Abstract
Campylobacter jejuni, an important food-borne human pathogen, is increasingly resistant to antimicrobials. Natural transformation is considered to be a main mechanism for mediating the transfer of genetic materials encoding antibiotic resistance determinants in C. jejuni, but direct evidence for this notion is still lacking. In this study, we determined the role of Cj1211 in natural transformation and in the development of antibiotic resistance in C. jejuni. Insertional mutagenesis of Cj1211, a Helicobacter pylori ComH3 homolog, abolished natural transformation in C. jejuni. In vitro coculture of C. jejuni strains carrying either kanamycin or tetracycline resistance markers demonstrated the development of progenies that were resistant to both antibiotics, indicating that the horizontal transfer of antibiotic resistance determinants actively occurs in mixed Campylobacter populations. A mutation of Cj1211 or the addition of DNase I in culture media completely inhibited the formation of progenies that were resistant to both antibiotics, indicating that the horizontal transfer of the resistance determinants is mediated by natural transformation. Interestingly, the mutation of Cj1211 also reduced the frequency of emergence of spontaneous mutants that were resistant to fluoroquinolone (FQ) and streptomycin but did not affect the outcome of FQ resistance development under FQ treatment, suggesting that natural transformation does not play a major role in the emergence of FQ-resistant Campylobacter strains during treatment with FQ antimicrobials. These results define Cj1211 as a competence factor in Campylobacter, prove the role of natural transformation in the horizontal transfer of antibiotic resistance determinants in Campylobacter, and provide new insights into the mechanism underlying the development of FQ-resistant Campylobacter strains.
Campylobacter jejuni is a gram-negative organism and a major food-borne human pathogen that causes more than 2 million cases of campylobacteriosis in the United States each year (38). Infection with Campylobacter often results in diarrhea, fever, and abdominal cramps. Post-Campylobacter infection is also associated with Guillain-Barré syndrome, an acute demyelinating disease of the peripheral nervous system (27, 67). Antibiotic therapy is necessary when Campylobacter infections are prolonged or severe or when they occur in immunocompromised hosts. However, Campylobacter resistance to various antibiotics is on the rise and has become a concern for public health (20, 44).
Bacteria acquire antibiotic resistance by mutations and horizontal gene transfer (9, 34, 40, 52). Mutations in the genes encoding antibiotic targets help bacteria counteract the attack by antibiotics. For example, point mutations in gyrA, rpoB, and rpsL result in resistance to fluoroquinolone, rifampin, and streptomycin, respectively (34). Horizontal gene transfer is mediated by transduction, conjugation, and natural transformation (40). Transduction is carried out by bacteriophages, while conjugation is mediated by self-replicating plasmids and requires cell-to-cell contact (18, 52). Unlike transduction and conjugation, natural transformation does not require bacteriophages or cell-to-cell contact. Instead, natural transformation involves the uptake of free DNA from the surrounding environment by some competent bacterial species and subsequent homologous recombination to incorporate the internalized DNA into the recipient chromosome (8, 15, 52). Most bacteria possess at least one of the mechanisms for the horizontal transfer of genetic materials (52).
Campylobacter is capable of conjugation and natural transformation (53, 59). Conjugative transfer of antibiotic resistance, often resistance to tetracycline, was reported in Campylobacter in culture media and in infected animals (2, 48, 53, 56). Natural transformation is well recognized in Campylobacter and has facilitated the genetic manipulation of Campylobacter strains for research purposes (24, 59, 60). The natural competence of Campylobacter is widely regarded as an important mechanism to generate genetic diversity of this bacterium by mediating horizontal gene transfer (10, 12, 17, 32, 62). It appears that both natural transformation and conjugation potentially play significant roles in the transfer of antibiotic resistance determinants in Campylobacter. Bacteriophages infecting different Campylobacter strains have been reported (1, 21, 30), but their role in mediating horizontal gene transfer in Campylobacter is unknown.
Bacterial competence for natural transformation requires the function of a series of proteins involved in DNA binding, fragmentation, degradation, and transport (8, 14). During the process of natural transformation, the transforming DNA must pass through the hydrophobic membrane barrier to enter the cytosol, and this transmembrane passage is mediated by cytoplasmic membrane channel proteins (8, 15). In both gram-negative and gram-positive bacteria, several competence proteins such as Helicobacter pylori ComE3, Neisseria gonorrhoeae ComA, Streptococcus pneumoniae CelB, Haemophilus influenzae Rec-2, and Bacillus subtilis ComEC have been reported to play a critical role in natural transformation by facilitating the transfer of extracellular DNA to the cytosol (16, 25, 41, 46, 66). In B. subtilis, ComEC functions as homodimers in the membrane and forms an aqueous channel through which transforming DNA enters the cytosol (13). In H. pylori, the ComE3 protein (a ComEC homolog) is speculated to be a membrane transporter involved in DNA transfer (66). Several competence factors in C. jejuni have been identified, including RecA, involved in DNA recombination (23); VirB10, encoded by the pVir plasmid (3, 4); ComEA (Cj0011c), involved in DNA binding (29); and several proteins of the type II secretion pathway (61) and the N-linked protein glycosylation pathway (35). In spite of these advances in our understanding of the natural transformation of Campylobacter, little is known about the detailed mechanisms involved in the uptake of DNA through the outer membrane, processing of DNA in the periplasm, and transfer of DNA from the periplasm to the cytosol. Based on the genomic sequence of C. jejuni strain NCTC 11168 (43), Cj1211 shares significant amino acid sequence homology (33% identity; E = 5e-34) with H. pylori ComE3, a putative membrane channel protein involved in the transfer of DNA (66). However, the role of Cj1211 in the natural transformation of C. jejuni is unknown. In addition, the contribution of natural transformation to the horizontal transfer of antibiotic resistance determinants and to the emergence of fluoroquinolone (FQ) resistance in C. jejuni has not been formally demonstrated. In this study, we conducted both in vitro and in vivo experiments to address these questions.
MATERIALS AND METHODS
Bacterial strains and culturing conditions.
C. jejuni NCTC 11168 and its derivatives were used in this study (Table 1). The strains were grown in Mueller-Hinton (MH) medium under microaerobic (5% O2, 10% CO2, and 85% N2) conditions at 42°C. When needed, the culture media were supplemented with kanamycin (50 μg ml−1), tetracycline (5 μg ml−1), or chloramphenicol (10 μg ml−1).
TABLE 1.
C. jejuni strains and plasmids used in this study
| C. jejuni strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| NCTC 11168 | Wild type | 43 |
| JB101 | NCTC 11168 derivative; Cj0011c::cat | 29 |
| JB201 | NCTC 11168 derivative; Cj1211::cat | This study |
| JB201C | JB201 harboring pJB1211 | This study |
| JB201P | JB201 harboring pRY108 | This study |
| JB202 | NCTC 11168 derivative; hipO::kan | This study |
| BQ304 | NCTC 11168 derivative; dcuA::tet(O) | Our laboratory |
| JB203 | NCTC 11168 derivative; Cj1211::cat hipO::kan | This study |
| JB204 | NCTC 11168 derivative; Cj1211::cat dcuA::tet(O) | This study |
| Plasmids | ||
| pUOA18 | E. coli-Campylobacter shuttle plasmid; Cmr | 58 |
| pMW10 | E. coli-Campylobacter shuttle plasmid; Kanr | 63 |
| pRY108 | E. coli-Campylobacter shuttle plasmid; Kanr | 65 |
| pJB1211 | pRY108 derivative; pRY108::Cj1211 | This study |
Construction of various mutants.
A Cj1211 mutant was constructed by inserting the cat gene into the middle of Cj1211. A 1,555-bp fragment containing Cj1211 was PCR amplified with VentR DNA polymerase (New England Biolabs, Ipswich, MA) using primers Cj1211_F and Cj1211_R (Table 2). The PCR product was cloned into SmaI-digested pUC19. pUC19 containing Cj1211 was digested with SwaI and ligated with cat, which was amplified from pUOA18 (58) with primers Cat_F and Cat_R (Table 2). The orientation of the inserted cat gene was confirmed by PCR. The plasmid was electroporated into C. jejuni NCTC 11168, and the Cj1211::cat mutant was selected on MH agar plates containing chloramphenicol (10 μg ml−1). The Cj1211 mutant was confirmed by PCR and was designated JB201 in this study (Table 1).
