Abstract
A new primer set was designed to specifically amplify ca. 1,100 bp of aoxB genes encoding the As(III) oxidase catalytic subunit from taxonomically diverse aerobic As(III)-oxidizing bacteria. Comparative analysis of AoxB protein sequences showed variable conservation levels and highlighted the conservation of essential amino acids and structural motifs. AoxB phylogeny of pure strains showed well-discriminated taxonomic groups and was similar to 16S rRNA phylogeny. Alphaproteobacteria-, Betaproteobacteria-, and Gammaproteobacteria-related sequences were retrieved from environmental surveys, demonstrating their prevalence in mesophilic As-contaminated soils. Our study underlines the usefulness of the aoxB gene as a functional marker of aerobic As(III) oxidizers.
Arsenic (As) exists mainly in two toxic soluble forms, arsenite, As(III), and arsenate, As(V), with the latter tending to associate with some oxyhydroxides and clay minerals. The bacterial oxidation of As(III) can thus contribute to a natural attenuation of As contamination by decreasing As bioavailability. These properties have recently been used to develop a bioprocess for removing As from a mining effluent by using the activity of As-metabolizing bacteria indigenous to the contaminated site (4). The feasibility of such a process depends on a good knowledge of the ability of the indigenous microflora to oxidize As(III) and requires reliable methods for detecting, identifying, and monitoring As(III) oxidizers in the environment.
More than 50 phylogenetically diverse As(III)-oxidizing strains distributed among 25 genera have been isolated from various environments so far. Bacterial aerobic As(III) oxidation is performed by a dedicated enzyme, the As(III) oxidase (1, 36, 40), which belongs to the dimethyl sulfoxide (DMSO) reductase of the molybdenum family (9). In Alcaligenes faecalis, it is an α1β1 heterodimer comprising a large subunit incorporating a molybdenum center and a [3Fe-4S] cluster and a small subunit incorporating a Rieske-type [2Fe-2S] cluster (9). Genes encoding these subunits are cotranscribed as an operon and have been successively characterized in Herminiimonas arsenicoxydans (26), Rhizobium sp. strain NT-26 (36), and Agrobacterium tumefaciens (21). They have also been found in the genome of Chloroflexus aurantiacus, on a plasmid in Thermus thermophilus, in two aerobic thermophilic As(III) oxidizers, and in the genome of strains for which the ability to oxidize As(III) has not been experimentally proven (27).
Due to the polyphyly of As(III)-oxidizing bacteria, the aoxB gene encoding the catalytic subunit of the enzyme seems to be a valuable molecular marker for investigating its ecology and the potential of As(III) oxidation in the environment. To this end, a recent study described primers targeting the first quarter of the aoxB gene to detect its presence and expression in the environment and suggested that the gene is widely distributed among the Bacteria and also is widespread in soil-water systems containing As (16).
In our present study, we designed new primers to extend the genetic information to the first half of the aoxB gene. We then explored the genetic diversity of this gene in order (i) to identify conserved structural and functional domains, (ii) to compare AoxB and 16S rRNA phylogenies to evaluate whether phylogenetic information about As(III)-oxidizing bacteria can be inferred from the aoxB gene, and (iii) to assess the composition of As(III)-oxidizing communities in environmental diversity surveys.
Primer evaluation and validation.
Primers aoxBM1-2F (5′-CCACTTCTGCATCGTGGGNTGYGGNTA-3′, positions 66 to 92 in the H. arsenicoxydans aoxB open reading frame) and aoxBM3-2R (5′-TGTCGTTGCCCCAGATGADNCCYTTYTC-3′, positions 1150 to 1177) were designed on the most distantly located conserved regions with the CODEHOP program (32) from nine AoxB protein sequences from the Bacteria (Alcaligenes faecalis [GenBank accession no. AY297781], Herminiimonas arsenicoxydans [accession no. AF509588], “Thiomonas arsenivorans” [accession no. EU304260], Agrobacterium tumefaciens [accession no. DQ151549], Rhizobium sp. strain NT26 [accession no. AY345225], Chloroflexus aurantiacus [accession no. NZ_AAAH01000321], and Thermus thermophilus [accession no. NC_000854]), and the Archaea (Aeropyrum pernix [accession no. NC_000854] and Sulfolobus tokodaii [accession no. NC_003106]). The primers target the first part (amino acids 22 to 32, H. arsenicoxydans AoxB numbering) of the CX2CX3CX70S motif (CHFCIVGCGYH) required for binding a [3Fe-4S] cluster and the consensus motif YEKGIIWGN (amino acids 383 to 391).
