Abstract
Comparative genetic maps of Papuan Saccharum officinarum L. (2n = 80) and S. robustum (2n = 80) were constructed by using single-dose DNA markers (SDMs). SDM-framework maps of S. officinarum and S. robustum were compared with genetic maps of sorghum and maize by way of anchor restriction fragment length polymorphism probes. The resulting comparisons showed striking colinearity between the sorghum and Saccharum genomes. There were no differences in marker order between S. officinarum and sorghum. Furthermore, there were no alterations in SDM order between S. officinarum and S. robustum. The S. officinarum and S. robustum maps also were compared with the map of the polysomic octoploid S. spontaneum ‘SES 208’ (2n = 64, x = 8), thus permitting relations to homology groups (“chromosomes”) of S. spontaneum to be studied. Investigation of transmission genetics in S. officinarum and S. robustum confirmed preliminary results that showed incomplete polysomy in these species. Because of incomplete polysomy, multiple-dose markers could not be mapped for lack of a genetic model for their segregation. To coalesce S. officinarum and S. robustum linkage groups into homology groups (composed of homologous pairing partners), they were compared with sorghum (2n = 20), which functioned as a synthetic diploid. Groupings suggested by comparative mapping were found to be highly concordant with groupings based on highly polymorphic restriction fragment length polymorphism probes detecting multiple SDMs. The resulting comparative maps serve as bridges to allow information from one Andropogoneae to be used by another, for breeding, ecology, evolution, and molecular biology.
Saccharum L. is part of a polyploid complex within the Andropogoneae (1). Cultivated forms of Saccharum spp. (sugarcane) are most notably used for sugar and alcohol production worldwide, especially in the tropics. Polyploidy in Saccharum is widespread and has been largely responsible for the genetic and taxonomic complexity that has, until recently, suppressed genetic dissection of the sugarcane genome and comprehension of phylogenetic relations (reviewed in ref. 2). Recent studies using DNA markers have established ploidy level of one individual from a wild species, S. spontaneum (2n = 64, from India), revealing polysomic inheritance and octoploidy (thus, x = 8) (3–5). Results obtained with genetic markers have been independently confirmed by using fluorescent in situ hybridization with rDNA probes (6, 7). Molecular systematic studies have revealed that maternally inherited genomes of Saccharum and sorghum diverged recently (8). In the absence of known extant taxa that can be considered diploid relatives of sugarcane, Al-Janabi et al. (8) argued that sorghum fill their place. Comparative mapping of grasses (9–25) has revealed conservation of gene order in recent studies (reviewed in ref. 22). This knowledge likely will have a significant impact on plant breeding strategies and germplasm characterization, conservation, and use, with significant implications for world agriculture.
Detailed and multiple comparisons between maize and sorghum, which also are part of the Andropogoneae, have been conducted (12–15). These have shown that there is a large amount of conservation between these two genomes, which likely diverged before sugarcane diverged from sorghum (8, 15, 26, 27). In addition, Grivet et al. (28, 29) made preliminary comparisons between cultivated Saccharum and maize by using Saccharum accessions that have come from breeding programs. Saccharum spp. bred for human use are known to suffer various types of chromosome abnormalities and rearrangements (30).
To further investigate the phylogenetic relationship of sorghum and Saccharum as well as to realize the potential benefits of comparative mapping, we constructed single-dose DNA marker (SDM) linkage maps of Papuan (2n = 80) forms of S. officinarum and S. robustum by using anchor loci that had been mapped in sorghum, maize, and, in some cases, S. spontaneum. The domesticated species, S. officinarum, is thought to have been derived primarily from S. robustum in Papua New Guinea (31). Thus, comparing S. robustum with S. officinarum should give insight into domestication processes and key agriculturally important regions of their genomes (32, 33). We also investigated the inheritance of quantitative traits in these two species (C.T.G., R. W. Doerge, G.R.S., R. J. Honeycutt, and B.W.S.S., unpublished results).
MATERIALS AND METHODS
Plant Materials and DNA Manipulations.
Plant materials were kindly provided by the Hawaiian Sugar Planters’ Association (Aiea, HI). The population consisted of 100 individuals produced by crossing S. officinarum ‘LA Purple’ as a female with S. robustum ‘Mol 5829’. Cytological evaluation of the population showed that parents and progeny displayed strict bivalent pairing at meiosis and had 2n = 80 chromosomes, as described previously (32). Genomic DNAs were extracted according to the method of Honeycutt et al. (34).
DNA Markers.
Restriction fragment length polymorphisms (RFLPs). DNA restriction, electrophoresis, blotting, Southern hybridization, and autoradiography were performed as described (4). One hundred and ninety probes were surveyed against parental DNA blots digested with DraI, EcoRI, HindIII, and XbaI to identify polymorphisms.