TABLE 2.
Primers used in this study
| Primer | Sequence (5′-3′)a |
|---|---|
| Cj1211_F | GCTGCAGGAGTTTTAAGTTCTCAAGGAAAA |
| Cj1211_R | GAGCAAATCTCCATAGTTTATAAGATGCAAAAATAA |
| Cat_F | CTAACTTCCGGTGAAGGATATCTAGAAAAA |
| Cat_R | GCCCATTCTATAGATATATGGATCCGCGCGC |
| hipO_F | CACAATATGCCTTTTGGTAGCGATAAGAAA |
| hipO_R | CGCAAGTTTACAGAATTTACAAGAACTTTTTG |
| Kan_F | CTTATCAATATATCCATGGAATGGGCAAAGCAT |
| Kan_R | GATAGAACCATGGATAATGCTAAGACAATCACTAAA |
| Cj1211_UF | CATCATTTTGCTGCTTTTCTAGATCATTGCTA (XbaI) |
| Cj1211_UR | AAAAGAGTAAGAAAAAGAATTCCATAGAGACATTAATAGCCTTTAAAAAAATATTATCTTATGTATG |
| Cj1211_DF | ATGTCTCTATGGAATTCTTTTTCTTACTCTTTT |
| Cj1211_DR | GGATTTAGAATTCTTGGATTATTTATCTTTTTGGG (EcoRI) |
| Cj1211RT_R | GATAATTCCATGGCTCAAAAATTGATTCTTGCTTAAA |
Restriction sites embedded in the primers are underlined and indicated in parentheses.
To facilitate the measurement of the horizontal transfer of antibiotic resistance determinants in Campylobacter, two C. jejuni NCTC 11168 mutants harboring different antibiotic markers were constructed. The hipO mutant was constructed by the insertion of the aphA3 gene, encoding kanamycin resistance, into hipO. For this purpose, the DNA region containing the hipO gene was amplified with Ex Taq (TaKaRa Bio Inc., Japan) using primers hipO_F and hipO_R (Table 2) and cloned into pGEM-T (Promega, Madison, WI). pGEM-T::hipO was digested with NheI and blunt ended with Klenow fragment (TaKaRa Bio Inc.). This construct was ligated with aphA3 DNA, which was amplified with VentR DNA polymerase (New England Biolabs) from pMW10 (63) by use of primers Kan_F and Kan_R (Table 2). The resulting plasmid was used as a suicide vector and introduced into C. jejuni by electroporation. The transformants were selected on MH agar plates supplemented with kanamycin (30 μg ml−1). The hipO mutant was confirmed by PCR and named JB202 (Table 1). C. jejuni strain BQ304, in which the tet(O) gene (encoding tetracycline resistance) was inserted into dcuA (B. Guo and Q. Zhang, unpublished data), was also used in this study (Table 1). The hipO mutant and the dcuA mutant were further used to generate JB203 and JB204 (Table 1). JB203 was generated by the transformation of JB202 using genomic DNA of JB201, and JB204 (Table 1) was constructed by transforming BQ304 with the genomic DNA of JB201. Detailed information on these mutants is described in Table 1. The selection of the hipO and dcuA mutants was based on three criteria. First, the inactivation of these genes must not influence the natural transformation of C. jejuni. Second, the growth of one mutant should not suppress the growth of the other in bacterial cocultures. Third, these mutants must carry different antibiotic resistance markers. The hipO mutant was previously used for a natural transformation study of Campylobacter by other investigators (10). The dcuA mutant carried an antibiotic marker different from that of the hipO mutant, and JB201 and was readily available in our laboratory. Both the hipO and dcuA mutants did not affect the natural transformation in C. jejuni (see Fig. S1 in the supplemental material) and met the criteria mentioned above.
Complementation of the Cj1211 mutant.
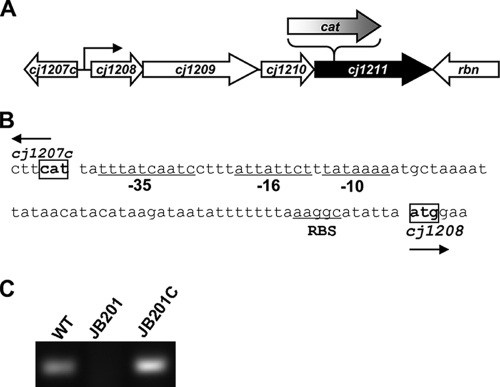
Cj1211 and its three upstream genes appear to form an operon (Fig. 1A). There is a predicted promoter in the intergenic region between Cj1207c and Cj1208 (Fig. 1B), and this promoter was fused with Cj1211 for complementation of the Cj1211 mutant. A 173-bp intergenic region between Cj1207c and Cj1208 was amplified with primers Cj1211_UF and Cj1211_UR (Table 2). The entire coding sequence of Cj1211 was PCR amplified with primers Cj1211_DF and Cj1211_DR (Table 2). Primer Cj1211_UR was designed to contain a sequence (33 bp) overlapping that of primer Cj1211_DF (Table 2). After the purification of PCR products with the QIAQuick PCR purification kit (Qiagen, Valencia, CA), these two PCR products were PCR ligated by taking advantage of the overlapped sequence as an annealing region. The ligated product was digested by XbaI and EcoRI and then cloned into pRY108, which is an Escherichia coli-C. jejuni shuttle plasmid (65). pRY108 harboring Cj1211 was transferred into JB201 cells by conjugation (39), which resulted in the creation of JB201C (Table 1). As shown in Fig. 1C, the complementation restored the expression of Cj1211 in JB201C cells. RT-PCR was performed with primers Cj1211_DF and Cj1211RT_R (Table 2).
FIG. 1.
Transcription unit of Cj1211. (A) Genomic organization of Cj1211 and its flanking genes. Open reading frames and their transcription directions are indicated by boxed arrows. The location of the cat insertion in JB201 is indicated with a bracket above Cj1211. The predicted RpoD promoter upstream of Cj1208 is indicated with a curved arrow. (B) Predicted promoter sequences upstream of Cj1208. The −10, −16, and −35 regions and the ribosomal binding site (RBS) are underlined. The start codons of Cj1208 and Cj1207c are boxed. (C) RT-PCR analysis of the transcription of Cj1211 in wild-type NCTC 11168 (WT), JB201, and JB201C.
Natural transformation.
Natural transformation was done according to a method described previously (59). The donor DNA was isolated from the Cj0011c mutant (designated JB101 in this study) and BQ304 (Table 1). Both mutant strains were readily available in our laboratory, and their antibiotic resistance markers, aphA3 and tet(O), confer resistance to kanamycin and tetracycline, respectively, making them suitable for use as donor DNA in transformation experiments. Transformants were selected on MH agar plates containing tetracycline (5 μg ml−1) or kanamycin (30 μg ml−1). The transformation frequency was defined as the number of transformants per 1 μg of DNA divided by the total bacterial number. The transformation experiment with BQ304 DNA was done twice, while the experiment with JB101 DNA was repeated three times. Each transformation experiment was performed in triplicate.
Assay of DNA binding and uptake.
DNA binding and uptake were assessed as previously described (61). Briefly, 1.2 μg of chromosomal DNA from C. jejuni NCTC 11168 was nick translated using the Roche (Basel, Switzerland) nick translation kit in the presence of 40 μCi [α-33P]dCTP as recommended by the manufacturer. The 33P-labeled DNA was washed by ethanol precipitation and resuspended in TE (10 mM Tris, 1 mM EDTA [pH 8.0]). C. jejuni strains were grown on MH agar plates overnight. C. jejuni cells were washed three times with MH broth and resuspended to an optical density at 600 nm (OD600) of 0.5. Labeled DNA (100 ng) was added to 1 ml of bacterial suspension. After microaerobic incubation at 37°C for 30 min, half of the bacterial suspension (500 μl) was treated with DNase I (100 μg ml−1) for 10 min at room temperature. The DNase I-treated and nontreated suspensions were washed three times and resuspended in MH broth. Each sample was mixed with 5 ml Ecoscint H liquid scintillation fluid (National Diagnostics, Atlanta, GA) by vigorous shaking for 15 s. Beta emission from 33P was detected by automated liquid scintillation (Beckman Coulter, Fullerton, CA). The radioactivities from DNase I-treated and nontreated samples indicate the levels of DNA uptake and binding, respectively. Triplicate treatments were prepared for each experiment, and the experiment was repeated three times.