Specific amplifications (35 cycles, annealing temperature of 52°C, 1 min 10 s elongation time) of ca. 1,100 bp were obtained for the following 21 chemoautotrophic and chemoheterotrophic As(III)-oxidizing strains belonging to Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, and Chloroflexi. T. arsenivorans b6T (3), Leptothrix sp. strain S1.1 (4), Variovorax sp. strain 4.2 (4), H. arsenicoxydans ULPAs1T (43), Acidovorax sp. strain 75, Acinetobacter sp. strain 33, Alcaligenes sp. strain YI013H, Alcaligenes sp. strain T12RB, Aminobacter sp. strain 86, Burkholderia sp. strain YI019A, Limnobacter sp. strain 83, Pseudomonas sp. strains 1, 46, 72, 73, 89, and D2OHCJ, and Ralstonia sp. strain 22 were isolated in our laboratories from As-contaminated environments (see Table S1 in the supplemental material). Thiomonas sp. strain WJ68 and Thiomonas sp. strain NO115 were isolated from mining sites (7, 15). Chloroflexus aurantiacus DSM635 was isolated from hot springs (30). A less-specific amplification was obtained for the archaeon Aeropyrum pernix DSM11879. The detection of distinct genera thus indicated the ability to detect diverse taxa of this metabolic group with the newly designed primers. No amplification was obtained from the 10 non-As(III)-oxidizing bacteria used as negative controls, of which 6 harbor a molybdenum enzyme of the DMSO reductase family (i.e., Rhodobacter sphaeroides, Desulfitobacterium hafniense, Bacillus selenitireducens, Geobacter metallireducens, Escherichia coli, and Halomonas denitrificans).
Sensitivity tests showed that as little as 8.3 pg of template DNA of H. arsenicoxydans was needed for aoxB gene amplification. Knowing that the 3.4-Mb genome of H. arsenicoxydans contains only one aoxB gene (27), this would represent approximately 2.2 × 103 copies of the aoxB gene. The sensitivity was lower than that of H. arsenicoxydans for the phylogenetically distant C. auriantiacus strain, with the detection limit being 1.7 × 104 aoxB gene copies.
Protein and nucleotide sequence analyses.
Analyses were conducted on 29 sequences of As(III)-oxidizing isolates, excluding recently published short sequences (16) and sequences from whole-genome data for which there is no experimental evidence that the corresponding strains oxidize As(III).
Examination of nucleotide sequence alignments using DnaSP version 4 software (33) revealed a high proportion of variable sites (82%), of which 74% were parsimony informative. Conservation degrees ranged from 48.6% to 92.3%. The second codon positions were clearly better conserved. The third codon positions were the most variable (99%), and saturation analysis using DAMBE program version 4.0.50 (44) showed that saturation occurred at this position. The number of substitution sites was higher for phylogenetically distant strains than for closely related ones. Nucleotide substitution causing amino acid substitution was more frequent than synonymous substitution, which accounted for only 23.7% of all substitutions.
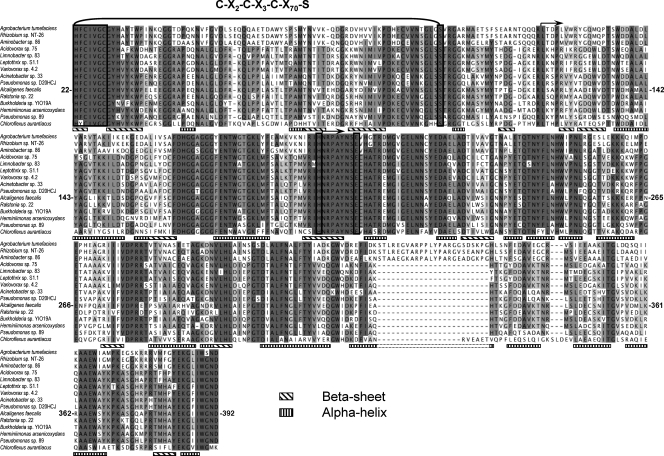
Comparative analysis of the deduced AoxB protein sequences showed variable conservation levels (38.4 to 96.8% identity), comparable to data obtained for other functional genes, e.g., those involved in sulfur oxidation, nitrous oxide reduction, and sulfite reduction (28, 37, 42). The 15 partial AoxB sequences presented in Fig. 1 showed 22% identity on 372 amino acids. Three of the four domains of the AoxB subunit described for A. faecalis (9) were retrieved in the aligned region (the fourth domain extends outside the sequences presented here). Domain I extends from residue 21 to residue 119 (according to A. faecalis protein sequence numbering) and binds the Rieske subunit and the [3Fe-4S] cluster coordinated by the motif C21-X2-C24-X3-C28-X70-S99. Except for amino acids present in the X70 region, a high conservation was observed, particularly between residues 21 and 31 and between residues 86 and 103. A highly conserved region between residues 166 and 261 encompassed domains II (residues 120 to 195) and III (residues 196 to 392) and contains several structural motifs, such as alpha-helices and residues like H195 and E203 implicated in the substrate-binding site of the