Abstract
During 2003, 40 carbapenem-resistant Pseudomonas aeruginosa clinical isolates collected in a Mexican tertiary-care hospital were screened for metallo-β-lactamase production. Thirteen isolates produced IMP-15, and 12 had a single pulsed-field gel electrophoresis pattern. The blaIMP-15 gene cassette was inserted in a plasmid-borne integron with a unique array of gene cassettes and was named In95.
Metallo-β-lactamase (MβL) production is an emerging mechanism of carbapenem resistance among enteric and nonfermenting gram-negative bacilli (11, 25). Five acquired MβL classes (IMP, VIM, SPM, GIM, and SIM) have been identified in various host organisms, most commonly, Pseudomonas aeruginosa, Acinetobacter species, and species of the family Enterobacteriaceae (4, 15, 17, 22, 28). MβL genetic determinants are usually associated with class 1 integron structures that may reside on mobile genetic elements, such as plasmids and transposons (10, 29).
Previous reports from the SENTRY Antimicrobial Surveillance Program have identified the SPM-1, IMP-16, VIM-2, and IMP-1 MβLs among P. aeruginosa, Acinetobacter spp., and Pseudomonas fluorescens isolates collected in South America (24). The same group has identified P. aeruginosa strains producing IMP-18 in Mexico. The gene encoding this MβL was found to be carried in a class 1 integron named In96 (7). Reports from North America are still rare; however, VIM-2, IMP-7, IMP-18, and VIM-7 (1, 14, 23) have been identified in isolates from the United States and Canada. Recently, IMP-15 was identified in Kentucky in a P. aeruginosa isolate obtained from a patient who had previously been hospitalized in Mexico (19).
In the present study, we report on the characterization of P. aeruginosa clinical isolates producing the IMP-15 MβL from a Mexican tertiary-care hospital.
(This work was presented in part at the 45th Annual Meeting of the Infectious Diseases Society of America, 2007, San Diego, CA.)
A total of 255 nonduplicate P. aeruginosa isolates recovered from clinical specimens at the Hospital Civil de Guadalajara Fray Antonio Alcalde, Jalisco, Mexico, from January to December 2003 were initially tested for their antimicrobial susceptibilities by the broth microdilution method (Dade MicroScan Inc., Sacramento, CA). Fifty-six (22%) of these isolates were resistant to carbapenems, and 40 of them were available for further characterization. These 40 isolates were tested for their antimicrobial susceptibilities by agar dilution, and the results were interpreted according to the guidelines of the Clinical and Laboratory Standards Institute (5). Their resistance profiles were as follows: imipenem, 100%; meropenem, 87%; ceftazidime, 61%; aztreonam, 24%, piperacillin, 19%; piperacillin-tazobactam, 14%, amikacin, 51%; gentamicin, 54%; and ciprofloxacin, 56%.
Molecular typing of the 40 carbapenem-resistant isolates was performed by pulsed-field gel electrophoresis (PFGE) (12, 27). Analysis of the restriction patterns showed the presence of one clone (clone A) with two subtypes that included 12 isolates. The remaining 28 isolates had unique PFGE patterns (data not shown).
The 40 carbapenem-resistant isolates were tested for MβL production by the double-disk synergy test (16) and Etest MBL (AB Biodisk, Solna, Sweden). In addition, they were tested by PCR with blaVIM- and blaIMP-specific primers (Table 1). Only 13 isolates displayed an MβL phenotype and yielded positive amplicons with the blaIMP-specific primers. Twelve of the 13 isolates belonged to clone A (Table 2). These 13 isolates were also screened for class 1 integrons by using primers targeting the 5′ and 3′ conserved sequences (CSs) (2, 18), yielding products of 5.4 and 1.4 kb. The amplification products were digested with the restriction endonucleases DraIII and HaeI and always showed identical restriction profiles (data not shown), suggesting that all isolates harbored two integrons of identical structure.
TABLE 1.