Choice of RFLP probes was based on existing sorghum, maize, and S. spontaneum genetic maps (see below). Every attempt was made to cover the sorghum genome completely and to enrich the intra-Saccharum comparisons. RFLP probes used were heterologous maize genomic clones and maize cDNAs previously mapped in maize and sorghum. Sugarcane (S. officinarum) genomic DNA probes (SG), as well as S. officinarum cDNA probes from buds (CSB), cell culture (CSC), and roots (CSR), which were previously mapped in Saccharum spontaneum ‘SES 208’ (4, 5), also were used. Candidate gene probes (35) from sucrose metabolism and transport pathways were mapped: smp-1, a sugarcane membrane protein supposed to be a glucose transporter (36); sps-1, sucrose phosphate synthase from maize (37); SS-1, maize sucrose synthase (38), and HBr-1, a maize phosphoglucomutase-encoding probe (kindly provided by S. Briggs, Pioneer Hi-Bred International, Johnston, IA).
Arbitrarily primed PCR.
Two concentrations (25 and 50 ng) of the parental genomic template DNAs and one (25 ng) of each progeny were subjected to thermal cycling in a System 9600 Cycler (Perkin–Elmer) by using the arbitrarily primed PCR protocol of Sobral and Honeycutt (39). Procedures for agarose gel electrophoresis, recording, and scoring of amplified products were described by Al-Janabi et al. (3). Some of the arbitrarily primed PCR products were amplified with [α32]P-dCTP, resolved in 5% polyacrilamide/50% urea gels in 1× Tris-borate-EDTA and visualized by autoradiography at room temperature for 1–3 days. Ten-mers of arbitrary sequence (Operon Technologies, Alameda, CA) and four RY-repeat 12-mer (CG6–5′-’TCGCTGCGGCGG-3′, CG7–5′-CTGCGGTCGCGG-3′, CG8–5′-CAGCCGTAGCGG-3′, and CG9–5′-CCGCGACTGCGG-3′) were screened against the mapping parents.
Selective restriction fragment amplification.
Amplified fragment length polymorphisms (AFLPs; ref. 40) were generated by using commercially available AFLP kits. Two hundred and fifty nanograms of genomic DNA of the parents and progeny were completely and simultaneously restricted with EcoRI and MseI. Restricted genomic DNA fragments were ligated to EcoRI and MseI adapters, diluted 1:10, and preamplified by using AFLP primers, each having one selective nucleotide. The preamplification products were diluted 1:50 and used as a template for selective amplification by using combinations of MseI- and EcoRI-specific primers, each containing three selective nucleotides. EcoRI-selective primers were labeled with [γ32]P-dATP before amplification. The thermal profile for both steps of amplification, primer labeling, and selective primer combinations were as recommended by the manufacturer. Selectively amplified products were resolved by electrophoresis in denaturing polyacrylamide gel as described for arbitrarily primed PCR.
Genetic Mapping of Saccharum.
Segregating polymorphisms were analyzed for dosage by using a χ2 test (P < 0.05), as described (2, 41). SDM linkage relationships were estimated by using Kosambi’s map function in mapmaker 2.0 for Macintosh (42) as described (3). In a cross between two heterozygous individuals, many SDMs will be present in one individual and absent in the other, as in a test-cross configuration. Therefore, meiosis and gametic segregation can be followed directly in each individual. Thus, this is a double-pseudo-test-cross mapping strategy. SDMs were analyzed by using a minimum lod of 7.0 and a maximum recombination fraction (θ) of θ < 0.25 (41), from which linkage groups were determined. Linkage groups were then ordered by using multipoint analyses. The best possible order was always accepted for comparative mapping purposes even when the lod score supporting the order was not large. Linkages in repulsion phase were determined as described (2, 3).
Comparative Mapping.
Only RFLP markers were used for comparisons, after linkage groups were determined by using all marker types. The maize linkage information was based on the maize map published in Maize Cooperative Newsletter and the University of Missouri, Columbia, Maize RFLP Map (http://teosinte.agron.missouri.edu). Sorghum linkage information and its comparison to maize linkage maps was compiled by using the studies of Whitkus et al. (12), Melake-Berhan et al. (14), Pereira et al. (43), Chittenden et al. (44), Lin et al. (20), Paterson et al. (21), and unpublished data graciously provided by Michael Lee (Department of Agronomy, Iowa State University). The names used herein for sorghum linkage groups are according to Chittenden et al. (44), Lin et al. (20), and Paterson et al. (21). UMC and Brookhaven National Laboratory probes were used as anchors to place the Iowa State University clones mapped by Pereira et al. (43) onto an inferred composite sorghum map. Comparisons with S. spontaneum were based on daSilva et al. (5) and used their nomenclature for homology groups.