enzyme. Interestingly, the HNRPAYNSE motif is exactly conserved in all bacterial As(III) oxidases. The conserved S99 and A199 residues are important in the demarcation of the catalytic subunit of As(III) oxidase with the other members of the DMSO reductase family of molybdenum enzymes (9). A lower amino acid conservation was observed in the other parts of the protein, although the presence of multiple conservative substitutions suggests that the physicochemical properties of these amino acids play a role in the structure and/or the functioning of the As(III) oxidase enzyme. Finally, regions that contain no structural element showed a low amino acid conservation. For example, the C. aurantiacus sequence shows six additional amino acids between residues 330 and 331. In addition, the three Alphaproteobacteria, namely A. tumefaciens, Rhizobium sp. strain NT26, and an Aminobacter sp., harbor 24 additional amino acids in this region. This further supports the low amino acid conservation observed throughout the As(III) oxidase protein sequence in these microorganisms compared to that of the Betaproteobacteria protein sequences. Taken together, our observations suggest that the As(III) oxidases identified here show moderate amino acid conservation but share several structural and functional domains similar to those found in A. faecalis AoxB (Fig. 1), supporting the important role of these domains in As(III) oxidase activity.
FIG. 1.
Protein sequence alignment and putative secondary structure of As(III) oxidases. The Alcaligenes faecalis sequence and its secondary structure were retrieved from the PDB database (http://www.rcsb.org/pdb/welcome.do). Other As(III) oxidase protein sequences were deduced from nucleotide aoxB sequences. Sequence alignment was carried out by CLUSTAL W (39). Amino acid residue numbers are defined according to the A. faecalis protein sequence, which starts at a histidine residue at position 22. Residues identical to those of the A. faecalis sequence are shown in gray-scale boxes. Residues known to play a role in As(III) oxidase activity are framed in black. Black arrows at positions 120 and 196 indicate the beginning of domains 2 and 3, respectively. The beta-sheet and alpha-helix are shown according to the three-dimensional structure of A. faecalis protein (9).
Deduced protein sequences of closely related species were generally better conserved than nucleotide sequences. For example, Alcaligenes species exhibited sequence identity values ranging from 80.2 to 96.8% for proteins and from 76.6 to 92.2% for nucleotides. AoxB sequences within the same subclass of Proteobacteria showed generally higher identity values (e.g., between 72.6% and 96% for Alphaproteobacteria). The highest divergences in protein sequences were observed between distantly related bacterial lineages, i.e., Proteobacteria versus the nonproteobacterial Thermus and Chloroflexus strains (identities between 38.4% and 50.7%), and Alphaproteobacteria (except Hydrogenophaga sp. strain CL3; see below) versus Betaproteobacteria and Gammaproteobacteria (identities between 46.7 and 53.5%). In contrast, nucleotide sequences were better conserved than protein sequences for distantly related species. Similar events have been reported for other functional genes, e.g., those involved in denitrification and sulfur oxidation (5, 28). In the case of aoxB genes, these observations can be explained by saturation at the third codon position for comparison of distant strains. The observed divergences suggest that the aoxB gene may have a long evolutionary history and support the hypothesis of an early common ancestor (24). Rhine et al. (31) recently reported that autotrophic aoxB genes formed a phylogenetic group distinct from heterotrophic aoxB genes, with the exception of A. tumefaciens aoxB genes, and suggested that the two groups evolved separately from a common ancestor. However, our study demonstrated that the aoxB gene of the autotrophic T. arsenivorans is phylogenetically affiliated with heterotrophic aoxB genes and not with genes of autotrophic As(III) oxidizers. Further data on aoxB gene sequences from pure heterotrophic and autotrophic strains will thus be needed to elucidate the evolution of this gene.