Sequences of primers used in this study
| Target | Primer name | Sequence (5′ to 3′) | Amplicon size (bp) | Annealing temp (°C) | Reference or source |
|---|---|---|---|---|---|
| blaIMP | IMP-F | GGAATAGAGTGGCTTAATTC | 275 | 58 | This study |
| IMP-R | GCCAAGCTTCTATATTTGCG | ||||
| blaVIM | VIM-F | GTGTTTGGTCGCATATCGC | 380 | 58 | This study |
| VIM-R | CGCAGCACCAGGATAGAAG | ||||
| intI1 | intl1a | CGTTCCATACAGAAGCTGG | 1 | ||
| 5′ CSb | GGCATCCAAGCAGCAAG | 60 | 17 | ||
| 3′ CSb | AAGCAGACTTGACCTGA | 17 | |||
| aadA1 | aadA1-F | ATGAGGGAAGCGGTGATCG | 792 | 60 | This study |
| aadA1-R | TTATTTGCGGACTACCTTG | ||||
| aacA4 | aacA4-F | ATGACTGAGCATGACCTTG | 508 | 56 | This study |
| aacA4-R | TGCGTGTTCGCTCGAATGCC | ||||
| aadA6 | aadA6-Fb | ATGAGTAACGCAGTACCCGC | 634 | 54 | This study |
| aadA6-Rb | CCCCAGTGGCAACGATATCC | ||||
| aacA7 | aacA7-F | ATGGATAGTTCGCCGCTCGT | 362 | 58 | This study |
| aacA7-R | GAGGCGAATTCGGTGCATCC | ||||
| qacH | qacH-F | CTGGCTCTTTCTGGCTATTG | 325 | 60 | This study |
| qacH-R | TCAATGTGCGCTGACCTTGG | ||||
| oxa2 | oxa2-F | ATGGCAATCCGAATCTTCGC | 828 | 60 | This study |
| oxa2-R | TTATCGCGCAGCGTCCGAGT | ||||
| orfD | orfD-Fb | CAGTATCTCAAACGCTGTG | 223 | 56 | This study |
| orfD-Rb | AATGTTAGAGCCAGAAGCC | ||||
| qacEΔ1 | L1a | GCCCTACACAAATTGGGAGA | 381 | 64 | 12 |
| R1a | AACACCGTCACCATGGCGCCG | 12 |
Primers used for PCR amplification and sequencing of intI1 (partial sequence) and qacEΔ1 genes of 5.4- and 1.4-kb class 1 integrons.
Primers used for PCR amplification and sequencing of the variable region of 1.4-kb class 1 integron.
TABLE 2.
Features of the blaIMP-15-producing Pseudomonas aeruginosa clinical isolates
| Isolate no. | Date of isolation (day/mo/yr) | Warda | Origin | PFGE clone | Plasmid size(s) (kb)b | MIC (μg/ml)c
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IMP | MER | CAZ | ATM | PIP | TZP | AMK | GEN | CIP | ||||||
| 4667 | 13/02/2003 | ICU | Blood | A | 30 | >128 | 128 | >128 | 32 | 64 | 64 | >128 | 128 | 32 |
| 4677 | 06/06/2003 | CVS | Blood | A | 30 | >128 | 128 | >128 | 16 | 64 | 64 | >128 | >128 | 64 |
| 4682-1 | 07/07/2003 | S | Secretion | A | 70, 30 | 128 | 128 | >128 | 32 | 64 | 64 | >128 | >128 | 32 |
| 4696 | 29/09/2003 | ICU | Catheter | A | 30 | 128 | 128 | >128 | 32 | 64 | 64 | >128 | >128 | 64 |
| 4698 | 11/10/2003 | ICU | Urine | A | 70, 30 | >128 | 128 | >128 | 16 | 64 | 64 | >128 | >128 | 64 |
| 4706 | 19/10/2003 | ICU | Urine | A | 30 | 32 | 128 | >128 | 16 | 64 | 64 | 64 | >128 | 64 |
| 4658 | 12/01/2003 | IM | Secretion | A1 | 30 | 128 | 128 | >128 | 16 | 64 | 64 | >128 | 128 | 32 |
| 4679 | 26/06/2003 | ICU | Pleural fluid | A1 | 30 | 128 | 128 | >128 | 16 | 64 | 64 | >128 | 128 | 64 |
| 4688 | 09/09/2003 | PS | Secretion | A1 | 30 | >128 | 128 | >128 | 16 | 64 | 64 | >128 | >128 | >128 |
| 4703 | 03/06/2003 | IM | Secretion | A1 | 30 | >128 | 128 | >128 | 16 | 64 | 64 | 128 | >128 | 64 |
