyl-CoA/malonyl-CoA/NADPH) of 1.0:7.3:13.8, respectively, a relationship that is close to the expected ratio for palmitic acid synthesis (2). Chromatographic analysis of the phenacyl esters of the radiolabeled fatty acids showed myristic acid to be the major fatty acid produced by the reconstituted hFAS with palmitic and stearic acids being minor components (Fig. 5). The formation of myristic acid as the major fatty acid over palmitic acid in this reconstituted system may be caused by the increased access of the TE partial activity to the fatty acyl chains bound to ACP, resulting in premature hydrolysis of the myristoyl group before its elongation into the palmitoyl derivative. One reason for this increased access of the acyl group to TE could be the relaxed or relatively loose association of the domains, which might occur because of the shortening of the interdomain component of the FAS complex. Another reason may be the presence of the TRX polypeptide at the N terminus of human FAS domain I (Fig. 1). Both conditions might have caused the premature release of the fatty acyl derivatives when they reached the C14 chain length, which is the shortest acyl chain length group that the TE is able to hydrolyze (26).
Figure 3.
Functional map and organization of the subunit proteins of FAS. The two subunits with their domains I, II, and III are drawn in an antiparallel arrangement (subunit division) so that two sites of palmitate synthesis are constructed (functional division). KS, β-ketoacyl synthase; DH, β-hydroxyacyl dehydratase; ER, enoyl reductase; KR, β-ketoacyl reductase. The wavy line represents the 4′-phosphopantetheine prosthetic group of ACP. (Inset) Model of chicken liver FAS based on small-angle neutron-scattering studies (2, 5). [This figure is adapted and the Inset is reprinted with permission from S.J.W. (2), Copyright 1989, American Chemical Society.]
Figure 4.
NADPH oxidation by reconstituted hFAS. The reaction mixture (1 ml) contained potassium phosphate (50 mM; pH 6.5), DTT (10 mM), NADPH (300 μM), and acetyl-CoA (50 μM). At time 0, a mixture of 50 μg (≈0.3 nmol) each of TRX-hFAS-dI and TRX-hFAS-dII-III was added, and the absorbance at 340 nm was recorded. At the indicated times, 100 μM of [2-14C]malonyl-CoA (2.5 × 105 cpm) was added, and the measurement was continued for 30 min. At that time, the reaction was stopped by adding 50 μl of NaOH (3 M). The mixture was then acidified by adding H2SO4, and the fatty acids were extracted in pentane and counted (22).
Figure 5.
Analysis of phenacyl esters of radiolabeled fatty acids produced by the reconstituted FAS. The reaction was carried out as described in the legend of Fig. 4 by using [2-14C]malonyl-CoA. The extracted fatty acids were derivatized with 2-bromoacetophenone, subjected to HPLC analysis by using a Vydac C18 column, and eluted with a linear gradient of acetonitrile as described (17).