Protein sequences are usually used for reconstructing the phylogeny of protein-encoding genes (6, 37). In addition, protein sequences appeared less noisy and more resolutive than nucleotide sequences for large-scale (phylum) analysis. Further reconstruction of As(III) oxidase phylogeny has thus been based on protein sequences.
Comparison of AoxB and 16S rRNA gene phylogenies.
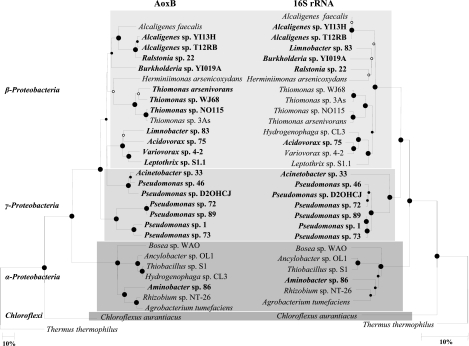
AoxB and 16S rRNA neighbor-joining (34) trees were constructed from unambiguous residues on distances estimated by the Kimura (22) and Jukes and Cantor (20) methods, respectively. The AoxB tree reconstructed the major taxonomic levels and was similar to the 16S rRNA tree (Fig. 2). Indeed, the C. aurantiacus AoxB sequence is clearly separated from the AoxB proteobacterial sequences, with a strong bootstrap support (100%). The Alphaproteobacteria and Betaproteobacteria/Gammaproteobacteria AoxB sequences form distinct phylogenetic branches, supported by 100% bootstrap values. In addition, sequences from species of the same genus, such as Thiomonas or Alcaligenes, cluster together.
FIG. 2.
Neighbor-joining phylogenetic trees showing the relationships between partial AoxB protein (351-residue) and partial 16S rRNA (1,139-nucleotide) sequences of bacterial As(III) oxidizers. Bacteria whose sequences were determined in this study are in bold. The Thermus thermophilus AoxB sequence was used as the outgroup. Circles at the branch nodes represent bootstrap percentages for 100 replicates (10): large filled circles, 95 to 100%; small filled circles, 75 to 95%; open circles, 50 to 75%. Scale bars correspond to 10 mutations per 100 residues. Phylogenetic programs were implemented in the TREECON package (41). Parsimony analysis (11) gave similar topologies.
However, a detailed comparison of the two trees revealed some discrepancies. For example, the pseudomonads are divided into two groups in the AoxB-based tree. Group 1 forms a distinct phylogenetic branch supported by a high bootstrap value (100%), while group 2 forms a separate branch of sequences more closely related to those from the Betaproteobacteria. The major inconsistency is the position of the betaproteobacterium Hydrogenophaga sp. strain CL3 within Alphaproteobacteria, which suggests the possibility of lateral gene transfer. However, two Hydrogenophaga spp. were properly positioned among the Betaproteobacteria on an AoxB tree constructed from shorter sequences (16). However, strain CL3 is also branching within Alphaproteobacteria when a tree is built from short sequences (data not shown). Collection of aoxB sequences from other Hydrogenophaga isolates would help to determine whether or not this conspicuous event is limited to the Hydrogenophaga species. Interestingly, the aoxB gene is plasmid carried in Thermus thermophilus HB8 and is carried by a genomic island in A. faecalis (38) and H. arsenicoxydans (27), indicating that lateral transfer of the gene can occur. These events have also been demonstrated for other functional genes (12, 23), particularly for arsC, a gene involved in the As cycle and in encoding the cytoplasmic As(V) reductase (18). Consequently, AoxB sequences must be used with caution to infer phylogenetic information about environmental As(III)-oxidizing bacteria. A greater collection of aoxB genes will undoubtedly increase confidence in the AoxB phylogeny.
AoxB diversity in environmental samples.
Diversity surveys of aoxB genes were conducted for evaluating the pertinence of the newly developed assay to assess the composition of As(III) oxidizers in the environment. Gene libraries were constructed (TA cloning kit; Invitrogen, Carlsbad, CA) on (i) an As-contaminated industrial soil (T12R) obtained from a former cokery (19), (ii) an enrichment of this soil under As(III)-oxidizing chemoheterotrophic conditions, and (iii) an enrichment of this soil under As(III)-oxidizing chemoautotrophic conditions. Enrichments were made from a 100-fold-diluted (wt/vol) T12R soil suspension in CasO1 selective medium (2) containing 200 mg liter−1 of As(III) and supplemented with 5 mM Na lactate, 5 mM acetate, and 0.2% yeast extract under heterotrophic conditions; As(III) oxidation was measured as described by Battaglia-Brunet et al. (2). DNA was extracted from 1 g of soil and 50 ml of enrichment cultures using the MoBio UltraClean soil extraction kit (MoBio, Solana Beach, CA).