DNA exchange in bacterial cocultures.
The coculture experiment was done to investigate the role of Cj1211 in mediating DNA transfer during bacterial growth. Cultures of C. jejuni grown overnight on MH agar plates were collected and resuspended in fresh MH broth to an OD600 of 0.05. For measuring DNA transfer in the Cj1211+ background, where C. jejuni contains the intact Cj1211, a bacterial suspension of JB202 (Kanr) was mixed with an equal volume of BQ304 (Tetr). The tubes were cultured microaerobically at 37°C with shaking (200 rpm). Samples were taken at 0, 3, 8, and 24 h and plated onto MH agar plates containing kanamycin (30 μg ml−1), tetracycline (5 μg ml−1), or both. To measure DNA transfer in the Cj1211− background, where Cj1211 was inactivated, equal volumes of JB203 (Kanr Cmr) and JB204 (Tetr Cmr) were mixed, and the mixture was cultured and sampled as described above. In some experiments, bacterial cultures were washed with MH broth prior to preparation of the mixture to remove free DNA in the inocula. This washing step was done by centrifugation (13,000 × g for 1 min) and was repeated five times. The washed bacterial cells were then mixed for the coculture experiment. To determine the involvement of free DNA in the transfer, DNase I was added to the mixed culture at a concentration of 30 μg ml−1 in some experiments. The enzyme was added at the beginning of the coculture experiment. Each experiment was performed with three technical replicates and repeated three times.
Frequency of spontaneous mutations in the Cj1211 mutant.
To determine the effect of the Cj1211 mutation on the emergence of spontaneous mutants, cultures of JB201 and wild-type NCTC 11168 grown overnight on MH agar plates were resuspended in MH broth to an OD600 of 0.07. The bacterial suspensions were incubated at 37°C with shaking (200 rpm) for 4 h (OD600 ≈ 0.3), and the bacterial suspensions were spread onto MH agar plates containing ciprofloxacin (1 μg ml−1) (4× the MIC) or streptomycin (3 μg ml−1) (3× the MIC) to assess the numbers of spontaneous mutants that were resistant to ciprofloxacin or streptomycin. Total bacterial counts were also determined by plating onto MH agar plates without antibiotics. The frequency of spontaneous mutation was calculated by dividing the bacterial count on MH agar plates containing antibiotics by the total bacterial count.
Development of FQ-resistant (FQr) mutants under in vitro antibiotic treatment.
Cultures of wild-type NCTC 11168 and JB201 grown overnight were resuspended in MH broth to an OD600 of 0.05. The bacterial suspensions were cultured microaerobically at 37°C with shaking (200 rpm). Afer 8 h of incubation, ciprofloxacin was added to the media at a final concentration of 1 μg ml−1, and the cultures were continuously incubated for another 40 h. At 0, 3, 8, 24, and 48 h after the start of the incubation, bacterial samples (100 μl each) were taken and spread onto MH agar plates containing ciprofloxacin (1 μg ml−1) to enumerate the number of FQr mutant. The total bacterial number was also counted by serial dilutions of the samples and plating onto MH agar plates without antibiotics.
Development of FQr mutants under in vivo antibiotic treatment.
Five-day-old chickens were divided into two groups, and each group consisted of 10 chickens. One group was inoculated with wild-type NCTC 11168, and the other was inoculated with JB201. Each chicken received approximately 1 × 10 6 CFU of C. jejuni, which were freshly grown in MH broth overnight. Before inoculation, the absence of Campylobacter in the chickens was confirmed by culturing cloacal swabs on MH agar plates containing Campylobacter-selective supplements (SR0232E and SR0117E; Oxoid). Six days after the inoculation, enrofloxacin treatment was initiated, and the antibiotic was given in drinking water (50 mg liter−1) for 5 days. The medicated water was prepared and replaced daily. Fecal samples were collected with cloacal swabs, diluted with MH broth, and plated onto MH agar plates containing Campylobacter-selective supplements. In addition, each sample was also plated onto MH agar plates containing ciprofloxacin (4 μg ml−1) to count FQr isolates. Representative isolates from each group were randomly selected, used in PCR to confirm the Cj1211 mutation, and tested for MICs of ciprofloxacin using the Etest (AB Biodisk, Solna, Sweden). The differences in the emergence of FQr mutants between the wild type and JB201 at each sampling time point were compared with a Student's t test.
RESULTS
Identification of Cj1211 as a potential competence protein.
According to the genomic sequence of C. jejuni NCTC 11168 (43), Cj1211 is predicted to be an integral membrane protein and shares 33% identity and 53% similarity to H. pylori ComE3. However, it has low overall homology (less than 15% identity) with other functionally defined ComE3 orthologs such as B. subtilis ComEC, N. gonorrhoeae ComA, S. pneumoniae CelB, and H. influenzae Rec-2. Despite the low overall homology with other ComE3 orthologs, the putative competence domain, which was previously described for B. subtilis ComEC (13), was relatively conserved (23% identity and 39% similarity with B. subtilis ComEC, 19% identity and 36% similarity with N. gonorrhoeae ComA, and 38% identity and 58% similarity with H. pylori ComE3) (Fig. 2). C. jejuni Cj1211 has 419 amino acids, which is similar in size to the ComE3 (417 amino acids) of H. pylori. However, other ComE3 homologs are larger, with approximately 700 amino acids. A motif search (http://smart.embl-heidelberg.de/) showed that the C-terminal regions of B. subtilis ComEC, N. gonorrhoeae ComA, and H. influenzae Rec-2 contain a lactamase B domain (7), whereas this domain was absent in C. jejuni Cj1211, H. pylori ComE3, and S. pneumoniae CelB, suggesting possible functional variations among these proteins. Cj1211 was annotated as an integral membrane protein (43). Predictions by multiple programs including PSORTb v.2.0 (http://www.psort.org/psortb/index.html), CELLO v.2.5 (http://cello.life.nctu.edu.tw/), PSLpred (http://www.imtech.res.in/raghava/pslpred/), and Gneg-PLoc (http://202.120.37.186/bioinf/Gneg/) suggested that Cj1211 is a possible inner membrane protein (data not shown). Other C. jejuni strains and Campylobacter species such as C. jejuni RM1221, C. jejuni 81116, C. coli, C. upsaliensis, C. lari, C. curvus, C. concisus, and C. fetus possess proteins homologous to Cj1211 (data not shown), suggesting that Cj1211 is highly conserved among campylobacters. Interestingly, the published genomic sequences of C. jejuni 81-176 (NCBI accession number NC_008787) and C. jejuni doylei 269.97 (accession number NC_009707) did not identify a Cj1211 homolog. However, analysis of the genomic sequences indicated that these two strains possess the DNA sequences encoding proteins homologous to Cj1211 (99% and 97% amino acid identities, respectively). The Cj1211 homologs are located between CJJ81176_1224 and yihY in C. jejuni 81-176 and between JJD26997_0518 and JJD26997_0519 in C. jejuni doylei 269.97. The reason that Cj1211 was not annotated in these two strains was unknown.
FIG. 2.
Sequence alignment of the putative competence domain of Cj1211 with other ComE3 (ComEC) homologs. B. sub, B. subtilis ComEC (NCBI accession number BAA12454.1); N. gon, N. gonorrhoeae ComA (accession number YP_207438.1); H. pyl, H. pylori ComE3 (accession number NP_208153.1); C. jej, C. jejuni Cj1211 (accession number NP_282358.1). The competence domain is based on a report by Draskovic and Dubnau (13).
The Cj1211 gene appears to be positioned in an operon with three upstream genes (Cj1208, Cj1209, and Cj1210) (Fig. 1A). Cj1208 and Cj1209 overlap by 80 nucleotides, Cj1210 and Cj1211 overlap by 1 nucleotide, and Cj1209 and Cj1210 are separated by 8 nucleotides. Cj1208 and Cj1209 encode proteins of unknown functions, while Cj1210 is a probable membrane protein belonging to the DedA protein family whose function is unknown (43). There is a putative promoter upstream of Cj1208. Based on the consensus RpoD promoter sequence of C. jejuni (63), the −10, −16, and −35 regions of the promoter were identified (Fig. 1B). The fact that this promoter was able to express Cj1211 in JB201C (Fig. 1C) indicates that it is functional in C. jejuni.