| 4659 | 20/01/2003 | ICU | Blood | A2 | 30 | >128 | 128 | >128 | 16 | 64 | 64 | >128 | >128 | 64 |
| 4680 | 04/07/2003 | ICU | Catheter | A2 | 70, 30 | 128 | 128 | >128 | 16 | 64 | 64 | >128 | 128 | 32 |
| 4663 | 03/02/2003 | IM | Urine | NRd | 20 | 32 | >128 | >128 | 8 | 64 | 64 | 4 | 128 | 32 |
CVS, cardiovascular surgery; ICU, intensive care unit; S, surgery; IM, internal medicine; PS, plastic surgery.
Strains 4663 and 4703 each contain two additional plasmids of 1 and 3 kb.
IMP, imipenem; MER, meropenem; CAZ, ceftazidime; ATM, aztreonam; PIP, piperacillin; TZP, piperacillin-tazobactam; AMK, amikacin; GEN, gentamicin; CIP, ciprofloxacin.
NR, nonrelated.
A representative strain (strain 4677) from clone A was selected for further characterization of the MβL gene and the class 1 integrons. The 5.4- and 1.4-kb amplicons were separated by agarose gel electrophoresis, purified, and used for reamplification by PCR. Shotgun cloning of the 5.4-kb fragment was performed with the Zero Background cloning system (Invitrogen, Carlsbad, CA), according to the manufacturer's guidelines. The genetic library was sequenced by the chain termination method with a BigDye Terminator kit (Applied Biosystems, Foster City, CA), and analyses were carried out on an ABI Prism 3100 analyzer (Applied Biosystems). A total of 96 quality DNA sequences were obtained and assembled by using Phred-Phrap-Consed software (8). Sequence analysis revealed an integron containing seven gene cassettes (Fig. 1) that carried an aminoglycoside acetyltransferase-encoding gene, aacA7 (3), in the first position. This cassette was followed by a blaIMP-15 cassette identical to that deposited in GenBank (GenBank accession number AY553333). The MβL gene cassette was located upstream of an array of gene cassettes containing qacH, aacA4, aadA1, blaOXA-2, and another copy of aadA1, which was located upstream of qacEΔ1, which is usually found in the 3′ region of class 1 integrons (Fig. 1). The integron promoter region was sequenced and showed a single promoter (Pant; −35 sequence TGGACA and −10 sequence TAAGCT) that was previously characterized as a weak promoter sequence (6). The structure of this unique integron, named In95, was confirmed by PCR with various combinations of primers (Table 1).
FIG. 1.
Schematic representation of class 1 integron-containing clinical isolate P. aeruginosa 4677. In95 carried blaIMP-15 and six additional gene cassettes. The open reading frames are indicated by arrows; the attI site is indicated, and the attC sites (59-base element) are indicated by filled rectangles.
The structure of the 1.4-kb class 1 integron was revealed by overlapping PCR amplification and sequencing (Table 1). This additional conserved integron contained two gene cassettes, aadA6 and orfD, inserted between intI1 and qacEΔ1 and was identical to In51, which has been reported in P. aeruginosa strains from China and India (9, 21).