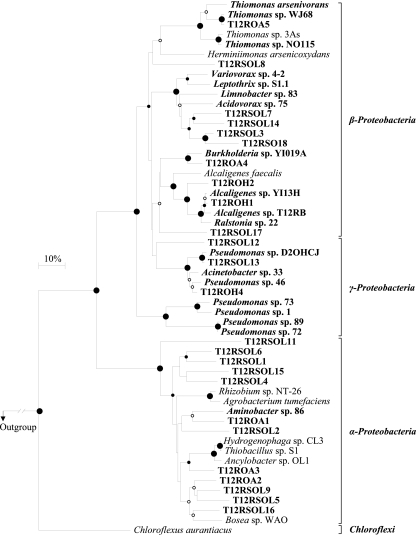
Of the 133 clones screened by RsaI/HaeIII restriction analyses, 62 inserts were sequenced and finally grouped into 28 operational taxonomic units (OTU) based on a 90% sequence identity. Only aoxB gene-related sequences were retrieved, and such exclusive specificity for the target gene has the advantage of reducing the stringency of annealing conditions, thus favoring greater sensitivity. All the environmental AoxB sequences contained the conserved residues predicted by Ellis et al. (9) as being essential for As(III) oxidase activity in A. faecalis, suggesting that these sequences produced functional enzymes. The environmental AoxB sequences were affiliated with sequences from strains belonging to the Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria, with high bootstrap values (Fig. 3).
FIG. 3.
Neighbor-joining phylogenetic tree of bacterial AoxB sequences retrieved from environmental surveys. T12RSOL, As-contaminated soil T12R; T12ROA, an As(III)-oxidizing chemoautotrophic enrichment of soil T12R; T12ROH, an As(III)-oxidizing chemoheterotrophic enrichment of soil T12R. Bacteria whose sequences were determined in this study are in bold. The Thermus thermophilus AoxB sequence was used as the outgroup. Circles at the branch nodes represent bootstrap percentages for 100 replicates (10): large filled circles, 95 to 100%; small filled circles, 75 to 95%; open circles, 50 to 75%. The scale bar corresponds to 10 mutations per 100 residues. All phylogenetic programs were implemented in the TREECON package (41).
A large diversity (18 OTU) was detected from the soil T12R. The library was dominated (58%) by Alphaproteobacteria-related sequences, among which 19% were related to a Bosea sp. Five OTU formed separate branches of known Alphaproteobacteria sequences. Betaproteobacteria-related OTU (17%) were mainly affiliated with an Acidovorax sp. Gammaproteobacteria-related OTU (25%) were mainly represented by group 2 pseudomonads. Interestingly, incubation of this soil under chemoheterotrophic As(III)-oxidizing conditions led to a decrease in diversity with the selection of betaproteobacterial Alcaligenes-related sequences (88%). Only 1 OTU showed a close relationship to the Gammaproteobacteria group 2 pseudomonads. Remarkably, no alphaproteobacterium was detected, although Alphaproteobacteria were dominant in the soil. When this soil was incubated under chemoautotrophic As(III)-oxidizing conditions, OTU related to sequences of the autotrophic As(III)-oxidizing Thiomonas (55%) and Burkholderia (17%), genera of the Betaproteobacteria, dominated the library. The Alphaproteobacteria-related OTU represented 32% of the library (mainly an Ancylobacter sp.). To date, the majority of known As(III)-oxidizing strains belonging to Alphaproteobacteria are autotrophs, suggesting a predominance of autotrophic metabolism among them (31). In contrast to the As(III)-oxidizing chemoheterotrophic enrichment, neither Gammaproteobacteria- nor Alcaligenes (Betaproteobacteria)-affiliated sequences were found. Our results underline the usefulness of functional molecular markers in diversity surveys for directly detecting the functionality of an environment without the need of cultivation.