Contribution of Cj1211 to natural transformation.
The sequence similarity of Cj1211 to ComE3 suggested that Cj1211 could be a competence protein. To examine this hypothesis, a Cj1211 mutant was constructed and used for natural transformation. The inactivation of Cj1211 did not change the bacterial growth rate in MH broth (data not shown). A mutation of Cj1211 resulted in a significant reduction (P < 0.01) in the natural transformation frequency compared with that of the wild type (Table 3). The reduction was at least 2,175-fold and 832-fold when Tetr DNA and Kanr DNA were used as the donor DNAs, respectively (Table 3). In fact, no transformants were observed with JB201 under the conditions used in this study. Complementation of JB201 with pJB1211 restored the transformation frequency to the level of the wild type, while the control vector (pRY108) did not have an effect on transformation (Table 3). Together, these results clearly showed that Cj1211 plays an important role in the natural transformation of C. jejuni.
TABLE 3.
Contribution of Cj1211 to natural transformation in C. jejuni
| Source of donor DNA | Mean transformation frequency ± SDa
|
|||
|---|---|---|---|---|
| Wild type | JB201 | JB201C | JB201P | |
| BQ304b | (1.74 ± 0.76) × 10−5 | <0.8 × 10−8 | (1.41 ± 0.91) × 10−5 | <3.9 × 10−8 |
| JB101c | (9.16 ± 7.34) × 10−6 | <1.1 × 10−8 | NT | NT |
Each frequency represents the means ± standard deviations of data from triplicate transformations from a single experiment. In all experiments, differences in transformation frequencies between the wild-type strain and JB201 were consistently observed. NT, not tested due to the presence of the Kanr gene in the complementing plasmid.
Conferring tetracycline resistance.
Conferring kanamycin resistance.
Mutation of Cj1211 did not affect DNA binding and uptake.
Since the mutagenesis of Cj1211 significantly impaired the natural transformability of C. jejuni, an assay of DNA binding and uptake was performed to determine which transformation step was affected by the Cj1211 mutation. The assay was performed with 33P-labeled genomic DNA of C. jejuni NCTC 11168. Compared with the wild-type strain, JB201 did not show significant changes in DNA binding and uptake (DNA binding values of 1,470.0 ± 92.6 cpm ml−1 for the wild type and 1,699.3 ± 111.6 cpm ml−1 for JB201 and DNA uptake values of 213.3 ± 48.1 cpm ml−1 for the wild type and 220.7 ± 18.6 cpm ml−1 for JB201). This result is in contrast to the finding with ComE3 of H. pylori, in which a mutation of CmeE3 caused a significant reduction in both DNA binding and uptake (66), but is similar to observations of N. gonorrhoeae and Pseudomonas stutzeri, for which mutations of their ComE3 orthologs did not affect DNA binding and uptake (16, 22).
Role of Cj1211 in mediating horizontal transfer of antibiotic resistance determinants.
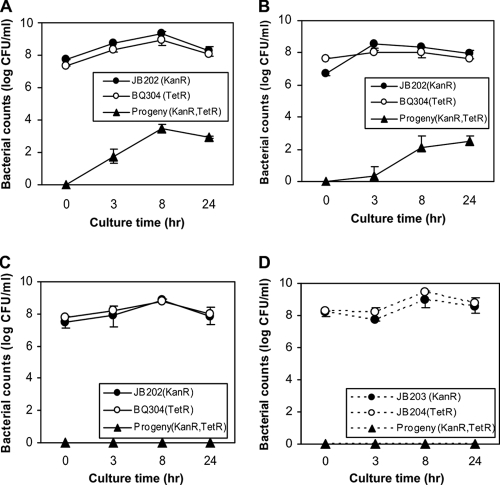
Previous studies have shown that a transfer of antibiotic resistance determinants occurred in bacterial cocultures of C. jejuni without supplementing extracellular DNA (10, 62), but the contribution of natural transformation to this process has not been formally demonstrated. Since the Cj1211 mutation abolished natural transformation in C. jejuni, we compared the transfer of antibiotic resistance markers in the Cj1211+ background to that in the Cj1211− background. The coculture of the Cj1211+ strains carrying either aphA3 (for kanamycin resistance) or tet(O) (for tetracycline resistance) generated progenies that were resistant to both antibiotics within a few hours (Fig. 3A). PCR analysis of the doubly resistant colonies (10 from each sampling at 3, 8, and 24 h) revealed that all of the tested colonies harbored the aphA3 and tet(O) genes inserted into hipO and dcuA, respectively (data not shown), confirming the horizontal transfer of the antibiotic resistance determinants in the coculture. Washing the bacterial inocula prior to coculture delayed (P = 0.00047) but did not abolish the emergence of doubly resistant progenies (Fig. 3B). The addition of DNase I into the coculture to a final concentration of 30 μg ml−1 completely prevented the appearance of doubly resistant bacterial cells (Fig. 3C). In contrast to the results obtained with the Cj1211+ strains, the coculture experiment using the Cj1211− strains (JB203 and JB204) did not result in the appearance of doubly resistant progenies (Fig. 3D). This observation was not due to changes in antimicrobial susceptibility because the Cj1211 mutation did not affect the MICs of kanamycin and tetracycline in JB203 and JB204 (data not shown). Furthermore, the acquisition of aphA3 or tet(O) rendered C. jejuni highly resistant to kanamycin (MIC > 256 μg ml−1) or tetracycline (MIC = 16 μg ml−1) in both the Cj1211+ and Cj1211− backgrounds. Together, these results strongly indicate that DNA exchange occurs actively in Campylobacter cocultures and that the transfer is mediated by free DNA and natural transformation. These findings also confirmed that Cj1211 plays a critical role in the natural transformation of C. jejuni and in the horizontal transfer of genetic materials including antibiotic resistance determinants.
FIG. 3.
Natural transformation-mediated transfer of antibiotic resistance determinants in Campylobacter cocultures. (A) Coculture of C. jejuni JB202 (Kanr) and BQ304 (Tetr) in MH broth with no antibiotics. The numbers of bacterial cells that were resistant to both kanamycin and tetracycline were determined on plates containing both antibiotics. (B) Effect of washing the inocula on the development of doubly resistant progenies. JB202 and BQ304 were washed with MH broth five times before being inoculated into the coculture. (C) Effect of DNase I treatment on the development of doubly resistant progenies. DNase I was added to the coculture (at a final concentration of 30 μg ml−1) of JB202 and BQ304. (D) Coculture of C. jejuni JB203 (Kanr) and JB204 (Tetr) in the absence of DNase I treatment. The Cj1211 gene in both strains was inactivated by insertional mutagenesis. See Table 1 for detailed information on the strains used in this experiment. The results show the means ± standard deviations of data from triplicate samples in a single experiment. Similar patterns of differences were observed in three independent experiments.
Role of Cj1211 in emergence of FQr mutants.
Campylobacter strains can develop resistance to certain antibiotics through spontaneous mutations in target genes. For instance, the resistance of Campylobacter to FQs is mediated by spontaneous mutations in GyrA, which occur at frequencies of 10−6 to 10−8 (64). However, the detected mutants could include both bona fide spontaneous mutants and transformants, because cell death naturally occurs in bacterial cultures, and the DNA released from dead FQr cells is conceivably available for the transformation of the wild-type cells. To determine if natural transformation influences the frequency of emergence of FQr mutants in Campylobacter, we compared the rates of mutant emergence in the wild type and JB201. Interestingly, JB201 exhibited a significant reduction (approximately 31-fold) in the frequency of emergence of mutants resistant to ciprofloxacin (Table 4). This reduction in mutation frequency was not due to the change in the susceptibility to ciprofloxacin because the FQr mutants from JB201 and the wild type had the same MICs, and the inactivation of Cj1211 in FQr mutants did not reduce the MIC of ciprofloxacin (data not shown). In order to confirm whether the reduced emergence of FQr mutants in JB201 was linked to natural transformation, the bacterial culture of the wild-type strain was treated with DNase I to remove free DNA. DNase I treatment has been commonly used to assess the function of natural transformation (42, 50, 51). DNase I was added to the bacterial culture to a final concentration of 100 μg ml−1, which was 3.3 times higher than the concentration used to inhibit the transfer of Tetr and Kanr by natural transformation (Fig. 3D). The DNase I treatment did not result in a significant reduction in the frequency of FQr mutants (Table 4). This result suggests that free DNA was not a major player in the emergence of FQr mutants, and the reduced emergence of FQr mutants in JB201 could not be explained by its defect in natural transformation. In addition, we examined the development of spontaneous mutants that are resistant to streptomycin, to which the resistance is associated with point mutations in the rpsL gene (34). JB201 showed approximately a 129-fold decrease in the frequency of emergence of streptomycin-resistant mutants (Table 4), suggesting that the Cj1211 mutation decreases the general spontaneous mutation rates in C. jejuni.