RESULTS
Colinearity of Sorghum and Saccharum Genetic Maps.
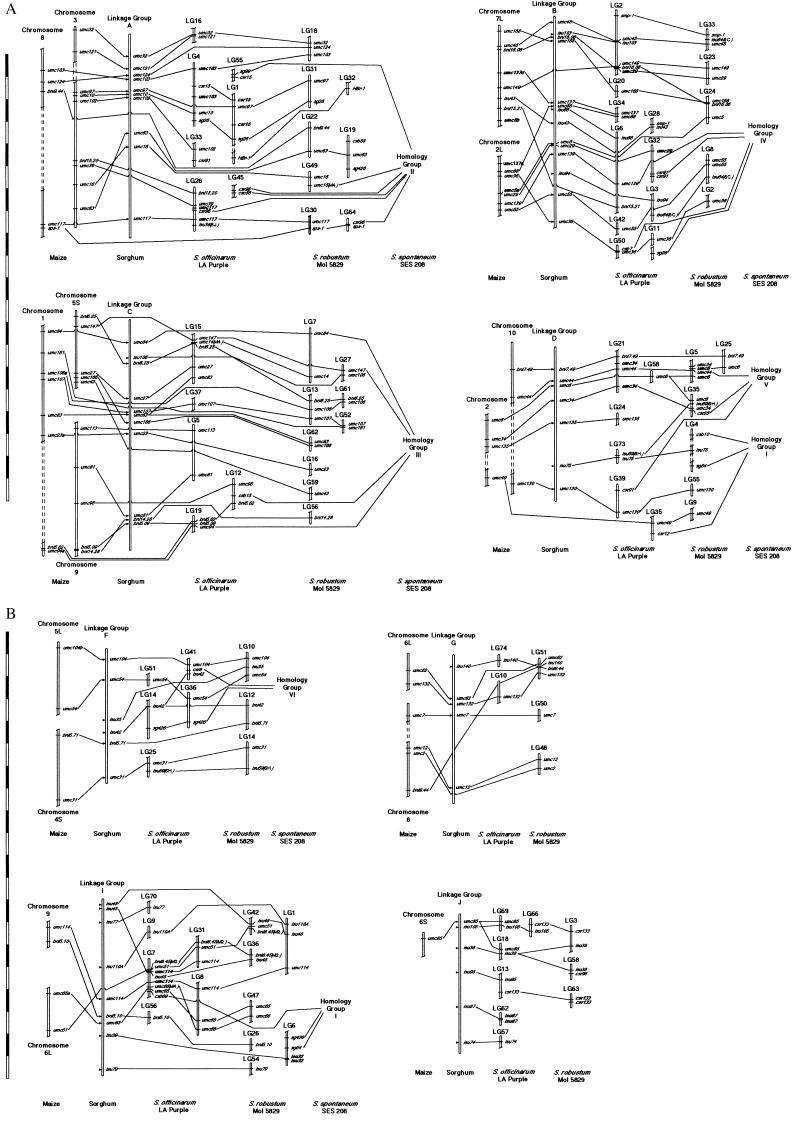
The most striking result of comparative mapping of Papuan Saccharum in relation to sorghum is that marker colinearity was nearly perfect (Fig. 1). Only four cases of change in marker order (inversions) of Saccharum in relation to sorghum were observed. Inversions were in linkage groups (LGs) 1, 10, 24, and 51 of S. robustum. No changes in marker order were seen from sorghum relative to S. officinarum. Because markers at θ < 3cM cannot be accurately ordered with this sample size, inversions detected in the LGs 1 and 10 may be real, whereas those of LGs 24 and 51 may be artifactual.
Figure 1.
Comparative genetic maps of maize, sorghum, and three species of Saccharum. DNA marker positions in maize were based on the UMC maize RFLP map (http://teosinte.agron.missouri.edu). Sorghum linkage groups (LGs) were named according to Chittendenn et al. (44). Sorghum marker positions were compiled based on Whitkus et al. (12), Pereira et al. (43), Lin et al. (20), and Paterson et al. (21); their map locations are indicated by an arrow. S. spontaneum ‘SES 208’ homeology groups (HGs) were based on daSilva et al. (5). DNA markers whose positions conflict with the sorghum comparisons have their location indicated in parenthesis, using “S” for sorghum and “M” for maize. Linkage groups were formed by using RFLP, AFLP, and arbitrarily primed PCR markers, but those shown are only the RFLP markers because they enabled the comparisons. Probes that have been mapped in a species have their position indicated with a line that traverses the chromosome or linkage group, whereas probes whose positions are unknown (i.e., not mapped) contain lines that pass behind the chromosome or linkage group. Maize probes unmapped in sorghum have arrows indicating their inferred position in sorghum (based on combining information from previous maps). Breaks in the LGs or chromosomes indicate discontinuity of the LGs or chromosomes. For maize chromosomes, S = short arm and L = long arm. (Bar = 10 cM.)