The exclusive detection of environmental AoxB sequences belonging to the phylum Proteobacteria is probably related to the fact that we studied a mesophilic environment, which is likely to contain a large majority of As(III) oxidizers from this phylum. To date, all but two of the As(III)-oxidizing bacteria isolated from mesophilic sites belong to the phylum Proteobacteria, and a recent study (16) has shown that 98% of aoxB-like sequences retrieved from mesophilic As-contaminated soils and sediments belong to the Proteobacteria. The only reports on the ability of mesophilic gram-positive bacteria to perform As(III) oxidation concerned Microbacterium lacticum (25) and Bacillus arsenoxydans (14), with the general physiology of the latter organism being similar to that of the betaproteobacterium A. faecalis (29). As(III)-oxidizing bacteria belonging to other phyla, such as Deinococcus-Thermus (13), Chloroflexi (24), and Aquificae (8), have been isolated only from geothermic sites. It is also noteworthy that As(III)-oxidizing Proteobacteria are able to colonize geothermic sites (17, 35).
Our study has demonstrated that the aoxB gene has the major features of a molecular marker. (i) The aoxB gene has been found in all the aerobic As(III)-oxidizing bacteria tested so far. (ii) Conserved regions across this gene have enabled the design of valuable primers. (iii) The studied region is sufficiently large to obtain genetic variation, allowing the discrimination of phylogenetic groups. These data are encouraging for the further use of the aoxB gene as a functional marker specific to aerobic As(III) oxidizers in environmental diversity surveys. Future work will focus on evaluating the link between As speciation and concentration, and aoxB gene diversity and abundance, in the environment.
Nucleotide sequence accession numbers.
The aoxB sequences have been deposited in the GenBank database under accession no. EU304260 to EU304278, EU304293 to EU304310, and EU304313 to EU304321, and the 16S rRNA gene sequences have been deposited under accession no. EU304279 to EU304292.
Supplementary Material
Acknowledgments
This is BRGM contribution no. 05252.
We thank the Agence de l'Environnement et de la Maîtrise de l'Energie (ADEME) and the French Ministry of Education and Research for financially supporting this research. The work was partly done within the framework of the Groupement de Recherche-Métabolisme de l'Arsenic chez les Prokaryotes (GDR2909-CNRS) and of the European Union FP6 Integrated Project AquaTerra (project no. GOCE 505428).
We are grateful to K. H. Hallberg for providing Thiomonas sp. strain WJ68 and Thiomonas sp. strain NO115. We also thank Marie-Claire Lett and Laurence Casalot for useful discussions.
Footnotes
Published ahead of print on 23 May 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Anderson, G. L., J. Williams, and R. Hille. 1992. The purification and characterization of arsenite oxidase from Alcaligenes faecalis, a molybdenum-containing hydroxylase. J. Biol. Chem. 267:23674-23682. [PubMed] [Google Scholar]
- 2.Battaglia-Brunet, F., M.-C. Dictor, F. Garrido, C. Crouzet, D. Morin, K. Dekeyser, M. Clarens, and P. Baranger. 2002. An arsenic(III)-oxidizing bacterial population: selection, characterization, and performance in reactors. J. Appl. Microbiol. 93:656-667. [DOI] [PubMed] [Google Scholar]
- 3.Battaglia-Brunet, F., C. Joulian, F. Garrido, M.-C. Dictor, D. Morin, K. Coupland, D. Barrie Johnson, K. B. Hallberg, and P. Baranger. 2006. Oxidation of arsenite by Thiomonas strains and characterization of Thiomonas arsenivorans sp. nov. Antonie van Leeuwenhoek 89:99-108. [DOI] [PubMed] [Google Scholar]
- 4.Battaglia-Brunet, F., Y. Itard, F. Garrido, F. Delorme, C. Crouzet, C. Greffié, and C. Joulian. 2006. A simple biogeochemical process removing arsenic from a mine drainage water. Geomicrobiol. J. 23:1-11. [Google Scholar]
- 5.Braker, G., J. Zhou, L. Wu, A. H. Devol, and J. M. Tiedje. 2000. Nitrite reductase genes (nirK and nirS) as functional markers to investigate diversity of denitrifying bacteria in Pacific northwest marine sediment communities. Appl. Environ. Microbiol. 66:2096-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Case, R. J., Y. Boucher, I. Dahllöf, C. Holmström, W. F. Doolittle, and S. Kjelleberg. 2007. Use of 16S rRNA and rpoB genes as molecular markers for microbial ecology studies. Appl. Environ. Microbiol. 73:278-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coupland, K., F. Battaglia-Brunet, K. B. Hallberg, M.-C. Dictor, F. Garrido, and D. B. Johnson. 2004. Oxidation of iron, sulfur and arsenic in mine waters and mine wastes: an important role for novel Thiomonas spp., p. 639-646. In M. Tsezos, A. Hatzikioseyian, and E. Remoudaki (ed.), Biohydrometallurgy: a sustainable technology in evolution. National Technical University of Athens, Zografou, Greece.