TABLE 4.
Effect of Cj1211 mutation and DNase I treatment on the emergence of spontaneous mutants that are resistant to FQ and streptomycin
| Type of spontaneous mutant | Mean spontaneous mutation frequency ± SDa
|
|
|---|---|---|
| Wild type | JB201 | |
| FQr mutantsb | (3.0 ± 1.5) × 10−7 | (9.8 ± 5.2) × 10−9 |
| FQr mutants with DNase I treatmentc | (1.9 ± 1.1) × 10−7 | NT |
| Strr mutantsd | (4.4 ± 3.0) × 10−6 | (3.4 ± 2.2) × 10−8 |
Means ± standard deviations of data for triplicate samples of a single experiment. NT, not tested; Str, streptomycin.
The concentration of ciprofloxacin used is 1 μg ml−1; the experiment was repeated five times.
DNase I was added to the bacterial suspension at a concentration of 100 μg ml−1; the experiment was repeated six times.
The concentration of streptomycin for plating was 3 μg ml−1; the experiment was repeated five times.
Effect of the Cj1211 mutation on development of FQr Campylobacter mutants under in vitro treatment.
Previously, it was shown that FQr Campylobacter strains rapidly developed from an FQ-susceptible population when treated with FQ antimicrobials (36, 37, 45). The selection of preexisting FQr mutants by the treatment is certainly involved in the development of resistance, but the role of natural transformation in resistance development is unknown. Since Cj1211 contributes to natural transformation and the emergence of spontaneous FQr mutants, we investigated if mutations of Cj1211 affected the development of FQr mutants when treated with ciprofloxacin. As shown in Fig. 4, the number of spontaneous FQr mutants was much lower in JB201 than that in the wild-type strain prior to exposure to ciprofloxacin. After ciprofloxacin treatment was initiated, the FQr populations from the wild type and JB201 were both increased. Although the number of FQr mutants from JB201 was lower than that from the wild-type strain during the first hours of treatment, the mutant numbers for both strains were similar after 40 h of antibiotic treatment. This result suggests that the development of FQr mutants under FQ treatment is dominated by selection and enrichment, and natural transformation does not affect the result of selection.
FIG. 4.
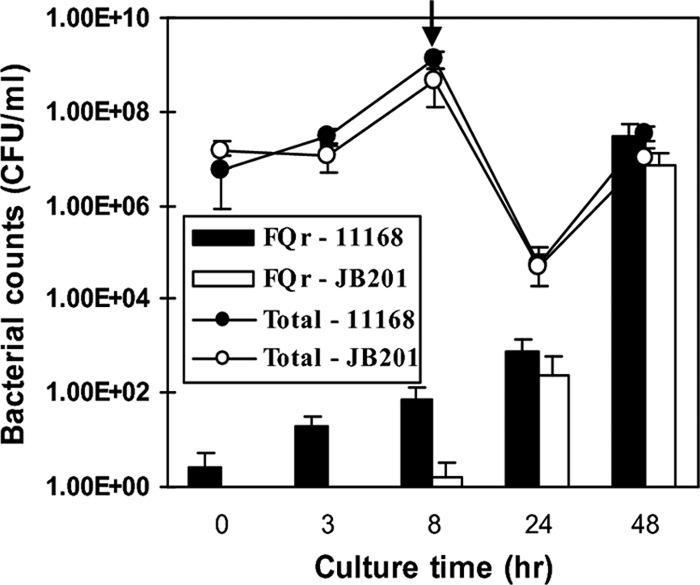
Effect of the Cj1211 mutation on the development of FQr mutants in cultures treated with ciprofloxacin. C. jejuni NCTC 11168 and JB201 were grown in separate culture tubes without ciprofloxacin for 8 h, and ciprofloxacin was then added to the cultures to a final concentration of 1 μg ml−1. The initiation time of ciprofloxacin treatment is indicated by an arrow. Open and solid circles indicate the total bacterial counts of JB201 and NCTC 11168, respectively, while the open and solid bars represented the numbers of FQr mutants of NCTC 11168 and JB201, respectively. The results show the means ± standard deviations of data for triplicate samples in a single experiment. Similar patterns of results were obtained in three independent experiments.
Effect of the Cj1211 mutation on the emergence of FQr mutants under antibiotic treatment in vivo.
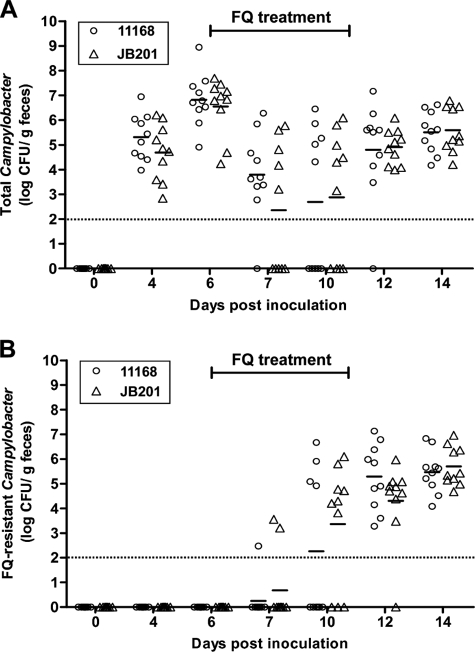
To determine the effect of the Cj1211 mutation on the emergence of FQr mutants under in vivo conditions, two groups of chickens were inoculated with the wild type or JB201 and treated with a therapeutic dose of enrofloxacin (50 mg liter−1 in drinking water). JB201 colonized chickens as efficiently as the wild type (Fig. 5A). Enrofloxacin treatment greatly decreased the colonization levels of both the wild type and JB201 but did not eliminate the two strains from the chickens (Fig. 5A). In fact, the Campylobacter populations rebounded after the initial decline. Prior to the treatment, no FQr mutants were detected in either group. One day after the initiation of antibiotic treatment, FQr mutants could be detected in a few of the treated chickens (Fig. 5B). The numbers of FQr mutants and chickens carrying FQr Campylobacter steadily increased during the treatment. At the last sampling time, all Campylobacter strains in the chickens were FQr (Fig. 5A and B). There were no significant differences (P > 0.05) between the two groups in the levels of total Campylobacter and FQr mutants. This finding indicated that the Cj1211 mutation did not affect the development of FQr mutants in chickens treated with enrofloxacin, which was consistent with the result from the in vitro study (Fig. 4). Representative Campylobacter colonies selected from each group were examined for ciprofloxacin MIC by Etest. Except for the isolates collected on day 7 (1 day after initiation of treatment), the majority of the tested isolates from the later sampling times had ciprofloxacin MICs of >32 μg ml−1, and a few had ciprofloxacin MICs of 4 μg ml−1. In addition, some isolates from the JB201-inoculated group were tested by PCR, and all of the tested isolates carried the insertional mutation in Cj1211, indicating that the mutation was stably maintained during in vivo colonization.
FIG. 5.
Effect of the Cj1211 mutation on the development of FQr mutants in chickens treated with enrofloxacin. Two groups of chickens (10 birds/group) were inoculated with wild-type NCTC 11168 (circles) and JB201 (triangles), respectively. (A) Total Campylobacter numbers detected in the chickens. (B) Numbers of FQr Campylobacter CFU detected in the chickens. The period of FQ treatment (from 6 to 11 days postinfection) is indicated with a bar above each panel. Each symbol represents the Campylobacter number in a single bird. Some symbols are superimposed. The mean of each group is indicated by a horizontal bar. The detection limit is approximately 100 CFU/g feces and is indicated by a dotted line.