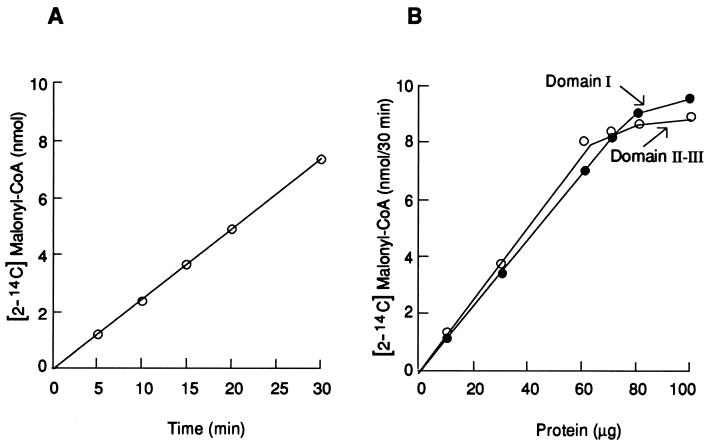
The synthesis of fatty acids by the reconstituted functional site was proportional to the time of incubation, as demonstrated by the rate of oxidation of NADPH (Fig. 4) and by the incorporation of [2-14C]malonyl-CoA into fatty acids (Fig. 6A). Moreover, when the amount of TRX-hFAS-dI was constant and an increasing amount of TRX-hFAS-dII-III was added, the rate of fatty acid synthesis, as measured by the incorporation of [2-14C]malonyl-CoA within 30 min, was proportional to the amount of TRX-hFAS-dII-III added (Fig. 6B). Conversely, if the amount of TRX-hFAS-dII-III remained constant and an increasing amount of TRX-hFAS-dI was added, there was a proportional increase in the rate of fatty acid synthesis that plateaued when the amount of TRX-hFAS-dI approximately equaled that of TRX-hFAS-dII-III (Fig. 6B). Because the partial purities of the two recombinant domain proteins are nearly the same, as are their molecular weights, nearly equal amounts of each component were needed to reconstitute the active fatty acid-synthesizing site and, therefore, give a constant rate of synthesis. This observation was expected based on our model of FAS as depicted in Fig. 3 (2, 20–22).
Figure 6.
Kinetics of the reconstituted system and its dependence on the two halves of the FAS subunit. (A) Time course of the incorporation of [2-14C]malonyl-CoA into fatty acids. Each reaction mixture contained the same components as described in the legend of Fig. 4 plus 100 nmol of [2-14C]malonyl-CoA (2.5 × 105 cpm). The reaction was started by adding a premixed solution of 0.3 nmol each of TRX-hFAS-dI and TRX-hFAS-dII-III, and the mixture was incubated at 25°C. At the indicated times, the reaction was stopped by adding NaOH, and the fatty acids were extracted and counted as described in the legend of Fig. 4. (B) Dependence of fatty acid synthesis on the presence of both TRX-hFAS-dI and TRX-hFAS-dII-III. Each reaction mixture contained the same components as described in the legend of Fig. 4 plus 100 nmol of [2-14C]malonyl-CoA (2.5 × 105 cpm). In the experiment labeled Domain I, 50 μg (≈0.3 nmol) of TRX-hFAS-dII-III and increasing amounts of TRX-hFAS-dI protein were added to each reaction mixture. In the experiment labeled Domains II and III, 50 μg (≈0.3 nmol) of TRX-hFAS-dI and increasing amounts of TRX-hFAS-dII-III protein were added to the reaction mixture. The tubes were incubated at 25°C for 30 min. The reaction was stopped by adding NaOH, and the fatty acids were extracted and counted as described in the legend to Fig. 4.
The ketosynthase component of TRX-hFAS-dI has an active Cys-SH to which the acyl group is attached prior to its elongation by the C2 unit derived from the malonyl-S-pantetheinyl derivative of the ACP component of TRX-hFAS-dII-III (2, 19–21). As expected (3), treating domain I with iodoacetamide caused alkylation of the Cys-SH of the ketosynthase component, thereby reducing fatty acid synthesis (Table 2). When the native FAS was treated with the bifunctional reagent 1,3-dibromo-2-propanone, FAS activity was inhibited because of alkylation of the sulfhydryl groups of the -SH groups at the ketosynthase sites of the FAS subunits; these thiol groups were identified as those of Cys-SH and pantetheine-SH (19, 21). Furthermore, the reaction of these two thiol components with 1,3-dibromo-2-propanone resulted in cross-linking of the two subunits of the FAS dimer. These studies and others (18–20) led us to conclude that the two FAS subunits are arranged antiparallel to each other so that the pantetheine residue of the ACP of one subunit is juxtaposed opposite a Cys-SH of the ketosynthase of the second subunit, forming in the active FAS homodimer two such sites for fatty acid synthesis. Treating the reconstituted fatty acid-synthesizing complex with 1,3-dibromo-2-propanone inhibited fatty acid synthesis (Table 2) by cross-linking the Cys-SH of the ketosynthase partial activity of domain I with the cysteamine-SH of the 4′-phosphopantetheine prosthetic group of the ACP component of domains II and III resulted in the formation of a protein with a molecular mass of 300 kDa (data not shown).