- 8.Donahoe-Christiansen, J., S. D'Imperio, C. R. Jackson, W. P. Inskeep, and T. R. McDermott. 2004. Arsenite-oxidizing Hydrogenobaculum strain isolated from an acid-sulfate-chloride geothermal spring in Yellowstone National Park. Appl. Environ. Microbiol. 70:1865-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis, P. J., T. Conrads, R. Hille, and P. Kuhn. 2001. Crystal structure of the 100 kDa arsenite oxidase from Alcaligenes faecalis in two crystal forms at 1.64 Å and 2.03 Å. Structure 9:125-132. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 12.Friedrich, M. W. 2002. Phylogenetic analysis reveals multiple lateral transfers of adenosine-5′-phosphosulfate reductase genes among sulfate-reducing microorganisms. J. Bacteriol. 184:278-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gihring, T. M., G. K. Druschel, R. B. McCleskey, R. J. Hamers, and J. F. Banfield. 2001. Rapid arsenite oxidation by Thermus aquaticus and Thermus thermophilus: field and laboratory investigations. Environ. Sci. Technol. 35:3857-3862. [DOI] [PubMed] [Google Scholar]
- 14.Green, H. H. 1918. Isolation and description of a bacterium causing oxidation of arsenite to arsenate in cattle-dipping baths. Rep. Dir. Vet. S. Afr. 6:593-599. [Google Scholar]
- 15.Hallberg, K. B., and D. B. Johnson. 2003. Novel acidophiles isolated from moderately acidic mine drainage waters. Hydrometallurgy 71:139-148. [Google Scholar]
- 16.Inskeep, W. P., R. E. Macur, N. Hamamura, T. P. Warelow, S. A. Ward, and J. M. Santini. 2007. Detection, diversity and expression of aerobic bacterial arsenite oxidase genes. Environ. Microbiol. 9:934-943. [DOI] [PubMed] [Google Scholar]
- 17.Jackson, C. R., H. W. Langner, J. Donahoe-Christiansen, W. P. Inskeep, and T. R. McDermott. 2001. Molecular analysis of microbial community structure in an arsenite-oxidizing acidic thermal spring. Environ. Microbiol. 3:532-542. [DOI] [PubMed] [Google Scholar]
- 18.Jackson, C. R., and S. L. Dugas. 2003. Phylogenetic analysis of bacterial and archaeal arsC gene sequences suggests an ancient, common origin for arsenate reductase. BMC Evol. Biol. 23:3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joubert, A. V. P., L. Lucas, F. Garrido, C. Joulian, and M. Jauzein. 2007. Effect of temperature, gas phase composition, pH and microbial activity on As, Zn, Pb and Cd mobility in selected soils in the Ebro and Meuse Basins in the context of global change. Environ. Pollut. 148:749-758. [DOI] [PubMed] [Google Scholar]
- 20.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, NY.