DISCUSSION
This study clearly showed that Cj1211 shares significant sequence similarity to ComE3, possesses the competence domain shared by the ComEC (ComE3) homologs (Fig. 2), and is a key player in the natural transformation of C. jejuni (Table 3). Based on these findings, we consider Cj1211 to be an inner membrane transporter protein involved in the transfer of DNA across the membrane of C. jejuni. Interestingly, the mutation of Cj1211 did not yield measurable differences in DNA binding and uptake (see above). In gram-positive bacteria such as B. subtilis and S. pneumoniae, the ComEC mutation abolished DNA uptake but did not reduce DNA binding (6, 25). Instead, DNA accumulated on the bacterial surface of the ComEC mutants (6, 49). In gram-negative bacteria, the mutagenesis of ComEC (ComE3) homologs resulted in conflicting results about DNA binding and uptake. Although the H. pylori ComE3 mutant exhibited a significant reduction in both DNA binding and uptake (66), the mutation of ComE3 orthologs in N. gonorrhoeae and P. stutzeri significantly impaired the natural transformation of these bacteria without affecting DNA binding and uptake (16, 22). The discrepancy in DNA uptake between gram-negative and gram-positive bacteria may be explained by the differences in the membrane structures and transformation processes between these two groups of bacteria. In gram-positive bacteria, the mutation of ComEC would directly impair DNA uptake by blocking DNA transfer from the external space to the bacterial cytoplasm. However, in gram-negative bacteria, ComE3 homologs are inner membrane proteins, and the mutation of these proteins should still allow DNA to be transferred to the periplasmic space. Despite the inability of ComE3 mutants to transfer the transforming DNA from the periplasm to the cytosol, the total amount of DNA within bacterial cells may still be comparable to that in wild-type strains. This possibility is likely, because the DNA uptake assay measures the total radioactivity within bacterial cells without differentiating the DNA in the periplasmic space from the DNA in the cytosol. Thus, the commonly used DNA uptake method may not be sensitive enough to determine the role of ComE3 homologs in DNA internalization into the cytosol of gram-negative bacteria.
In our previous study, it was shown that C. jejuni possesses a ComEA homolog (Cj0011c), which is a periplasmic DNA receptor binding to both double-stranded and single-stranded DNA with a greater affinity for double-stranded DNA than for single-stranded DNA (29). The mutation of Cj0011c resulted in about a 10-fold reduction in the transformation frequency (29). An early study showed that the mutation of recA completely abolished the transformation of this bacterium, indicating that recA plays a critical role in the natural transformation of Campylobacter, probably by influencing the recombination step (23). Mutation of VirB10, an N-glycosylated protein encoded by the pVir plasmid, decreased the transformation frequency in C. jejuni 81-176 (4, 35). In addition, mutation of the N-linked protein glycosylation pathway resulted in a significant reduction (10,000-fold) in transformation frequency in C. jejuni (35). Recently, transposon mutagenesis identified a dozen of genes involved in natural transformation (61), several of which are involved in DNA uptake. Findings in our study identified a new competence protein that likely forms an inner membrane channel for the passage of transforming DNA into the cytosol.
Horizontal gene transfer contributes to the intercellular exchange of genetic information and is mediated by natural transformation, conjugation, and transduction (52, 55). Natural transformation has long been recognized in Campylobacter and is regarded as being a main mechanism for mediating genetic exchange and diversity in Campylobacter (10, 32, 59, 61, 62). Previously, it was shown that the exchange of chromosomally encoded antibiotic resistance determinants occurred in Campylobacter cocultures and in chicken gut colonized by Campylobacter (10, 62). Although natural transformation was suspected to be a major player in genetic exchange, its role in mediating the horizontal transfer of antibiotic resistance was not formally demonstrated. In this study, we showed the key role of natural transformation in mediating DNA transfer occurring in Campylobacter cocultures (Fig. 3). This conclusion is based on the fact that the addition of DNase I to the coculture and the mutation of Cj1211 abolished the development of doubly resistant mutants (Fig. 3C and 3D). The free DNA available for natural transformation in the cocultures was likely released from Campylobacter cells. However, it was unknown if the free DNA was from dead cells or was secreted by growing cells. Since Campylobacter is widely present in animals and birds, and mixed infection by multiple Campylobacter strains can occur in animal hosts, it is conceivable that natural transformation directly contributes to the spread of antibiotic resistance among different Campylobacter strains.
FQ antimicrobials are important for the clinical treatment of campylobacteriosis. However, Campylobacter has become increasingly resistant to FQ antimicrobials. In Campylobacter strains, resistance to FQs is conferred by gyrase mutations and the function of the multidrug efflux pump CmeABC (19, 36, 57). Campylobacter does not have ParC and ParE, and single point mutations in the gyrA gene can confer high-level resistance to FQ antimicrobials (36, 44, 45, 47, 57). FQ resistance occurs at a rate as high as 10−6 from an FQ-susceptible population (64), but it is technically difficult to directly know if the detected mutants are spontaneous mutants, transformation-generated mutants, or both. To address this question, we added DNase I to the bacterial cultures, and this treatment did not result in a significant change in the frequency of emergence of FQr mutants (Table 4), indicating that natural transformation does not play a major role in the emergence of FQr mutants. Interestingly, the insertional mutagenesis of Cj1211 substantially reduced the frequency of emergence of FQr mutants (Table 4), but the reduction cannot be explained by the defect of the Cj1211 mutant in natural transformation because DNase treatment did not influence the rate of mutant emergence. However, it is possible that Cj1211, a predicted inner membrane protein, may interact with other cytoplasmic proteins involved in DNA transformation and recombination and thus affect both spontaneous mutation and transformation in Campylobacter. In B. subtilis, competence proteins (ComGA, ComFA, YwpH, and RecA) colocalize at the bacterial poles (26, 31), suggesting spatial proximity or protein interactions between membrane-associated competence proteins and cytoplasmic proteins (e.g., RecA). A recent study indeed indicated that membrane-associated competence proteins (ComEC, ComFA, ComGA, and ComEA) interact with cytoplasmic soluble proteins (RecA, DprA, SsbB, and YjbF) and that a deletion of one competence protein destabilizes the partner proteins and the functional complex in B. subtilis (33). If Cj1211 forms a functional complex with other cytoplasmic proteins in C. jejuni, such as RecA, which is involved in both natural transformation and spontaneous mutation (5, 8, 54), the loss of Cj1211 may affect the function of RecA and consequently influence the spontaneous mutation rates. This possibility is high because the mutagenesis of Cj1211 also reduced the emergence of streptomycin-resistant mutants (Table 4), suggesting that the general mutation rate was decreased in the Cj1211 mutant. How Cj1211 interacts with other competence proteins in C. jejuni will be examined in future studies.
Multiple studies showed that when Campylobacter-infected animals were treated with FQ antimicrobials, FQr Campylobacter mutants rapidly emerged and populated the treated animals (11, 28, 36, 37). The rapid emergence of FQr mutants under selective pressure may have contributed to the high prevalence of FQr Campylobacter strains on a worldwide scale. The development of FQr Campylobacter from an FQ-susceptible population under FQ treatment certainly involves the selection of preexisting spontaneous mutants, but whether natural transformation is involved in the process is unknown. By taking advantage of the Cj1211 mutant, which is defective in natural transformation, we conducted both in vitro experiments and animal studies to determine if natural transformation affects the development of FQr mutants under selection pressure. Prior to exposure to ciprofloxacin, the number of naturally occurring FQr mutants in the bacterial population was quite low (Fig. 4). Once the antibiotic treatment was initiated, the majority of the FQ-susceptible bacterial cells were killed, and a large amount of free DNA would be released from the dead cells into the culture supernatant. It is conceivable that the free DNA would be available to transform the FQr cells, but the transformants would be quickly killed by ciprofloxacin. Thus, the continuous presence of ciprofloxacin in the cultures eventually eliminated the impact of natural transformation and enriched the FQr population (Fig. 4). The in vitro findings were confirmed by the animal treatment study, which showed that the dynamics of development of FQr Campylobacter were similar between the wild-type strain and JB201 when the chickens were treated with enrofloxacin (Fig. 5). Together, these results suggest that natural transformation does not have a major impact on the development of FQr Campylobacter strains under FQ-selective pressure.