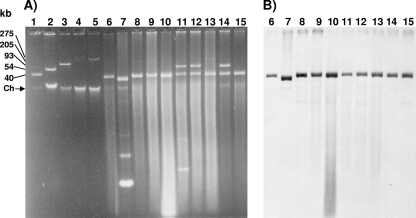
The plasmid contents of the IMP-15-producing isolates were analyzed by the method of Kieser (13). All 12 isolates of clone A harbored a plasmid of 30 kb. Three of these isolates (isolates 4680, 4682-1, and 4698) also harbored a second plasmid of 70 kb. The IMP-15-producing strain (strain 4663) showing a PFGE pattern distinct from that of clone A and carried a 20-kb plasmid and smaller plasmids (Fig. 2A; Table 2). Southern blotting with a blaIMP-15-specific DNA probe generated by PCR amplification with primers IMP-F and IMP-R (275 bp; Table 1) and labeled nonradioactively (ECL direct nucleic acid labeling and detection system; GE Healthcare, Piscataway, NJ) revealed that blaIMP-15 was carried on the 30-kb plasmid in all isolates of clone A and in the 20-kb plasmid in the genetically distinct isolate (Fig. 2B).
FIG. 2.
Plasmid profile and Southern hybridization analysis of blaIMP-15. (A) Plasmid profiles of the MβL-producing P. aeruginosa isolates. Plasmids were prepared from 10 isolates and were subjected to agarose gel electrophoresis. (B) Southern hybridization analysis. The 275-bp fragment amplified from blaIMP-15 by PCR was used as the DNA probe. Lanes: 1, plasmid R6K; 2, plasmid RP4; 3, plasmid R1; 4, pMG229; 5, pUD21; 6, strain 4658; 7, strain 4663; 8, 4667; 9, strain 4677; 10, strain 4679; 11, strain 4680; 12, strain 4682-1; 13, strain 4696; 14, strain 4698; 15, strain 4703. Ch, chromosomal DNA.
Plasmid preparations of P. aeruginosa 4677 and 4663 were transformed by electroporation into Escherichia coli DH10B and P. aeruginosa PU21 as described by Smith and Iglewski (26), and the recipient strains were plated onto LB agar supplemented with ceftazidime (1 μg/ml) or imipenem (4 μg/ml). Conjugation experiments were performed in liquid medium, as described by Miller (20). The two clinical isolates used for transformations were mated with E. coli J53-2 and P. aeruginosa PAO1, and the conjugation mixture was plated on LB plates supplemented with rifampin (100 μg/ml) and ceftazidime or imipenem at the same concentration used in the transformation experiments. Neither transfer experiment yielded colonies, suggesting that these plasmids were nontransferable under these experimental conditions.
The gene encoding IMP-15 was previously described in Thailand in a class 1 integron with a different gene cassette array (GenBank accession no. AY553333); however, the characterization of the isolates carrying blaIMP-15 was not reported in the literature. Interestingly, an IMP-15-producing P. aeruginosa isolate obtained from a patient with wound drainage was recovered at University of Kentucky HealthCare in August 2005. This patient had previously been hospitalized in Mexico (in March 2005) (19). Molecular analysis of the class 1 integron encoding blaIMP-15 (In95) from that patient showed that it was identical to the one reported in this work (19). These results suggest that the In95 class 1 integron could be broadly disseminated in Mexican hospitals.
Nucleotide sequence accession number.
The sequence of integron In95 carrying blaIMP-15 reported in this study has been deposited in the GenBank database and has been assigned accession number EF184216.
Acknowledgments
This work was supported by grant 37195-M from CONACYT and grant SALUD-2003-C01-009.
We thank T. Rojas and F. Reyna for excellent laboratory assistance. We thank Mariana Castanheira (JMI Laboratories) and Michael Dunn (Centro de Ciencias Genómicas, UNAM), Cuernavaca, Morelos, Mexico, for reviewing the manuscript.
Footnotes
Published ahead of print on 19 May 2008.