A very stringent two-point lod score (seven) was chosen because we would prefer to err on the conservative side. However, lowering the lod score for two-point analysis from seven to four was consistent with comparative mapping to sorghum (data not shown), with one exception. In S. officinarum LGs 4 and 9, homeologous to sorghum LGs A and I, respectively, there was discordance when the two-point analysis was done by using a minimum lod score of 4.0, because LGs 4 and 9 were united at the lower score.
Details and complete representations of S. officinarum and S. robustum genetic maps are described elsewhere (C.T.G., R. J. Honeycutt, G.R.S., and B.W.S.S., unpublished results). Briefly, 271 polymorphisms were generated by the RFLP probes in S. officinarum, and 268 were generated in S. robustum. Of those, 176 and 173 were SDMs (P < 0.05), respectively. Arbitrarily primed PCR, by using 64 primers, generated 126 (S. officinarum) and 78 (S. robustum) polymorphisms, of which 73 and 54 were SDMs, respectively. Finally, for AFLP markers, 12 primer combinations yielded 135 (S. officinarum) and 107 (S. robustum) polymorphisms, resulting in 96 and 74 SDMs, respectively.
Determination of Homologous Groups by Comparative Mapping and Highly Polymorphic SDMs.
In a diploid relative of a polysomic octoploid (with bivalent pairing), we expect that each chromosome from the diploid will identify eight homeologs in the octoploid. We used sorghum linkage groups to identify homeology groups (HGs) in S. officinarum and S. robustum, based on the presence of at least one RFLP probe previously mapped in maize and sorghum. The sorghum-determined HGs were also compared with other results (Table 1). There were two differences in relation to Grivet et al. (29), however, that were concordant with S. spontaneum HGs (5) and with maize–sorghum comparisons (12, 21). The composite sugarcane LGs II and III (29) were linked into one group by our data (homeology with sorghum LG A), which was confirmed by a homeologous relationship with S. spontaneum HG 2 (5). In addition, maize–sorghum comparisons relate LG A from sorghum to maize chromosomes 3 and 8. It is noteworthy that Grivet et al. (29) also suggested the possibility of composite groups II and III being united. However, locus umc7, placed by Grivet et al. (29) on the extremity of composite LG II, was mapped onto LG G of sorghum by Whitkus et al. (12) and by our work, although there are no neighboring anchor probes to resolve homeology of this region with respect to sorghum–Saccharum comparisons (Fig. 1). Likewise, composite sugarcane LGs I and VI were homeologous to sorghum LG C and S. spontaneum HG 3, and to maize chromosomes 1, 5, and 9. We did not find LGs in Saccharum that were homeologous to composite LG V; this likely was caused by a lack of common probes. Finally, the composite LG IX could be homeologous to sorghum LG I based on umc114 or sorghum LG C based on umc81. Because other markers in S. officinarum LG 7 support homeology with sorghum LG I and parts of LG C in sorghum are homeologous to maize chromosome 9, which also shares homeology with sorghum LG I (21), we concluded that composite group IX is homeologous to sorghum LG I.
Table 1.
Relationships of sorghum, maize, and Saccharum linkage groups and chromosomes
| Sorghum LG* | Maize chromosomes or arms† | Composite sugarcane LGs‡ | S. spontaneum HG§ |
|---|---|---|---|
| A | 3 and 8 | II and III | 2 |
| B | 2L and 7L | X | 4 |
| C | 1, 5S, and 9 | I and VI | 3 |
| D | 2S and 10 | VIII | 1 and 5 |
| F | 4S and 5L | VII | 6 |
| G | 6L and 8¶ | IV | |
| I | 6 and 9 | IX | 1 |
| J | 6S‖ | Ungrouped |
Groupings based on sorghum LGs were compared with those obtained by using highly polymorphic probes (4, 5). Highly polymorphic probes have been used in S. spontaneum to organize linkage groups into HGs (4). In that case, resulting HG assignments were concordant with HGs independently assembled by using multiple-dose restriction fragments (2, 5). In S. officinarum, 14 probes detected more than one SDM, for a total of 54 SDMs (3.86 per probe). Of these 54 SDMs, only 6 suggested discordant groupings, in relation to those observed by comparison with sorghum. However, two (sg26 on LG 11 and csr91 on LG 39) of the six SDMs suggesting discordant groupings were concordant with assignments made by comparison with S. spontaneum (see below). The remaining four SDMs had locations that remained discordant with sorghum and could not be reconciled through comparisons with S. spontaneum. The four loci were: isu38 (LG 26), csr96 (LG 70), csr91 (LG 33), and umc132 (LG 56). These four loci may represent cases of duplicated loci on nonhomeologous linkage groups that do not pair during meiosis. In S. robustum, 21 probes were highly polymorphic, detecting 46 SDMs (2.19 per probe). Six of these SDMs were detected by using probes (sg54, sg426, and bnl9.44) not mapped in sorghum but that detected LGs in more than one sorghum-determined homeology group. Of the remaining 40 SDMs, 2 (from isu64) were found on LGs 33 and 8, homeologous to sorghum LG B (Fig. 1), even though isu64 was mapped onto the end of sorghum LG C (43), and one other (csr96 on LG 58) could not be reconciled with the S. spontaneum-determined HGs.