In conclusion, our study identified Cj1211 as being a new competence factor in C. jejuni and proved the role of natural transformation in the horizontal transfer of antibiotic resistance determinants. In addition, we showed that Cj1211 influences the emergence of spontaneous FQr mutants, but this effect appears to be independent of its role in natural transformation. Furthermore, we showed that natural transformation does not play a significant role in the development of FQr Campylobacter strains resulting from FQ treatments. These findings provide new insights into the mechanisms of natural transformation and antibiotic resistance development in Campylobacter.
Supplementary Material
Acknowledgments
We thank Baoqing Guo for constructing the BQ304 mutant used in this study.
This work was supported by National Institutes of Health grant RO1DK063008 and National Research Initiative competitive grant 2007-35201-18278 from the USDA Cooperative State Research, Education, and Extension Service.
Footnotes
Published ahead of print on 27 May 2008.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Atterbury, R. J., P. L. Connerton, C. E. Dodd, C. E. Rees, and I. F. Connerton. 2003. Isolation and characterization of Campylobacter bacteriophages from retail poultry. Appl. Environ. Microbiol. 69:4511-4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avrain, L., C. Vernozy-Rozand, and I. Kempf. 2004. Evidence for natural horizontal transfer of tetO gene between Campylobacter jejuni strains in chickens. J. Appl. Microbiol. 97:134-140. [DOI] [PubMed] [Google Scholar]
- 3.Bacon, D. J., R. A. Alm, D. H. Burr, L. Hu, D. J. Kopecko, C. P. Ewing, T. J. Trust, and P. Guerry. 2000. Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect. Immun. 68:4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bacon, D. J., R. A. Alm, L. Hu, T. E. Hickey, C. P. Ewing, R. A. Batchelor, T. J. Trust, and P. Guerry. 2002. DNA sequence and mutational analyses of the pVir plasmid of Campylobacter jejuni 81-176. Infect. Immun. 70:6242-6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergé, M., I. Mortier-Barrière, B. Martin, and J. P. Claverys. 2003. Transformation of Streptococcus pneumoniae relies on DprA- and RecA-dependent protection of incoming DNA single strands. Mol. Microbiol. 50:527-536. [DOI] [PubMed] [Google Scholar]
- 6.Bergé, M., M. Moscoso, M. Prudhomme, B. Martin, and J. P. Claverys. 2002. Uptake of transforming DNA in gram-positive bacteria: a view from Streptococcus pneumoniae. Mol. Microbiol. 45:411-421. [DOI] [PubMed] [Google Scholar]
- 7.Carfi, A., S. Pares, E. Duee, M. Galleni, C. Duez, J. M. Frere, and O. Dideberg. 1995. The 3-D structure of a zinc metallo-beta-lactamase from Bacillus cereus reveals a new type of protein fold. EMBO J. 14:4914-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, I., P. J. Christie, and D. Dubnau. 2005. The ins and outs of DNA transfer in bacteria. Science 310:1456-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies, J. 1994. Inactivation of antibiotics and the dissemination of resistance genes. Science 264:375-382. [DOI] [PubMed] [Google Scholar]
- 10.de Boer, P., J. A. Wagenaar, R. P. Achterberg, J. P. van Putten, L. M. Schouls, and B. Duim. 2002. Generation of Campylobacter jejuni genetic diversity in vivo. Mol. Microbiol. 44:351-359. [DOI] [PubMed] [Google Scholar]
- 11.Delsol, A. A., J. Sunderland, M. J. Woodward, L. Pumbwe, L. J. Piddock, and J. M. Roe. 2004. Emergence of fluoroquinolone resistance in the native Campylobacter coli population of pigs exposed to enrofloxacin. J. Antimicrob. Chemother. 53:872-874. [DOI] [PubMed] [Google Scholar]
- 12.Dorrell, N., J. A. Mangan, K. G. Laing, J. Hinds, D. Linton, H. Al Ghusein, B. G. Barrell, J. Parkhill, N. G. Stoker, A. V. Karlyshev, P. D. Butcher, and B. W. Wren. 2001. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 11:1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Draskovic, I., and D. Dubnau. 2005. Biogenesis of a putative channel protein, ComEC, required for DNA uptake: membrane topology, oligomerization and formation of disulphide bonds. Mol. Microbiol. 55:881-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubnau, D. 1999. DNA uptake in bacteria. Annu. Rev. Microbiol. 53:217-244. [DOI] [PubMed] [Google Scholar]
- 15.Dubnau, D., and R. Provvedi. 2000. Internalizing DNA. Res. Microbiol. 151:475-480. [DOI] [PubMed] [Google Scholar]
- 16.Facius, D., and T. F. Meyer. 1993. A novel determinant (comA) essential for natural transformation competence in Neisseria gonorrhoeae and the effect of a comA defect on pilin variation. Mol. Microbiol. 10:699-712. [DOI] [PubMed] [Google Scholar]
- 17.Fouts, D. E., E. F. Mongodin, R. E. Mandrell, W. G. Miller, D. A. Rasko, J. Ravel, L. M. Brinkac, R. T. DeBoy, C. T. Parker, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, S. A. Sullivan, J. U. Shetty, M. A. Ayodeji, A. Shvartsbeyn, M. C. Schatz, J. H. Badger, C. M. Fraser, and K. E. Nelson. 2005. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol. 3:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frost, L. S., R. Leplae, A. O. Summers, and A. Toussaint. 2005. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3:722-732. [DOI] [PubMed] [Google Scholar]
- 19.Ge, B., P. F. McDermott, D. G. White, and J. Meng. 2005. Role of efflux pumps and topoisomerase mutations in fluoroquinolone resistance in Campylobacter jejuni and Campylobacter coli. Antimicrob. Agents Chemother. 49:3347-3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibreel, A., and D. E. Taylor. 2006. Macrolide resistance in Campylobacter jejuni and Campylobacter coli. J. Antimicrob. Chemother. 58:243-255. [DOI] [PubMed] [Google Scholar]
- 21.Grajewski, B. A., J. W. Kusek, and H. M. Gelfand. 1985. Development of a bacteriophage typing system for Campylobacter jejuni and Campylobacter coli. J. Clin. Microbiol. 22:13-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graupner, S., and W. Wackernagel. 2001. Identification and characterization of novel competence genes comA and exbB involved in natural genetic transformation of Pseudomonas stutzeri. Res. Microbiol. 152:451-460. [DOI] [PubMed] [Google Scholar]
- 23.Guerry, P., P. M. Pope, D. H. Burr, J. Leifer, S. W. Joseph, and A. L. Bourgeois. 1994. Development and characterization of recA mutants of Campylobacter jejuni for inclusion in attenuated vaccines. Infect. Immun. 62:426-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guerry, P., R. Yao, R. A. Alm, D. H. Burr, and T. J. Trust. 1994. Systems of experimental genetics for Campylobacter species. Methods Enzymol. 235:474-481. [DOI] [PubMed] [Google Scholar]
- 25.Hahn, J., G. Inamine, Y. Kozlov, and D. Dubnau. 1993. Characterization of comE, a late competence operon of Bacillus subtilis required for the binding and uptake of transforming DNA. Mol. Microbiol. 10:99-111. [DOI] [PubMed] [Google Scholar]
- 26.Hahn, J., B. Maier, B. J. Haijema, M. Sheetz, and D. Dubnau. 2005. Transformation proteins and DNA uptake localize to the cell poles in Bacillus subtilis. Cell 122:59-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hughes, R. A., and D. R. Cornblath. 2005. Guillain-Barre syndrome. Lancet 366:1653-1666. [DOI] [PubMed] [Google Scholar]
- 28.Humphrey, T. J., F. Jorgensen, J. A. Frost, H. Wadda, G. Domingue, N. C. Elviss, D. J. Griggs, and L. J. Piddock. 2005. Prevalence and subtypes of ciprofloxacin-resistant Campylobacter spp. in commercial poultry flocks before, during, and after treatment with fluoroquinolones. Antimicrob. Agents Chemother. 49:690-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeon, B., and Q. Zhang. 2007. Cj0011c, a periplasmic single- and double-stranded DNA-binding protein, contributes to natural transformation in Campylobacter jejuni. J. Bacteriol. 189:7399-7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khakhria, R., and H. Lior. 1992. Extended phage-typing scheme for Campylobacter jejuni and Campylobacter coli. Epidemiol. Infect. 108:403-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kidane, D., and P. L. Graumann. 2005. Intracellular protein and DNA dynamics in competent Bacillus subtilis cells. Cell 122:73-84. [DOI] [PubMed] [Google Scholar]