REFERENCES
- 1.Aboufaycal, H., H. S. Sader, K. Rolston, L. M. Deshpande, M. Toleman, G. Bodey, I. Raad, and R. N. Jones. 2007. blaVIM-2 and blaVIM-7 carbapenemase-producing Pseudomonas aeruginosa isolates detected in a tertiary care medical center in the United States: report from the MYSTIC program. J. Clin. Microbiol. 45:614-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adrian, P. V., C. J. Thomson, K. P. Klugman, and S. G. Amyes. 2000. New gene cassettes for trimethoprim resistance, dfr13, and streptomycin-spectinomycin resistance, aadA4, inserted on a class 1 integron. Antimicrob. Agents Chemother. 44:355-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aubert, D., D. Girlich, T. Naas, S. Nagarajan, and P. Nordmann. 2004. Functional and structural characterization of the genetic environment of an extended-spectrum beta-lactamase blaVEB gene from a Pseudomonas aeruginosa isolate obtained in India. Antimicrob. Agents Chemother. 48:3284-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castanheira, M., M. A. Toleman, R. N. Jones, F. J. Schmidt, and T. R. Walsh. 2004. Molecular characterization of a beta-lactamase gene, blaGIM-1, encoding a new subclass of metallo-beta-lactamase. Antimicrob. Agents Chemother. 48:4654-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial susceptibility testing. Document M100-S18. Clinical and Laboratory Standards Institute, Wayne, PA.
- 6.Collis, C. M., and R. M. Hall. 1995. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob. Agents Chemother. 39:155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garza-Ramos, U., P. Tinoco, J. Silva-Sanchez, R. Morfin-Otero, E. Rodriguez-Noriega, G. Leon-Garnica, H. S. Sader, and R. N. Jones. 2008. Metallo-beta-lactamase IMP-18 is located in a class 1 integron (In96) in a clinical isolate of Pseudomonas aeruginosa from Mexico. Int. J. Antimicrob. Agents 31:78-80. [DOI] [PubMed] [Google Scholar]
- 8.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 9.Gu, B., M. Tong, W. Zhao, G. Liu, M. Ning, S. Pan, and W. Zhao. 2007. Prevalence and characterization of class I integrons among Pseudomonas aeruginosa and Acinetobacter baumannii isolates from patients in Nanjing, China. J. Clin. Microbiol. 45:241-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall, R. M., C. M. Collis, M. J. Kim, S. R. Partridge, G. D. Recchia, and H. W. Stokes. 1999. Mobile gene cassettes and integrons in evolution. Ann. N. Y. Acad. Sci. 870:68-80. [DOI] [PubMed] [Google Scholar]
- 11.Hirakata, Y., K. Izumikawa, T. Yamaguchi, H. Takemura, H. Tanaka, R. Yoshida, J. Matsuda, M. Nakano, K. Tomono, S. Maesaki, M. Kaku, Y. Yamada, S. Kamihira, and S. Kohno. 1998. Rapid detection and evaluation of clinical characteristics of emerging multiple-drug-resistant gram-negative rods carrying the metallo-beta-lactamase gene blaIMP. Antimicrob. Agents Chemother. 42:2006-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaufmann, M. E. 1998. Pulse-field gel electrophoresis. Methods Mol. Med. 15:17-31. [DOI] [PubMed] [Google Scholar]
- 13.Kieser, T. 1984. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid 12:19-36. [DOI] [PubMed] [Google Scholar]
- 14.Laupland, K. B., M. D. Parkins, D. L. Church, D. B. Gregson, T. J. Louie, J. M. Conly, S. Elsayed, and J. D. Pitout. 2005. Population-based epidemiological study of infections caused by carbapenem-resistant Pseudomonas aeruginosa in the Calgary Health Region: importance of metallo-beta-lactamase (MBL)-producing strains. J. Infect. Dis. 192:1606-1612. [DOI] [PubMed] [Google Scholar]