Relation of Papuan Saccharum (2n = 80) Forms to Indian S. spontaneum ‘SES 208’ (2n = 64).
Comparisons of SDM order between S. officinarum and S. robustum did not reveal any changes, as may be expected for two so closely related sympatric species. One SDM order change was observed between S. officinarum LG 7 and LG 8 (both homeologous to sorghum LG I); however, the distance between umc66 and umc65 in LG 28 was 2.0 cM. We were unable to detect homeologous LGs in relation to S. spontaneum HGs 7 and 8, probably because these HGs were composed of LGs with few markers, reducing the number of common markers between maize and S. spontaneum in these HGs.
Preferential Homolog Pairing in Papuan Saccharum.
There were 17/74 S. officinarum and 10/65 S. robustum LGs that displayed at least one SDM linked in repulsion phase, for a total of 46/287 (16.0%) and 24/208 (11.5%) SDMs, respectively. Detection of repulsion-phase linkages within both genomes with a sample of 100 individuals strongly suggests that both genomes are incompletely polysomic (2, 41).
Conservation of Quantitative Trait Loci (QTL) for Flowering Control in S. officinarum LG 7.
In LG 7 of S. officinarum, C.T.G. et al. (unpublished results) found that umc114e.2490 is strongly associated with short-day flowering. Herein, we showed that LG 7 was homeologous to sorghum LG I (orthologous to the Se1/Se3 region of rice chromosome 6). This region is also homeologous to a region of maize chromosome 9 that harbors QTLs that affect flowering in four maize populations (20) and in sorghum (43). LG 7 also revealed other QTLs in our analyses and, interestingly, also shared loci with maize chromosomes 2 and 4. Sorghum’s LG I also was found to have limited shared loci with maize chromosomes 1 (csu77), 4 (cdo344, umc47), and 10 (bcd147) (21). Repulsion-phase linkages were common in S. officinarum LG 7. Thus, S. officinarum LG 7 and sorghum LG I seem to represent genomic regions that have suffered large amounts of rearrangements in relation to maize and that harbor genes that are agronomically interesting to sugarcane breeders.
DISCUSSION
Colinearity Within the Andropogoneae.
Our results show striking colinearity between Saccharum and sorghum as predicted by prior phylogenetic analyses (8, 26). With the exception of two inversions, all comparisons between sorghum and Saccharum point to conservation between these genera. We predict that alleles cloned from sorghum based on map position usually will be orthologous to alleles from Saccharum. Map-based gene cloning in the Andropogoneae will benefit from sorghum’s small genome size (C = 0.8 pg or 750 Mbp; ref. 45). The finding of such high conservation between Saccharum and sorghum strongly suggests that a set of PCR-based anchor loci could be developed for use within Andropogoneae crops and their wild relatives (2). These anchors could be developed from cDNA sequencing/mapping projects and widely disseminated, thus creating a tool to permit easy detection of novel alleles from diverse germplasm. In addition, we have shown that it is possible to use comparative mapping to identify homologs in polyploids that correspond to homeologous chromosomes in diploids. Thus, comparative mapping by using PCR-based anchor loci also would serve as a nuclear phylogenetic tool.
Transmission Genetics in Saccharum.
Detailed genetic maps of S. spontaneum ‘SES 208’ (2n = 64) were used to demonstrate polysomic inheritance and octoploidy (2, 3, 5). Inference of octoploidy in SES 208 was based on lack of linkages in repulsion (41) and on the proportion of detectable SDM-to-MDM (multiple dose marker) classes (4, 5).