Table 2.
Effects of iodoacetamide and 1,3-dibromo-2-propanone on fatty acid synthesis by the reconstituted system
| Alkylating agent | Incorporation of [2-14C]malonyl-CoA into fatty acids, nmol
|
|
|---|---|---|
| Without treatment | With treatment | |
| Iodoacetamide* | 7.8 | 2.1 |
| 1,3-Dibromo-2-propanone† | 5.2 | 0.1 |
TRX-hFAS-dI (50 μg) was treated with iodoacetamide (0.5 mM) for 60 min at 0°C and then was added to a reaction mixture containing K phosphate (50 mM; pH 6.8), DTT (10 mM), NADPH (150 μM), acetyl-CoA (50 μM), [2-14C]malonyl-CoA (75 μM; 2 × 105 cpm), and TRX-hFAS-dII-III (50 μg) in a total volume of 1 ml. The reaction mixture was incubated for 30 min at 25°C. The radiolabeled fatty acids were extracted and counted as described in the legend of Fig. 4.
TRX-hFAS-dI (0.3 nmol) and TRX-hFAS-dII-III (0.3 nmol) were mixed in 100 μl of K phosphate (pH 6.8) and EDTA (1 mM), and 1 μmol of dibromopropanone was then added. The mixture was incubated at 0°C for 10 min and was then tested for FAS in a reaction mixture similar to the one used for the iodoacetamide-treated protein (see above).
In conclusion, our model of the animal (including human) FAS (Fig. 3) that was based on biochemical analyses, functional mapping, and neutron-scattering studies is supported by the data presented in this report. In this model, the head-to-tail arrangement of the two subunits generates two sites for long-chain fatty acid synthesis. Each site consists of partial activities of one subunit and complementary activities of the second subunit. The fact the active fatty acid-synthesizing site could be reconstituted by assembling the two halves of human FAS subunit—that is, the recombinant proteins containing domain I of the N-terminal half (TRX-hFAS-dI) and domains II and III of the C-terminal half (TRX-hFAS-dII-III), fully supports this model.
Acknowledgments
We thank Pamela Paradis Tice, ELS, for editing the manuscript. This work was supported by a grant from the National Institutes of Health (GM-19091).
ABBREVIATIONS
- FAS
fatty acid synthase
- AT
acetyl-CoA transacylase
- MT
malonyl-CoA transacylase
- ACP
acyl carrier protein
- TE
thioesterase
- hFAS
human FAS
- dI
dII, and dIII, FAS domains I, II, and III
- TRX
thioredoxin
References
- 1.Jayakumar A, Tai M-H, Huang W-Y, Al-Feel W, Hsu M, Abu-Elheiga L, Chirala S S, Wakil S J. Proc Natl Acad Sci USA. 1995;92:8695–8699. doi: 10.1073/pnas.92.19.8695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wakil S J. Biochemistry. 1989;28:4523–4530. doi: 10.1021/bi00437a001. [DOI] [PubMed] [Google Scholar]
- 3.Stoops J K, Ross P R, Arslanian M J, Aune K C, Wakil S J, Oliver R M. J Biol Chem. 1979;254:7418–7426. [PubMed] [Google Scholar]
- 4.Kitamoto T, Nishigai M, Ikai A, Ohashi K, Seyama Y. Biochim Biophys Acta. 1985;827:164–173. doi: 10.1016/0167-4838(85)90086-x. [DOI] [PubMed] [Google Scholar]