- 32.Kim, J. S., D. K. Carver, and S. Kathariou. 2006. Natural transformation-mediated transfer of erythromycin resistance in Campylobacter coli strains from turkeys and swine. Appl. Environ. Microbiol. 72:1316-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kramer, N., J. Hahn, and D. Dubnau. 2007. Multiple interactions among the competence proteins of Bacillus subtilis. Mol. Microbiol. 65:454-464. [DOI] [PubMed] [Google Scholar]
- 34.Lambert, P. A. 2005. Bacterial resistance to antibiotics: modified target sites. Adv. Drug Deliv. Rev. 57:1471-1485. [DOI] [PubMed] [Google Scholar]
- 35.Larsen, J. C., C. Szymanski, and P. Guerry. 2004. N-linked protein glycosylation is required for full competence in Campylobacter jejuni 81-176. J. Bacteriol. 186:6508-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo, N., O. Sahin, J. Lin, L. O. Michel, and Q. Zhang. 2003. In vivo selection of Campylobacter isolates with high levels of fluoroquinolone resistance associated with gyrA mutations and the function of the CmeABC efflux pump. Antimicrob. Agents Chemother. 47:390-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDermott, P. F., S. M. Bodeis, L. L. English, D. G. White, R. D. Walker, S. Zhao, S. Simjee, and D. D. Wagner. 2002. Ciprofloxacin resistance in Campylobacter jejuni evolves rapidly in chickens treated with fluoroquinolones. J. Infect. Dis. 185:837-840. [DOI] [PubMed] [Google Scholar]
- 38.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller, W. G., A. H. Bates, S. T. Horn, M. T. Brandl, M. R. Wachtel, and R. E. Mandrell. 2000. Detection on surfaces and in Caco-2 cells of Campylobacter jejuni cells transformed with new gfp, yfp, and cfp marker plasmids. Appl. Environ. Microbiol. 66:5426-5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Normark, B. H., and S. Normark. 2002. Evolution and spread of antibiotic resistance. J. Intern. Med. 252:91-106. [DOI] [PubMed] [Google Scholar]
- 41.Notani, N. K., J. K. Setlow, V. R. Joshi, and D. P. Allison. 1972. Molecular basis for the transformation defects in mutants of Haemophilus influenzae. J. Bacteriol. 110:1171-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oyarzabal, O. A., R. Rad, and S. Backert. 2007. Conjugative transfer of chromosomally encoded antibiotic resistance from Helicobacter pylori to Campylobacter jejuni. J. Clin. Microbiol. 45:402-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 44.Payot, S., J. M. Bolla, D. Corcoran, S. Fanning, F. Megraud, and Q. Zhang. 2006. Mechanisms of fluoroquinolone and macrolide resistance in Campylobacter spp. Microbes Infect. 8:1967-1971. [DOI] [PubMed] [Google Scholar]
- 45.Payot, S., A. Cloeckaert, and E. Chaslus-Dancla. 2002. Selection and characterization of fluoroquinolone-resistant mutants of Campylobacter jejuni using enrofloxacin. Microb. Drug Resist. 8:335-343. [DOI] [PubMed] [Google Scholar]
- 46.Pestova, E. V., and D. A. Morrison. 1998. Isolation and characterization of three Streptococcus pneumoniae transformation-specific loci by use of a lacZ reporter insertion vector. J. Bacteriol. 180:2701-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Piddock, L. J., V. Ricci, L. Pumbwe, M. J. Everett, and D. J. Griggs. 2003. Fluoroquinolone resistance in Campylobacter species from man and animals: detection of mutations in topoisomerase genes. J. Antimicrob. Chemother. 51:19-26. [DOI] [PubMed] [Google Scholar]
- 48.Pratt, A., and V. Korolik. 2005. Tetracycline resistance of Australian Campylobacter jejuni and Campylobacter coli isolates. J. Antimicrob. Chemother. 55:452-460. [DOI] [PubMed] [Google Scholar]
- 49.Provvedi, R., I. Chen, and D. Dubnau. 2001. NucA is required for DNA cleavage during transformation of Bacillus subtilis. Mol. Microbiol. 40:634-644. [DOI] [PubMed] [Google Scholar]
- 50.Sikorski, J., S. Graupner, M. G. Lorenz, and W. Wackernagel. 1998. Natural genetic transformation of Pseudomonas stutzeri in a non-sterile soil. Microbiology 144:569-576. [DOI] [PubMed] [Google Scholar]
- 51.Stewart, G. J., and C. D. Sinigalliano. 1990. Detection of horizontal gene transfer by natural transformation in native and introduced species of bacteria in marine and synthetic sediments. Appl. Environ. Microbiol. 56:1818-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Summers, A. O. 2006. Genetic linkage and horizontal gene transfer, the roots of the antibiotic multi-resistance problem. Anim. Biotechnol. 17:125-135. [DOI] [PubMed] [Google Scholar]
- 53.Taylor, D. E., S. A. De Grandis, M. A. Karmali, and P. C. Fleming. 1981. Transmissible plasmids from Campylobacter jejuni. Antimicrob. Agents Chemother. 19:831-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tessman, E. S., I. Tessman, P. K. Peterson, and J. D. Forestal. 1986. Roles of RecA protease and recombinase activities of Escherichia coli in spontaneous and UV-induced mutagenesis and in Weigle repair. J. Bacteriol. 168:1159-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomas, C. M., and K. M. Nielsen. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3:711-721. [DOI] [PubMed] [Google Scholar]
- 56.Velazquez, J. B., A. Jimenez, B. Chomon, and T. G. Villa. 1995. Incidence and transmission of antibiotic resistance in Campylobacter jejuni and Campylobacter coli. J. Antimicrob. Chemother. 35:173-178. [DOI] [PubMed] [Google Scholar]
- 57.Wang, Y., W. M. Huang, and D. E. Taylor. 1993. Cloning and nucleotide sequence of the Campylobacter jejuni gyrA gene and characterization of quinolone resistance mutations. Antimicrob. Agents Chemother. 37:457-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, Y., and D. E. Taylor. 1990. Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression, and cloning vector construction. Gene 94:23-28. [DOI] [PubMed] [Google Scholar]
- 59.Wang, Y., and D. E. Taylor. 1990. Natural transformation in Campylobacter species. J. Bacteriol. 172:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wassenaar, T. M., B. N. Fry, and B. A. van der Zeijst. 1993. Genetic manipulation of Campylobacter: evaluation of natural transformation and electro-transformation. Gene 132:131-135. [DOI] [PubMed] [Google Scholar]
- 61.Wiesner, R. S., D. R. Hendrixson, and V. J. DiRita. 2003. Natural transformation of Campylobacter jejuni requires components of a type II secretion system. J. Bacteriol. 185:5408-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilson, D. L., J. A. Bell, V. B. Young, S. R. Wilder, L. S. Mansfield, and J. E. Linz. 2003. Variation of the natural transformation frequency of Campylobacter jejuni in liquid shake culture. Microbiology 149:3603-3615. [DOI] [PubMed] [Google Scholar]
- 63.Wösten, M. M., M. Boeve, M. G. Koot, A. C. van Nuenen, and B. A. van der Zeijst. 1998. Identification of Campylobacter jejuni promoter sequences. J. Bacteriol. 180:594-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yan, M., O. Sahin, J. Lin, and Q. Zhang. 2006. Role of the CmeABC efflux pump in the emergence of fluoroquinolone-resistant Campylobacter under selection pressure. J. Antimicrob. Chemother. 58:1154-1159. [DOI] [PubMed] [Google Scholar]
- 65.Yao, R., R. A. Alm, T. J. Trust, and P. Guerry. 1993. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130:127-130. [DOI] [PubMed] [Google Scholar]
- 66.Yeh, Y. C., T. L. Lin, K. C. Chang, and J. T. Wang. 2003. Characterization of a ComE3 homologue essential for DNA transformation in Helicobacter pylori. Infect. Immun. 71:5427-5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yuki, N. 2005. Carbohydrate mimicry: a new paradigm of autoimmune diseases. Curr. Opin. Immunol. 17:577-582. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.