A preliminary study of S. officinarum ‘LA Purple’ and S. robustum ‘Mol 5829’ genomes suggested incomplete polysomy, based on the detection of repulsion-phase SDMs on a small number (44 individuals) of progeny from this cross (32). Our results confirmed this observation. Complete disomy (essentially “diploid inheritance”) does not present a problem for genetic mapping or QTL detection because existing approaches have been developed for disomic inheritance. Thus, polyploids that transmit their genes essentially as diploids can be mapped directly by using existing approaches.
Complete polysomy (or nonpreferential pairing) is a new and relatively more complex situation for both genetic and QTL mapping. Added complexity is a result of the existence of more than one pairing homolog at meiosis. In cases of complete polysomy, such as in ‘SES 208’, there is no preference for selection of pairing homologs during meiosis. Completely polysomic species or individuals may display bivalent, multivalent, or a mixture of bivalent and multivalent formation at meiosis. If pairing is strictly bivalent, as has been shown for most Saccharum, then recently developed (46) genetic mapping approaches can be utilized. Recent studies in S. spontaneum have provided both statistical tools (46) and practical approaches (2, 47) for genetic mapping in completely polysomic polyploids with strict bivalent pairing. Approaches for QTL mapping may use an SDM-marker framework map (C.T.G. et al., unpublished results); because SDMs are used, these approaches can be used in completely polysomic or intermediate situations.
Incomplete polysomy, or partial preferential pairing, thought to represent intermediate stages of “diploidization” of polyploids, presents new difficulties for genetic mapping and QTL detection. The difficulties are associated with the current lack of models to determine expected segregation ratios for different classes because of partial preferential homolog pairing. Specifically, classes of MDMs, extremely useful for genetic mapping in polysomic polyploids (2), cannot be mapped with existing approaches (46, 47). Genetic mapping models that considered the amount of preferential pairing observed from the SDM framework map as part of the data for determining segregation ratios to infer linkages among MDMs would be highly desirable.
Colinearity as a Tool for Identification of Homologs.
daSilva et al. (4, 5) used mapping of highly polymorphic probes (i.e., those that detected more than one SDM per probe) and mapping of MDMs (some classes of double-dose and triple-dose markers) to identify homeologous groups composed of potential pairing partners in ‘SES 208’. Mapping of MDMs cannot be used in its current form because of incomplete polysomy (see above). Use of highly polymorphic probes suffers from the complication of duplicated loci on nonpairing homologs, a feature common to many plant genomes. Therefore, results from mapping of highly polymorphic RFLP probes must be considered tentative.
An alternative method for determining which homologs are potential pairing partners during meiosis is the use of comparative mapping to a diploid relative or progenitor. However, in Saccharum and other high-chromosome-number polyploids of unknown origin, diploid relatives are either unknown or extinct. Thus, use of a diploid (disomic) that is known to be closely related could serve the same purpose. For sugarcane, this type of disomic workhorse is sorghum (2, 8, 26). Because sorghum is based on x = 10 chromosomes and Papuan S. officinarum/S. robustum also seem to be based on x = 10 (6), we would expect that for a completely polysomic 2n = 80 octoploid, eight homeologs would be identified for each sorghum chromosome in a saturated genetic map. In either case, larger population sizes (many hundreds, at least) are extremely helpful from the standpoint of mapping in polyploids.
Conservation of Phenotypes.
Sorghum LG I is orthologous to rice chromosome 6, which contains short-day flowering mutations Se1 and Se3 (48), and paralogous to the Ma1 flowering gene region of sorghum LG D. Ma1 is specifically regulated by photoperiod (49). We have shown that sorghum LG I is homeologous to S. officinarum LG 7, and that the same probe (umc114) is nearby (Fig. 1). In two crosses between S. officinarum and S. spontaneum, Paterson et al. (21) detected associations in short-day flowering near pSB188. In sorghum, pSB188 maps to LG D, which contains the Ma1 locus that is paralagous to Se1/Se3 (20). Unfortunately, this region of the sorghum genome was not well covered in our study. It is reassuring to find conservation of location of genes involved in orthologous phenotypes between sorghum and Saccharum. Phenotypic data validate the comparative mapping approaches and results in a way that is not possible simply with anonymous genetic markers.
Acknowledgments
We thank Michael McClelland for support beyond the call of duty, Rebecca Doerge and Ron Sederoff for continued encouragement, and the Incas for thinking in threes and Machu Pichu. We thank those who shared probes and other information; in particular, Michael Lee. We also thank Paul Moore and K. K. Wu for acquiring and supplying field data and cane tops for DNA extraction; Perkin–Elmer/ABI Division for AFLP kits for beta-testing; Rhonda Honeycutt and Keira Maiden for technical assistance; and Joshua Kohn, Ivor Royston, and the Sidney Kimmel Cancer Center for varied assistance. C.T.G. was supported by a fellowship from the Brazilian National Research Council of the Ministry of Science and Technology (MCT/CNPq/RHAE; 260007/95–1). B.W.S.S. and G.R.S. were supported in part by a grant from Copersucar Technology Center (Piracicaba, Brazil) and the Mauritius Sugar Industry Research Institute (Réduit, Mauritius).