- 5.Stoops J K, Wakil S J, Uberbacher E C, Bunick G J. J Biol Chem. 1987;262:10246–10251. [PubMed] [Google Scholar]
- 6.Mattick J S, Tsukamoto Y, Nickless J, Wakil S J. J Biol Chem. 1983;258:15291–15299. [PubMed] [Google Scholar]
- 7.Mattick J S, Nickless J, Mizugaki M, Yang C-Y, Uchiyama S, Wakil S J. J Biol Chem. 1983;258:15300–15304. [PubMed] [Google Scholar]
- 8.Wong H, Mattick J S, Wakil S J. J Biol Chem. 1983;258:15305–15311. [PubMed] [Google Scholar]
- 9.Tsukamoto Y, Wong H, Mattick J S, Wakil S J. J Biol Chem. 1983;258:15312–15322. [PubMed] [Google Scholar]
- 10.Tsukamoto Y, Wakil S J. J Biol Chem. 1988;263:16225–16229. [PubMed] [Google Scholar]
- 11.Chirala S S, Kasturi R, Pazirandeh M, Stolow D T, Huang W-Y, Wakil S J. J Biol Chem. 1989;264:3750–3757. [PubMed] [Google Scholar]
- 12.Amy C M, Witkowski A, Naggert J, Williams B, Randhawa Z, Smith S. Proc Natl Acad Sci USA. 1989;86:3114–3118. doi: 10.1073/pnas.86.9.3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kameda K, Goodridge A G. J Biol Chem. 1991;266:419–426. [PubMed] [Google Scholar]
- 14.Holzer K R, Liu W, Hammes G G. Proc Natl Acad Sci USA. 1989;86:4387–4391. doi: 10.1073/pnas.86.12.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beck K-I, Schreglmann R, Stathopulos I, Klein H, Hoch J, Schweizer M. DNA Sequence. 1992;2:359–386. doi: 10.3109/10425179209020817. [DOI] [PubMed] [Google Scholar]
- 16.Chirala S S, Huang W-Y, Jayakumar A, Sakai K, Wakil S J. Proc Natl Acad Sci USA. 1997;94:5588–5593. doi: 10.1073/pnas.94.11.5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jayakumar A, Huang W-Y, Raetz B, Chirala S S, Wakil S J. Proc Natl Acad Sci USA. 1996;93:14509–14514. doi: 10.1073/pnas.93.25.14509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joshi A K, Smith S. J Biol Chem. 1993;268:22508–22513. [PubMed] [Google Scholar]
- 19.Stoops J K, Wakil S J. J Biol Chem. 1981;256:5128–5133. [PubMed] [Google Scholar]
- 20.Stoops J K, Wakil S J. J Biol Chem. 1982;257:3230–3235. [PubMed] [Google Scholar]
- 21.Stoops J K, Wakil S J. Biochem Biophys Res Commun. 1982;104:1018–1024. doi: 10.1016/0006-291x(82)91351-1. [DOI] [PubMed] [Google Scholar]
- 22.Singh N, Wakil S J, Stoops J K. J Biol Chem. 1984;259:3605–3611. [PubMed] [Google Scholar]
- 23.Arslanian M J, Wakil S J. Methods Enzymol. 1975;35:59–65. doi: 10.1016/0076-6879(75)35138-0. [DOI] [PubMed] [Google Scholar]
- 24.Wakil S J, Stoops J K, Joshi V C. Annu Rev Biochem. 1983;52:537–579. doi: 10.1146/annurev.bi.52.070183.002541. [DOI] [PubMed] [Google Scholar]
- 25.Volpe J J, Vagelos P R. Physiol Rev. 1977;56:339–417. doi: 10.1152/physrev.1976.56.2.339. [DOI] [PubMed] [Google Scholar]
- 26.Pazirandeh M, Chirala S S, Huang W-Y, Wakil S J. J Biol Chem. 1989;264:18195–18201. [PubMed] [Google Scholar]