- 15.Lauretti, L., M. L. Riccio, A. Mazzariol, G. Cornaglia, G. Amicosante, R. Fontana, and G. M. Rossolini. 1999. Cloning and characterization of blaVIM, a new integron-borne metallo-beta-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 43:1584-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, K., Y. S. Lim, D. Yong, J. H. Yum, and Y. Chong. 2003. Evaluation of the Hodge test and the imipenem-EDTA double-disk synergy test for differentiating metallo-beta-lactamase-producing isolates of Pseudomonas spp. and Acinetobacter spp. J. Clin. Microbiol. 41:4623-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, K., J. H. Yum, D. Yong, H. M. Lee, H. D. Kim, J. D. Docquier, G. M. Rossolini, and Y. Chong. 2005. Novel acquired metallo-beta-lactamase gene, blaSIM-1, in a class 1 integron from Acinetobacter baumannii clinical isolates from Korea. Antimicrob. Agents Chemother. 49:4485-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levesque, C., L. Piche, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin, C. A., K. Morita, J. A. Ribes, L. M. Deshpande, H. S. Sader, and M. Castanheira. 2008. IMP-15-producing Pseudomonas aeruginosa strain isolated in a U.S. medical center: a recent arrival from Mexico. Antimicrob. Agents Chemother. 52:2289-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller, J. 1972. Experiments in molecular genetics, p. 82-85. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Naas, T., L. Poirel, and P. Nordmann. 1999. Molecular characterisation of In51, a class 1 integron containing a novel aminoglycoside adenylyltransferase gene cassette, aadA6, in Pseudomonas aeruginosa. Biochim. Biophys. Acta 1489:445-451. [DOI] [PubMed] [Google Scholar]
- 22.Osano, E., Y. Arakawa, R. Wacharotayankun, M. Ohta, T. Horii, H. Ito, F. Yoshimura, and N. Kato. 1994. Molecular characterization of an enterobacterial metallo-beta-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob. Agents Chemother. 38:71-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pitout, J. D., B. L. Chow, D. B. Gregson, K. B. Laupland, S. Elsayed, and D. L. Church. 2007. Molecular epidemiology of metallo-beta-lactamase-producing Pseudomonas aeruginosa in the Calgary Health Region: emergence of VIM-2-producing isolates. J. Clin. Microbiol. 45:294-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sader, H. S., M. Castanheira, R. E. Mendes, M. Toleman, T. R. Walsh, and R. N. Jones. 2005. Dissemination and diversity of metallo-beta-lactamases in Latin America: report from the SENTRY Antimicrobial Surveillance Program. Int. J. Antimicrob. Agents 25:57-61. [DOI] [PubMed] [Google Scholar]
- 25.Senda, K., Y. Arakawa, K. Nakashima, H. Ito, S. Ichiyama, K. Shimokata, N. Kato, and M. Ohta. 1996. Multifocal outbreaks of metallo-beta-lactamase-producing Pseudomonas aeruginosa resistant to broad-spectrum beta-lactams, including carbapenems. Antimicrob. Agents Chemother. 40:349-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith, A. W., and B. H. Iglewski. 1989. Transformation of Pseudomonas aeruginosa by electroporation. Nucleic Acids Res. 17:10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toleman, M. A., A. M. Simm, T. A. Murphy, A. C. Gales, D. J. Biedenbach, R. N. Jones, and T. R. Walsh. 2002. Molecular characterization of SPM-1, a novel metallo-beta-lactamase isolated in Latin America: report from the SENTRY Antimicrobial Surveillance Programme. J. Antimicrob. Chemother. 50:673-679. [DOI] [PubMed] [Google Scholar]
- 29.Walsh, T. R. 2005. The emergence and implications of metallo-beta-lactamases in gram-negative bacteria. Clin. Microbiol. Infect. 11(Suppl. 6):2-9. [DOI] [PubMed] [Google Scholar]