ABBREVIATIONS
- SDM
single-dose DNA marker
- SG
sugarcane (S. officinarum) genomic DNA probe
- AFLP
amplified fragment length polymorphism
- RFLP
restriction fragment length polymorphism
- LG
linkage group
- HG
homology group
- MDM
multiple dose markers
- QTL
quantitative trait loci
References
- 1.Clayton W D, Renvoize S A. Genera Graminum: Grasses of the World, Kew Bulletin Additional Series XIII. London: Her Majesty’s Stationary Office; 1986. [Google Scholar]
- 2.daSilva J, Sobral B W S. In: The Impact of Plant Molecular Genetics. Sobral B W S, editor. Boston: Birkhäuser; 1996. pp. 3–37. [Google Scholar]
- 3.Al-Janabi S M, Honeycutt R J, McClelland M, Sobral B W S. Genetics. 1993;134:1249–1260. doi: 10.1093/genetics/134.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.daSilva J, Sorrells M E, Burnquist W L, Tanksley S D. Genome. 1993;36:782–791. doi: 10.1139/g93-103. [DOI] [PubMed] [Google Scholar]
- 5.daSilva J, Honeycutt R J, Burnquist W L, Al-Janabi S M, Sorrells M E, Tanksley S D, Sobral B W S. Mol Breed. 1995;1:165–179. [Google Scholar]
- 6.D’Hont A, Rao P S, Feldmann P, Grivet L, Islam-Faridi N, Taylor P, Glaszmann J C. Theor Appl Genet. 1995;91:320–326. doi: 10.1007/BF00220894. [DOI] [PubMed] [Google Scholar]
- 7.D’Hont A, Grivet L, Feldmann P, Rao S, Berding N, Glaszmann J C. Mol Gen Genet. 1996;250:405–413. doi: 10.1007/BF02174028. [DOI] [PubMed] [Google Scholar]
- 8.Al-Janabi S M, McClelland M, Petersen C, Sobral B W S. Theor Appl Genet. 1994;88:933–944. doi: 10.1007/BF00220799. [DOI] [PubMed] [Google Scholar]
- 9.Doebley J, Stec A, Wendel J, Edwards M. Proc Natl Acad Sci USA. 1990;87:9888–9892. doi: 10.1073/pnas.87.24.9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doebley J, Stec A. Genetics. 1991;129:285–295. doi: 10.1093/genetics/129.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doebley J, Stec A. Genetics. 1993;134:559–570. doi: 10.1093/genetics/134.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whitkus R, Doebley J, Lee M. Genetics. 1992;132:1119–1130. doi: 10.1093/genetics/132.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Binelli G, Gianfranceschi L, P, Taramino G, Bussi C, Stenhouse J, Ottaviano E. Theor Appl Genet. 1992;84:10–16. doi: 10.1007/BF00223975. [DOI] [PubMed] [Google Scholar]
- 14.Melake-Berhan A, Hulbert S H, Butler L G, Bennetzen J L. Theor Appl Genet. 1993;86:598–604. doi: 10.1007/BF00838715. [DOI] [PubMed] [Google Scholar]
- 15.Hulbert S H, Richter T E, Axtell J D, Bennetzen J L. Proc Natl Acad Sci USA. 1990;87:4251–4255. doi: 10.1073/pnas.87.11.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moore G, Gale M D, Kurata N, Flavell R B. Bio/Technology. 1993;11:584–589. [Google Scholar]
- 17.Kurata N, Moore G, Nagamura Y, Foote T, Yano M, Minobe Y, Gale M. Bio/Technology. 1994;12:276–278. [Google Scholar]
- 18.Ahn S, Tanksley S D. Proc Natl Acad Sci USA. 1993;90:7980–7984. doi: 10.1073/pnas.90.17.7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahn S, Anderson J A, Sorrells M E, Tanksley S D. Mol Gen Genet. 1994;241:483–490. doi: 10.1007/BF00279889. [DOI] [PubMed] [Google Scholar]
- 20.Lin Y-R, Schertz K F, Paterson A H. Genetics. 1995;141:391–411. doi: 10.1093/genetics/141.1.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paterson A H, Lin Y-R, Li Z, Schertz K, Doebley J F, Pinson S R M, Liu S-C, Stansel J W, Irvine J E. Science. 1995;269:1714–1718. doi: 10.1126/science.269.5231.1714. [DOI] [PubMed] [Google Scholar]