- 21.Kashyap, D. R., L. M. Botero, W. L. Franck, D. J. Hassett, and T. R. McDermott. 2006. Complex regulation of arsenite oxidation in Agrobacterium tumefaciens. J. Bacteriol. 188:1081-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequence. J. Mol. Biol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 23.Klein, M., M. Friedrich, A. J. Roger, P. Hugenholtz, S. Fishbain, H. Abicht, L. L. Blackall, D. A. Stahl, and M. Wagner. 2001. Multiple lateral transfers of dissimilatory sulfite reductase genes between major lineages of sulfate-reducing prokaryotes. J. Bacteriol. 183:6028-6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lebrun, E., M. Brugna, F. Baymann, D. Muller, D. Lièvremont, M.-C. Lett, and W. Nitschke. 2003. Arsenite oxidase, an ancient bioenergetic enzyme. Mol. Biol. Evol. 20:686-693. [DOI] [PubMed] [Google Scholar]
- 25.Mokashi, S. A., and K. M. Paknikar. 2002. Arsenic (III) oxidizing Microbacterium lacticum and its use in the treatment of arsenic contaminated groundwater. Lett. Appl. Microbiol. 34:258-262. [DOI] [PubMed] [Google Scholar]
- 26.Muller, D., D. Lièvremont, D. D. Simeonova, J.-C. Hubert, and M.-C. Lett. 2003. Arsenite oxidase aox genes from a metal-resistant β-proteobacterium. J. Bacteriol. 185:135-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muller, D., C. Médigue, S. Koechler, V. Barbe, M. Barakat, E. Talla, V. Bonnefoy, E. Krin, F. Arsène-Ploetze, C. Carapito, M. Chandler, B. Cournoyer, S. Cruveiller, C. Dossat, S. Duval, M. Heymann, E. Leize, A. Lieutaud, D. Lièvremont, Y. Makita, S. Mangenot, W. Nitschke, P. Ortet, N. Perdrial, B. Schoepp, P. Siguier, D. D. Simeonova, Z. Rouy, B. Segurens, E. Turlin, D. Vallenet, A. Van Dorsselaer, S. Weiss, J. Weissenbach, M.-C. Lett, A. Danchin, and P. N. Bertin. 2007. A tale of two oxidation states: bacterial colonization of arsenic-rich environments. PLoS Genet. 3:e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petri, R., L. Podgorsek, and J. F. Imhoff. 2001. Phylogeny and distribution of the soxB gene among thiosulfate-oxidizing bacteria. FEMS Microbiol. Lett. 197:171-178. [DOI] [PubMed] [Google Scholar]
- 29.Phillips, S. E., and M. L. Taylor. 1976. Oxidation of arsenite to arsenate by Alcaligenes faecalis. Appl. Environ. Microbiol. 32:392-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pierson, B. K., and R. W. Castenholz. 1974. A phototrophic gliding filamentous bacterium of hot springs, Chloroflexus aurantiacus, gen. and sp. nov. Arch. Microbiol. 100:5-24. [DOI] [PubMed] [Google Scholar]
- 31.Rhine, E. D., S. M. Ni Chadhain, G. J. Zylstra, and L. Y. Young. 2007. The arsenite oxidase genes (aroAB) in novel chemoautotrophic arsenite oxidizers. Biochem. Biophys. Res. Commun. 354:662-667. [DOI] [PubMed] [Google Scholar]
- 32.Rose, T. M., E. R. Schultz, J. G. Henikoff, S. Pietrokovski, C. M. McCallum, and S. Henikoff. 1998. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 26:1628-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rozas, J., J. C. Sánchez-DelBarrio, X. Messeguer, and R. Rozas. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496-2497. [DOI] [PubMed] [Google Scholar]
- 34.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 35.Salmassi, T. M., J. J. Walker, D. K. Newman, J. R. Leadbetter, N. R. Pace, and J. G. Hering. 2006. Community and cultivation analysis of arsenite oxidizing biofilms at Hot Creek. Environ. Microbiol. 8:50-59. [DOI] [PubMed] [Google Scholar]
- 36.Santini, J. M., and R. N. vanden Hoven. 2004. Molybdenum-containing arsenite oxidase of the chemolithoautotrophic arsenite oxidizer NT-26. J. Bacteriol. 186:1614-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scala, D. J., and L. J. Kerkhof. 1999. Diversity of nitrous oxide reductase (nosZ) genes in continental shelf sediments. Appl. Environ. Microbiol. 65:1681-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silver, S., and L. T. Phung. 2005. Genes and enzymes involved in bacterial oxidation and reduction of inorganic arsenic. Appl. Environ. Microbiol. 71:599-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.vanden Hoven, R. N., and J. M. Santini. 2004. Arsenite oxidation by the heterotroph Hydrogenophaga sp. str. NT-14: the arsenite oxidase and its physiological electron acceptor. Biochim. Biophys. Acta 1656:148-155. [DOI] [PubMed] [Google Scholar]
- 41.Van de Peer, Y., and R. De Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 42.Wagner, M., A. J. Roger, J. L. Flax, G. A. Brusseau, and D. A. Stahl. 1998. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J. Bacteriol. 180:2975-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weeger, W., D. Lièvremont, M. Perret, F. Lagarde, J.-C. Hubert, M. Leroy, and M.-C. Lett. 1999. Oxidation of arsenite to arsenate by a bacterium isolated from an aquatic environment. Biometals 12:141-149. [DOI] [PubMed] [Google Scholar]
- 44.Xia, X., and Z. Xie. 2001. DAMBE: software package for data analysis in molecular biology and evolution. J. Hered. 92:371-373. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.