- 22.Bennetzen J L. In: The Impact of Plant Molecular Genetics. Sobral B W S, editor. Boston: Birkhäuser; 1996. pp. 71–85. [Google Scholar]
- 23.Shields R. Nature (London) 1993;365:297–298. [Google Scholar]
- 24.Helentjaris T. Proc Natl Acad Sci USA. 1993;90:8308–8309. doi: 10.1073/pnas.90.18.8308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennetzen J L, Freeling M. Trends Genet. 1993;9:259–261. doi: 10.1016/0168-9525(93)90001-x. [DOI] [PubMed] [Google Scholar]
- 26.Sobral B W S, Braga D P V, LaHood E S, Keim P. Theor Appl, Genet. 1994;87:843–853. doi: 10.1007/BF00221137. [DOI] [PubMed] [Google Scholar]
- 27.Hamby R K, Zimmer E A. Plant Syst Evol. 1988;160:29–37. [Google Scholar]
- 28.Grivet L, D’Hont A, Dufour P, Hamon P, Roques D, Glaszmann J C. Heredity. 1994;73:500–508. [Google Scholar]
- 29.Grivet L, D’Hont A, Roques D, Feldman P, Lanaud C, Glaszmann J C. Genetics. 1996;142:987–1000. doi: 10.1093/genetics/142.3.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price S. Bot Gaz. 1957;March:146–159. [Google Scholar]
- 31.Brandes E W. Natl Geo. 1929;56:253–332. [Google Scholar]
- 32.Al-Janabi S M, Honeycutt R J, Sobral B W S. Theor Appl Genet. 1994;89:959–963. doi: 10.1007/BF00224524. [DOI] [PubMed] [Google Scholar]
- 33.Sills G R, Bridges W, Al-Janabi S M, Sobral B W S. Mol Breed. 1995;1:355–363. [Google Scholar]
- 34.Honeycutt R J, Sobral B W S, Keim P, Irvine J E. Plant Mol Biol Rep. 1992;10:66–72. [Google Scholar]
- 35.Crandall K A. In: The Impact of Plant Molecular Genetics. Sobral B W S, editor. Boston: Birkhäuser; 1996. pp. 137–157. [Google Scholar]
- 36.Bugos R C, Thom M. Plant Physiol. 1993;102:1367. doi: 10.1104/pp.102.4.1367. (abstr.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Worrell A C, Bruneau J M, Summerfeit K, Boersig M, Voelker T A. Plant Cell. 1991;3:1121–1130. doi: 10.1105/tpc.3.10.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCarty D R, Shaw J R, Hannah L C. Proc Natl Acad Sci USA. 1986;83:9099–9103. doi: 10.1073/pnas.83.23.9099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sobral B W S, Honeycutt R J. Theor Appl Genet. 1993;86:105–112. doi: 10.1007/BF00223814. [DOI] [PubMed] [Google Scholar]
- 40.Zabeau, M. & Vos, P. (1993) Eur. Patent Appl. 0534858.
- 41.Wu K K, Burnquist W L, Sorrells M E, Tew T L, Moore P H, Tanksley S D. Theor Appl Genet. 1992;83:788–794. doi: 10.1007/BF00224274. [DOI] [PubMed] [Google Scholar]
- 42.Lander E S, Green P, Abrahamson J, Barlow A, Daly M J, Lincoln S E, Newburg L. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- 43.Pereira M G, Lee M, Bramel-Cox P, Woodman W, Doebley J, Whitkus R. Genome. 1994;37:236–243. doi: 10.1139/g94-033. [DOI] [PubMed] [Google Scholar]
- 44.Chittenden L M, Schertz K F, Lin Y-R, Wing R A, Paterson A H. Theor Appl Genet. 1994;87:925–933. doi: 10.1007/BF00225786. [DOI] [PubMed] [Google Scholar]
- 45.Arumuganathan K, Earle E D. Plant Mol Biol Rep. 1991;9:208–218. [Google Scholar]
- 46.Ripol M I. M.S. thesis. Ithaca, NY: Cornell University; 1994. [Google Scholar]
- 47.daSilva J. Ph.D. dissertation. Ithaca, NY: Cornell University; 1993. [Google Scholar]
- 48.MacKill D J, Salam M A, Wang Z Y, Tanksley S D. Theor Appl Genet. 1993;85:536–540. doi: 10.1007/BF00220910. [DOI] [PubMed] [Google Scholar]
- 49.Quinby J R, Karper R E. J Am Soc Agron. 1945;37:926–936. [Google Scholar]